Tenascin C in Lung Diseases
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
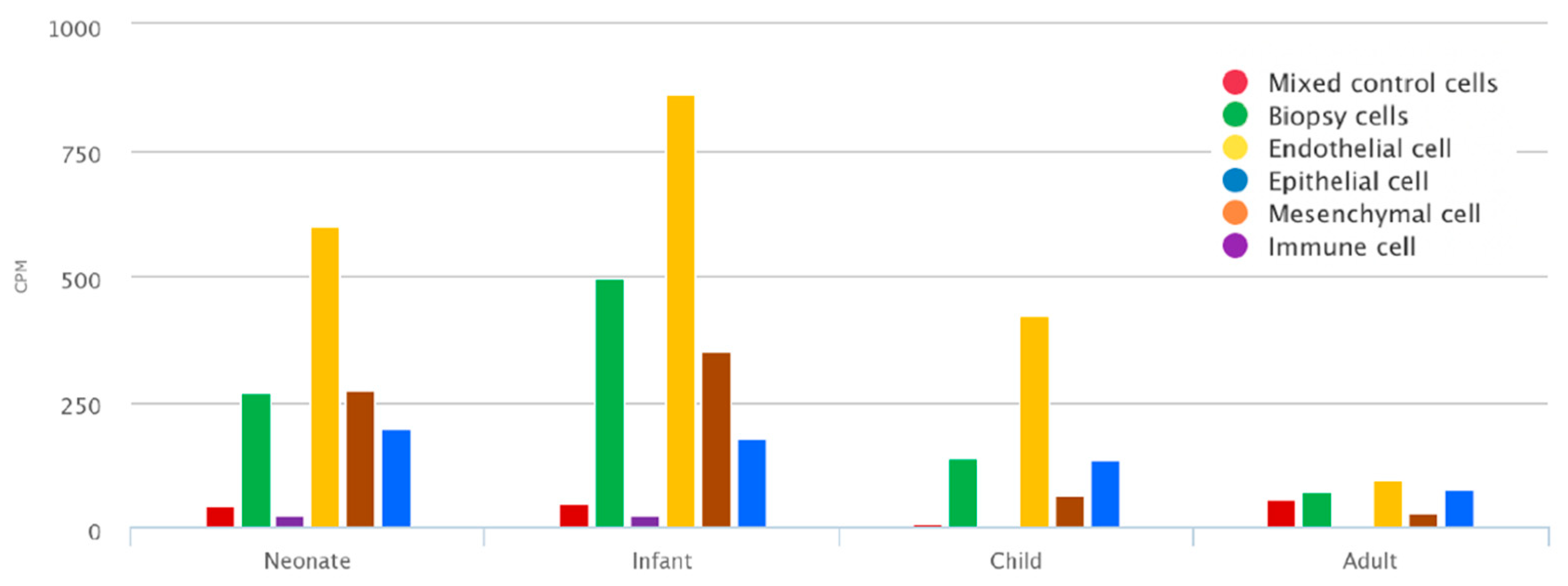
2.1. Mean TNC Expression during Lung Development Dataset
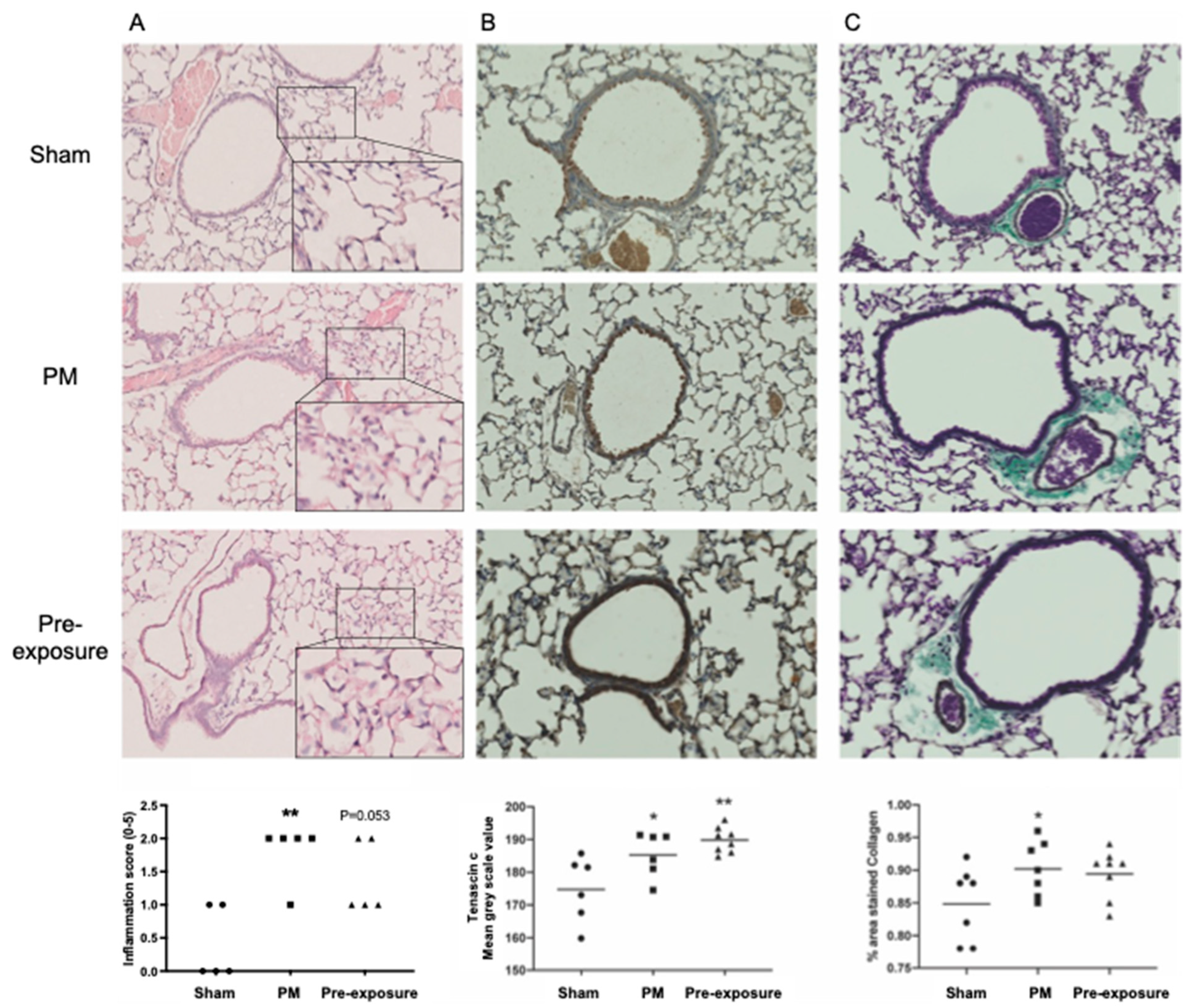
2.2. Traffic Related Particulate Matter (PM2.5) Exposure In Vivo
2.3. Histological Analysis
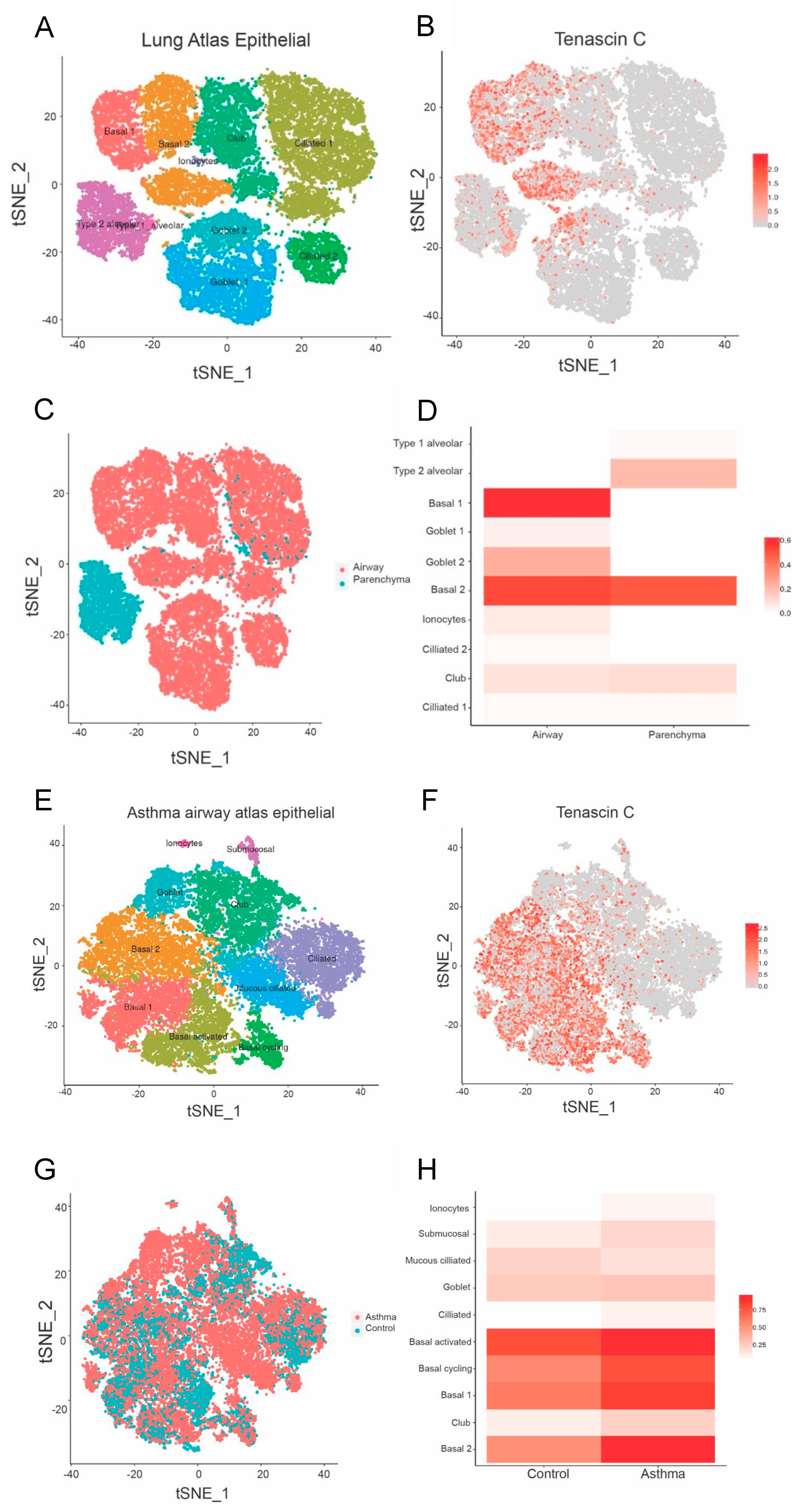
2.4. Asthma Dataset
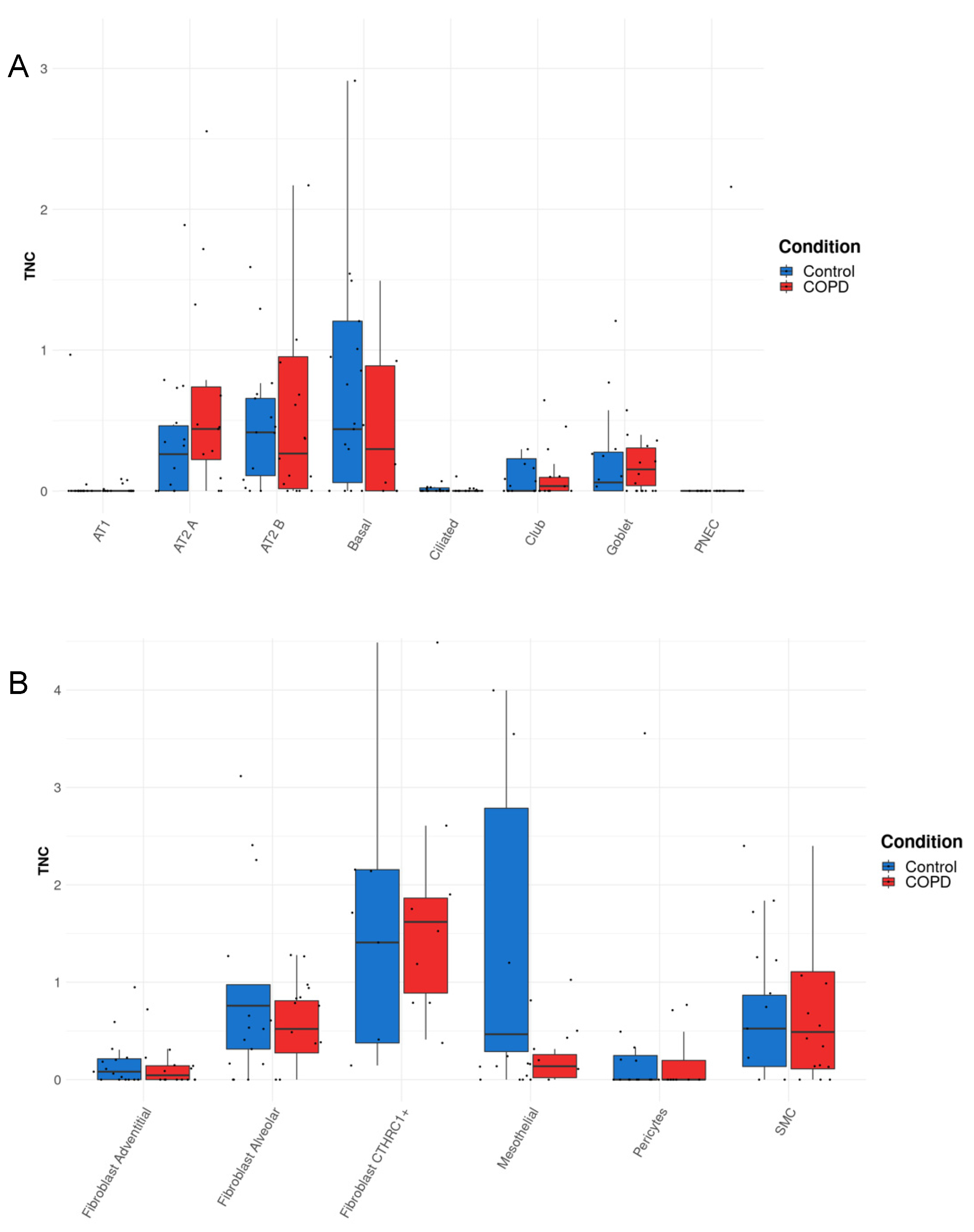
2.5. COPD Dataset
2.6. Lung Cancer Dataset
2.7. Interaction Network Construction
2.8. Statistical Analysis
3. Results
3.1. TNC Is Increased during Development and in Offspring from PM2.5-Exposed Dams
3.2. TNC Expression Is Higher in Basal Epithelial Cells Compared to Parenchyma in Healthy Airways, and in Asthmatic Samples Compared to Healthy Controls
3.3. TNC Expression Is Increased in AT2A Cells and Endothelial Cells (Peribronchial Vascular Endothelial Cells) in COPD Compared to Controls
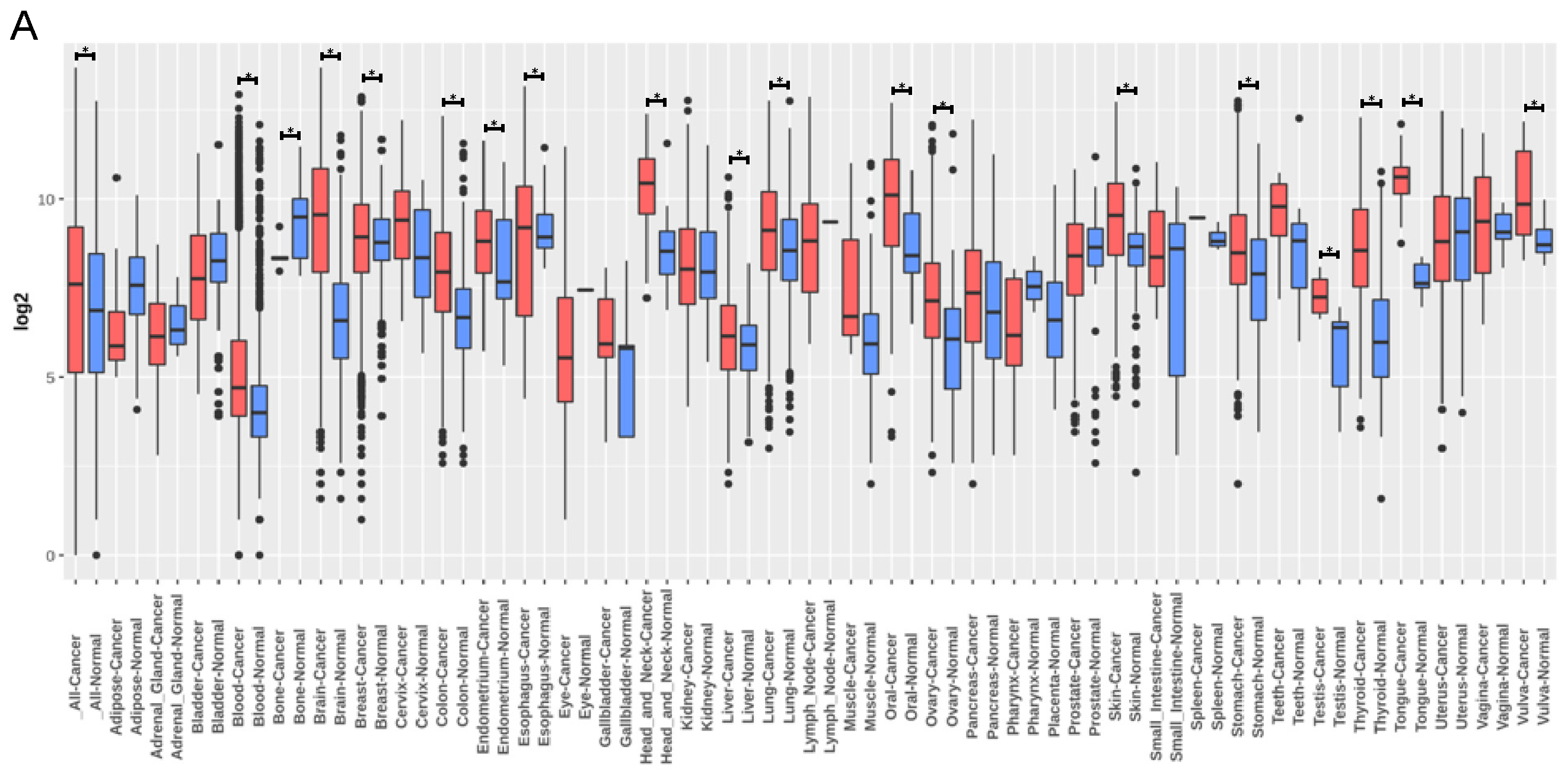
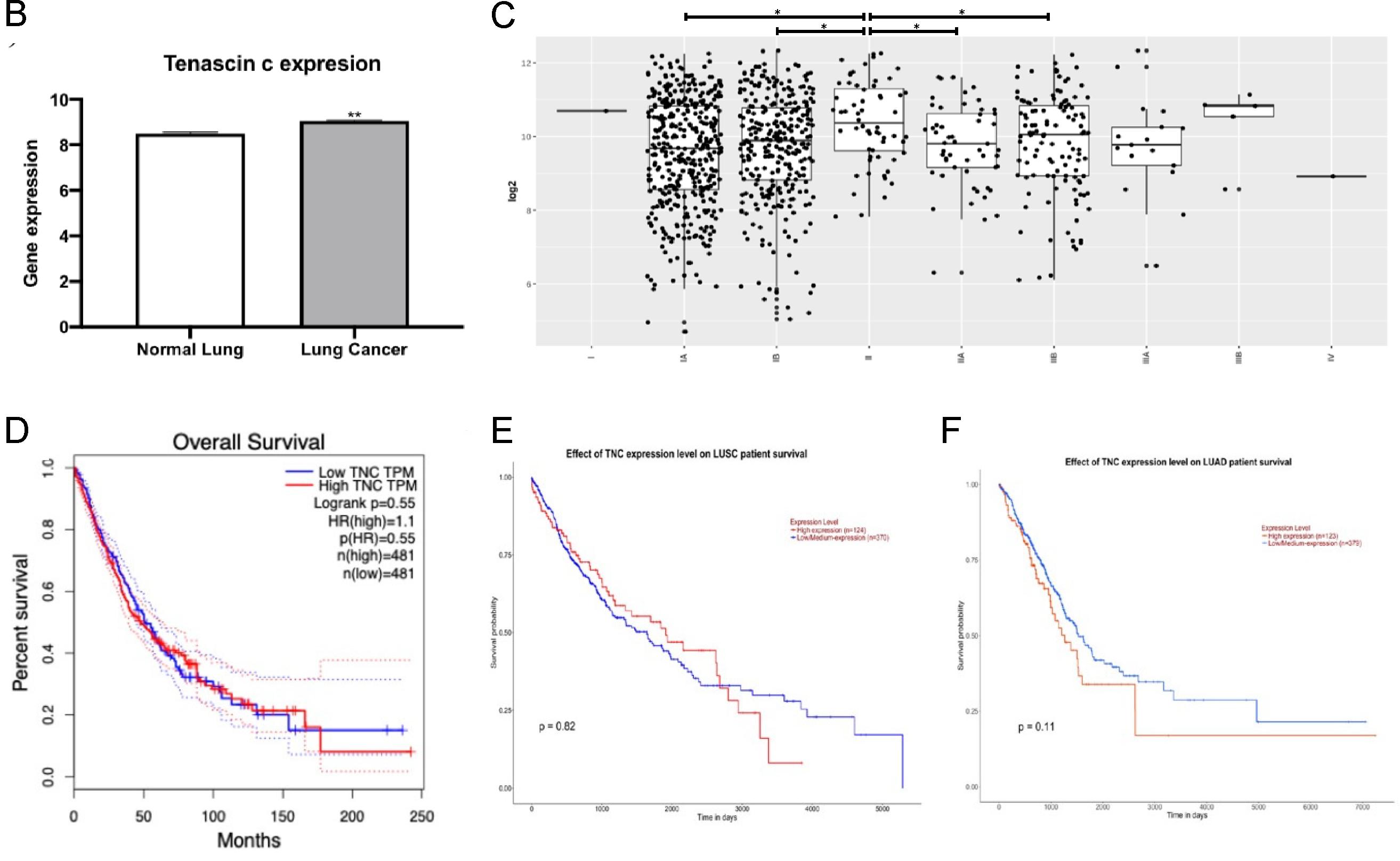
3.4. TNC Expression Is Increased in Lung Cancer but Does Not Correlate with Lung Cancer Survival
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bradshaw, A.D. Regulation of cell behavior by extracellular proteins. In Principles of Tissue Engineering; Elsevier: Amsterdam, The Netherlands, 2014; pp. 279–290. [Google Scholar]
- Koch, M.; Wehrle-Haller, B.; Baumgartner, S.; Spring, J.; Brubacher, D.; Chiquet, M. Epithelial synthesis of tenascin at tips of growing bronchi and graded accumulation in basement membrane and mesenchyme. Exp. Cell Res. 1991, 194, 297–300. [Google Scholar] [CrossRef] [PubMed]
- Midwood, K.S.; Chiquet, M.; Tucker, R.P.; Orend, G. Tenascin-C at a glance. J. Cell Sci. 2016, 129, 4321–4327. [Google Scholar] [CrossRef]
- Swindle, C.S.; Tran, K.T.; Johnson, T.D.; Banerjee, P.; Mayes, A.M.; Griffith, L.; Wells, A. Epidermal growth factor (EGF)-like repeats of human tenascin-C as ligands for EGF receptor. J. Cell Biol. 2001, 154, 459–468. [Google Scholar] [CrossRef] [PubMed]
- Mercado, M.L.; Nur-e-Kamal, A.; Liu, H.Y.; Gross, S.R.; Movahed, R.; Meiners, S. Neurite outgrowth by the alternatively spliced region of human tenascin-C is mediated by neuronal alpha7beta1 integrin. J. Neurosci. 2004, 24, 238–247. [Google Scholar] [CrossRef] [PubMed]
- San Martin, R.; Pathak, R.; Jain, A.; Jung, S.Y.; Hilsenbeck, S.G.; Pina-Barba, M.C.; Sikora, A.G.; Pienta, K.J.; Rowley, D.R. Tenascin-C and Integrin alpha9 Mediate Interactions of Prostate Cancer with the Bone Microenvironment. Cancer Res. 2017, 77, 5977–5988. [Google Scholar] [CrossRef]
- Chiovaro, F.; Chiquet-Ehrismann, R.; Chiquet, M. Transcriptional regulation of tenascin genes. Cell Adh. Migr. 2015, 9, 34–47. [Google Scholar] [CrossRef]
- Ali, M.K.; Kim, R.Y.; Brown, A.C.; Donovan, C.; Vanka, K.S.; Mayall, J.R.; Liu, G.; Pillar, A.L.; Jones-Freeman, B.; Xenaki, D.; et al. Critical role for iron accumulation in the pathogenesis of fibrotic lung disease. J. Pathol. 2020, 251, 49–62. [Google Scholar] [CrossRef]
- Gocheva, V.; Naba, A.; Bhutkar, A.; Guardia, T.; Miller, K.M.; Li, C.M.; Dayton, T.L.; Sanchez-Rivera, F.J.; Kim-Kiselak, C.; Jailkhani, N.; et al. Quantitative proteomics identify Tenascin-C as a promoter of lung cancer progression and contributor to a signature prognostic of patient survival. Proc. Natl. Acad. Sci. USA 2017, 114, E5625–E5634. [Google Scholar] [CrossRef]
- Yasuda, M.; Harada, N.; Harada, S.; Ishimori, A.; Katsura, Y.; Itoigawa, Y.; Matsuno, K.; Makino, F.; Ito, J.; Ono, J.; et al. Characterization of tenascin-C as a novel biomarker for asthma: Utility of tenascin-C in combination with periostin or immunoglobulin E. Allergy Asthma Clin. Immunol. 2018, 14, 72. [Google Scholar] [CrossRef]
- Alam, R.; Good, J.; Rollins, D.; Verma, M.; Chu, H.; Pham, T.H.; Martin, R.J. Airway and serum biochemical correlates of refractory neutrophilic asthma. J. Allergy Clin. Immunol. 2017, 140, 1004–1014.e13. [Google Scholar] [CrossRef]
- Flood-Page, P.; Menzies-Gow, A.; Phipps, S.; Ying, S.; Wangoo, A.; Ludwig, M.S.; Barnes, N.; Robinson, D.; Kay, A.B. Anti-IL-5 treatment reduces deposition of ECM proteins in the bronchial subepithelial basement membrane of mild atopic asthmatics. J. Clin. Investig. 2003, 112, 1029–1036. [Google Scholar] [CrossRef]
- Laitinen, A.; Altraja, A.; Kampe, M.; Linden, M.; Virtanen, I.; Laitinen, L.A. Tenascin is increased in airway basement membrane of asthmatics and decreased by an inhaled steroid. Am. J. Respir. Crit. Care Med. 1997, 156, 951–958. [Google Scholar] [CrossRef]
- Annoni, R.; Lancas, T.; Yukimatsu Tanigawa, R.; de Medeiros Matsushita, M.; de Morais Fernezlian, S.; Bruno, A.; Fernando Ferraz da Silva, L.; Roughley, P.J.; Battaglia, S.; Dolhnikoff, M.; et al. Extracellular matrix composition in COPD. Eur. Respir. J. 2012, 40, 1362–1373. [Google Scholar] [CrossRef]
- Zakarya, R.; Chan, Y.L.; Rutting, S.; Reddy, K.; Bozier, J.; Woldhuis, R.R.; Xenaki, D.; Van Ly, D.; Chen, H.; Brandsma, C.A.; et al. BET proteins are associated with the induction of small airway fibrosis in COPD. Thorax 2021, 76, 647–655. [Google Scholar] [CrossRef]
- Sun, Z.; Schwenzer, A.; Rupp, T.; Murdamoothoo, D.; Vegliante, R.; Lefebvre, O.; Klein, A.; Hussenet, T.; Orend, G. Tenascin-C Promotes Tumor Cell Migration and Metastasis through Integrin α9β1-Mediated YAP Inhibition. Cancer Res. 2018, 78, 950–961. [Google Scholar] [CrossRef]
- Takahashi, Y.; Sawada, G.; Kurashige, J.; Matsumura, T.; Uchi, R.; Ueo, H.; Ishibashi, M.; Takano, Y.; Akiyoshi, S.; Iwaya, T.; et al. Tumor-derived tenascin-C promotes the epithelial-mesenchymal transition in colorectal cancer cells. Anticancer Res. 2013, 33, 1927–1934. [Google Scholar]
- Wawrzyniak, D.; Grabowska, M.; Głodowicz, P.; Kuczyński, K.; Kuczyńska, B.; Fedoruk-Wyszomirska, A.; Rolle, K. Down-regulation of tenascin-C inhibits breast cancer cells development by cell growth, migration, and adhesion impairment. PLoS ONE 2020, 15, e0237889. [Google Scholar] [CrossRef]
- Du, Y.; Ouyang, W.; Kitzmiller, J.A.; Guo, M.; Zhao, S.; Whitsett, J.A.; Xu, Y. Lung Gene Expression Analysis Web Portal Version 3: Lung-at-a-Glance. Am. J. Respir. Cell Mol. Biol. 2021, 64, 146–149. [Google Scholar] [CrossRef]
- Du, Y.; Kitzmiller, J.A.; Sridharan, A.; Perl, A.K.; Bridges, J.P.; Misra, R.S.; Pryhuber, G.S.; Mariani, T.J.; Bhattacharya, S.; Guo, M.; et al. Lung Gene Expression Analysis (LGEA): An integrative web portal for comprehensive gene expression data analysis in lung development. Thorax 2017, 72, 481–484. [Google Scholar] [CrossRef]
- Du, Y.; Guo, M.; Whitsett, J.A.; Xu, Y. ‘LungGENS’: A web-based tool for mapping single-cell gene expression in the developing lung. Thorax 2015, 70, 1092–1094. [Google Scholar] [CrossRef]
- Wang, B.; Chan, Y.L.; Li, G.; Ho, K.F.; Anwer, A.G.; Smith, B.J.; Guo, H.; Jalaludin, B.; Herbert, C.; Thomas, P.S.; et al. Maternal particulate matter exposure impairs lung health and is associated with mitochondrial damage. Antioxidants 2021, 10, 1029. [Google Scholar] [CrossRef] [PubMed]
- Sauler, M.; McDonough, J.E.; Adams, T.S.; Kothapalli, N.; Barnthaler, T.; Werder, R.B.; Schupp, J.C.; Nouws, J.; Robertson, M.J.; Coarfa, C.; et al. Characterization of the COPD alveolar niche using single-cell RNA sequencing. Nat. Commun. 2022, 13, 494. [Google Scholar] [CrossRef] [PubMed]
- Park, S.J.; Yoon, B.H.; Kim, S.K.; Kim, S.Y. GENT2: An updated gene expression database for normal and tumor tissues. BMC Med. Genom. 2019, 12, 101. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.; Kang, B.; Li, C.; Chen, T.; Zhang, Z. GEPIA2: An enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019, 47, W556–W560. [Google Scholar] [CrossRef]
- Bhattacharyya, S.; Wang, W.; Morales-Nebreda, L.; Feng, G.; Wu, M.; Zhou, X.; Lafyatis, R.; Lee, J.; Hinchcliff, M.; Feghali-Bostwick, C.; et al. Tenascin-C drives persistence of organ fibrosis. Nat. Commun. 2016, 7, 11703. [Google Scholar] [CrossRef]
- Loffredo, L.F.; Coden, M.E.; Jeong, B.M.; Walker, M.T.; Anekalla, K.R.; Doan, T.C.; Rodriguez, R.; Browning, M.; Nam, K.; Lee, J.J. Eosinophil accumulation in postnatal lung is specific to the primary septation phase of development. Sci. Rep. 2020, 10, 4425. [Google Scholar] [CrossRef]
- Gremlich, S.; Cremona, T.P.; Yao, E.; Chabenet, F.; Fytianos, K.; Roth-Kleiner, M.; Schittny, J.C. Tenascin-C: Friend or Foe in Lung Aging? Front. Physiol. 2021, 12, 749776. [Google Scholar] [CrossRef]
- Roth-Kleiner, M.; Hirsch, E.; Schittny, J.C. Fetal Lungs of Tenascin-C–Deficient Mice Grow Well, but Branch Poorly in Organ Culture. Am. J. Respir. Cell Mol. Biol. 2004, 30, 360–366. [Google Scholar] [CrossRef]
- Goncalves, I.F.; Acar, E.; Costantino, S.; Szabo, P.L.; Hamza, O.; Tretter, E.V.; Klein, K.U.; Trojanek, S.; Abraham, D.; Paneni, F.; et al. Epigenetic modulation of tenascin C in the heart: Implications on myocardial ischemia, hypertrophy and metabolism. J. Hypertens. 2019, 37, 1861–1870. [Google Scholar] [CrossRef]
- Jung, C.-R.; Chen, W.-T.; Tang, Y.-H.; Hwang, B.-F. Fine particulate matter exposure during pregnancy and infancy and incident asthma. J. Allergy Clin. Immunol. 2019, 143, 2254–2262.e5. [Google Scholar] [CrossRef]
- Nakahara, H.; Gabazza, E.C.; Fujimoto, H.; Nishii, Y.; D’Alessandro-Gabazza, C.N.; Bruno, N.E.; Takagi, T.; Hayashi, T.; Maruyama, J.; Maruyama, K. Deficiency of tenascin C attenuates allergen-induced bronchial asthma in the mouse. Eur. J. Immunol. 2006, 36, 3334–3345. [Google Scholar] [CrossRef]
- Karjalainen, E.M.; Lindqvist, A.; Laitinen, L.A.; Kava, T.; Altraja, A.; Halme, M.; Laitinen, A. Airway inflammation and basement membrane tenascin in newly diagnosed atopic and nonatopic asthma. Respir. Med. 2003, 97, 1045–1051. [Google Scholar] [CrossRef]
- Rogers, N.; Clements, D.; Harrison, T.; Shaw, D.; Johnson, S. S11 expression of tenascin-C regulates airway smooth muscle derived matrix metalloproteinase-1 in asthma. Thorax 2012, 67, A8. [Google Scholar] [CrossRef]
- Brellier, F.; Hostettler, K.; Hotz, H.-R.; Ozcakir, C.; Çöloğlu, S.A.; Togbe, D.; Ryffel, B.; Roth, M.; Chiquet-Ehrismann, R. Tenascin-C triggers fibrin accumulation by downregulation of tissue plasminogen activator. FEBS Lett. 2011, 585, 913–920. [Google Scholar] [CrossRef]
- Parekh, K.; Ramachandran, S.; Cooper, J.; Bigner, D.; Patterson, A.; Mohanakumar, T. Tenascin-C, over expressed in lung cancer down regulates effector functions of tumor infiltrating lymphocytes. Lung Cancer 2005, 47, 17–29. [Google Scholar] [CrossRef]
- Sun, Z.; Velázquez-Quesada, I.; Murdamoothoo, D.; Ahowesso, C.; Yilmaz, A.; Spenlé, C.; Averous, G.; Erne, W.; Oberndorfer, F.; Oszwald, A. Tenascin-C increases lung metastasis by impacting blood vessel invasions. Matrix Biol. 2019, 83, 26–47. [Google Scholar] [CrossRef]



| Term ID | Term Description | Observed Gene Count | Background Gene Count | Strength | False Discovery Rate | |
|---|---|---|---|---|---|---|
| KEGG | hsa04512 | ECM-receptor interaction | 13 | 88 | 2.14 | 5.40 × 10−23 |
| hsa04510 | Focal adhesion | 13 | 198 | 1.79 | 3.94 × 10−19 | |
| hsa05412 | Arrhythmogenic right ventricular cardiomyopathy | 11 | 76 | 2.13 | 3.94 × 10−19 | |
| hsa05410 | Hypertrophic cardiomyopathy | 10 | 89 | 2.02 | 1.93 × 10−16 | |
| hsa05414 | Dilated cardiomyopathy | 10 | 95 | 1.99 | 2.85 × 10−16 | |
| Biological Process | GO:0007229 | Integrin-mediated signalling pathway | 11 | 94 | 2.04 | 2.66 × 10−16 |
| GO:0030198 | Extracellular matrix organization | 14 | 338 | 1.59 | 2.66 × 10−16 | |
| GO:0007155 | Cell adhesion | 16 | 925 | 1.21 | 3.71 × 10−14 | |
| GO:0031589 | Cell-substrate adhesion | 11 | 182 | 1.75 | 4.42 × 10−14 | |
| GO:0007160 | Cell-matrix adhesion | 10 | 127 | 1.87 | 1.22 × 10−13 | |
| Molecular Function | GO:0050840 | Extracellular matrix binding | 8 | 56 | 2.12 | 5.26 × 10−12 |
| GO:0005178 | Integrin binding | 9 | 147 | 1.76 | 4.48 × 10−11 | |
| GO:0050839 | Cell adhesion molecule binding | 11 | 538 | 1.28 | 2.24 × 10−9 | |
| GO:0001968 | Fibronectin binding | 5 | 27 | 2.24 | 1.36 × 10−7 | |
| GO:0005102 | Signalling receptor binding | 12 | 1581 | 0.85 | 7.11 × 10−6 | |
| Cellular Component | GO:0008305 | Integrin complex | 10 | 30 | 2.49 | 2.19 × 10−19 |
| GO:0005925 | Focal adhesion | 15 | 405 | 1.54 | 1.99 × 10−18 | |
| GO:0098797 | Plasma membrane protein complex | 15 | 547 | 1.41 | 9.63 × 10−17 | |
| GO:0070161 | Anchoring junction | 16 | 820 | 1.26 | 4.98 × 10−16 | |
| GO:0098802 | Plasma membrane signalling receptor complex | 11 | 169 | 1.78 | 2.29 × 10−15 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Donovan, C.; Bai, X.; Chan, Y.L.; Feng, M.; Ho, K.-F.; Guo, H.; Chen, H.; Oliver, B.G. Tenascin C in Lung Diseases. Biology 2023, 12, 199. https://doi.org/10.3390/biology12020199
Donovan C, Bai X, Chan YL, Feng M, Ho K-F, Guo H, Chen H, Oliver BG. Tenascin C in Lung Diseases. Biology. 2023; 12(2):199. https://doi.org/10.3390/biology12020199
Chicago/Turabian StyleDonovan, Chantal, Xu Bai, Yik Lung Chan, Min Feng, Kin-Fai Ho, Hai Guo, Hui Chen, and Brian G. Oliver. 2023. "Tenascin C in Lung Diseases" Biology 12, no. 2: 199. https://doi.org/10.3390/biology12020199
APA StyleDonovan, C., Bai, X., Chan, Y. L., Feng, M., Ho, K.-F., Guo, H., Chen, H., & Oliver, B. G. (2023). Tenascin C in Lung Diseases. Biology, 12(2), 199. https://doi.org/10.3390/biology12020199










