Unraveling the Genomic Association for Milk Production Traits and Signatures of Selection of Cattle in a Harsh Tropical Environment
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Location and Phenotypic Trait Recording
2.2. Genotyping and Quality Control
2.3. Estimation of Genomic Parameters for Milk and Milk Traits
2.4. Genome-Wide Associations and Gene Annotations
2.5. Selection Signature Analysis
3. Results
3.1. Genomic Heritabilities and Variance Component Estimates
3.2. Genome-Wide Associations and Potential Candidate Genes
3.3. Selection Signatures
3.4. Functional Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wit, D.; Rao, S.V.N.; Venkatasubramanian, V. Consequences of Crossbreeding Programme in India. Econ. Polit. Wkly. 1995, 30, A112–A116. [Google Scholar]
- Sinha, B.N. Taylor Cows of Patna. Indian Vet. J. 1951, 27, 272–276. [Google Scholar] [PubMed]
- Wakchaure, R.; Ganguly, S.; Para, P.; Praveen, P.; Kumar, A.; Sharma, S. Development of Crossbred Cattle in India: A Review. Int. J. Emerg. Technol. Adv. Eng. 2015, 5, 75–77. [Google Scholar]
- Pinto, A.; Yin, T.; Reichenbach, M.; Malik, P.K.; Schlecht, E.; König, S. Dairy Farms in an Urbanising Environment: Tradeoffs Between Productivity and Animal Welfare. In The Rural-Urban Interface; Springer: Berlin/Heidelberg, Germany, 2021; pp. 103–107. [Google Scholar] [CrossRef]
- Aliloo, H.; Mrode, R.; Okeyo, A.M.; Gibson, J.P. Ancestral Haplotype Mapping for GWAS and Detection of Signatures of Selection in Admixed Dairy Cattle of Kenya. Front. Genet. 2020, 11, 544. [Google Scholar] [CrossRef] [PubMed]
- Cole, J.B.; da Silva, M.V.G.B. Genomic Selection in Multi-Breed Dairy Cattle Populations. Rev. Bras. Zootec. 2016, 45, 195–202. [Google Scholar] [CrossRef]
- Chen, Z.; Yao, Y.; Ma, P.; Wang, Q.; Pan, Y. Haplotype-Based Genome-Wide Association Study Identifies Loci and Candidate Genes for Milk Yield in Holsteins. PLoS ONE 2018, 13, e0192695. [Google Scholar] [CrossRef]
- Singh, A.; Kumar, A.; Gondro, C.; Pandey, A.K.; Dutt, T.; Mishra, B.P. Genome Wide Scan to Identify Potential Genomic Regions Associated With Milk Protein and Minerals in Vrindavani Cattle. Front. Vet. Sci. 2022, 9, 760364. [Google Scholar] [CrossRef]
- Tajima, F. Statistical Method for Testing the Neutral Mutation Hypothesis by DNA Polymorphism. Genetics 1989, 123, 585–595. [Google Scholar] [CrossRef]
- Fay, J.C.; Wu, C.-I. Hitchhiking Under Positive Darwinian Selection. Genetics 2000, 155, 1405–1413. [Google Scholar] [CrossRef]
- Vy, H.M.T.; Kim, Y. A Composite-Likelihood Method for Detecting Incomplete Selective Sweep from Population Genomic Data. Genetics 2015, 200, 633–649. [Google Scholar] [CrossRef]
- Sabeti, P.C.; Reich, D.E.; Higgins, J.M.; Levine, H.Z.P.; Richter, D.J.; Schaffner, S.F.; Gabriel, S.B.; Platko, J.V.; Patterson, N.J.; McDonald, G.J.; et al. Detecting Recent Positive Selection in The Human Genome from Haplotype Structure. Nature 2002, 419, 832–837. [Google Scholar] [CrossRef] [PubMed]
- Voight, B.F.; Kudaravalli, S.; Wen, X.; Pritchard, J.K. A Map of Recent Positive Selection in the Human Genome. PLoS Biol. 2006, 4, e72. [Google Scholar] [CrossRef]
- Rothammer, S.; Seichter, D.; Förster, M.; Medugorac, I. A Genome-Wide Scan for Signatures of Differential Artificial Selection in Ten Cattle Breeds. BMC Genomics 2013, 14, 908. [Google Scholar] [CrossRef] [PubMed]
- Cheruiyot, E.K.; Bett, R.C.; Amimo, J.O.; Zhang, Y.; Mrode, R.; Mujibi, F.D.N. Signatures of Selection in Admixed Dairy Cattle in Tanzania. Front. Genet. 2018, 9, 607. [Google Scholar] [CrossRef]
- Li, R.; Li, C.; Chen, H.; Li, R.; Chong, Q.; Xiao, H.; Chen, S. Genome-wide Scan of Selection Signatures in Dehong Humped Cattle for Heat Tolerance and Disease Resistance. Anim. Genet. 2020, 51, 292–299. [Google Scholar] [CrossRef] [PubMed]
- Naderi, S.; Moradi, M.H.; Farhadian, M.; Yin, T.; Jaeger, M.; Scheper, C.; Korkuc, P.; Brockmann, G.A.; König, S.; May, K. Assessing Selection Signatures within and between Selected Lines of Dual-Purpose Black and White and German Holstein Cattle. Anim. Genet. 2020, 51, 391–408. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Mehrotra, A.; Gondro, C.; da Silva Romero, A.R.; Pandey, A.K.; Karthikeyan, A.; Bashir, A.; Mishra, B.P.; Dut, T.; Kumar, A. Signatures of Selection in Composite Vrindavani Cattle of India. Front. Genet. 2020, 11, 589496. [Google Scholar] [CrossRef]
- Taye, M.; Lee, W.; Caetano-Anolles, K.; Dessie, T.; Hanotte, O.; Mwai, O.A.; Kemp, S.; Cho, S.; Oh, S.J.; Lee, H.-K.; et al. Whole Genome Detection of Signature of Positive Selection in African Cattle Reveals Selection for Thermotolerance. Anim. Sci. J. 2017, 88, 1889–1901. [Google Scholar] [CrossRef]
- Pinto, A.; Yin, T.; Reichenbach, M.; Bhatta, R.; Schlecht, E.; König, S. Phenotypic Dairy Cattle Trait Expressions in Dependency of Social-Ecological Characteristics along Rural–Urban Gradients. Sustainability 2020, 12, 9021. [Google Scholar] [CrossRef]
- Mullakkalparambil Velayudhan, S.; Brügemann, K.; Pinto, A.; Yin, T.; Reichenbach, M.; Sejian, V.; Bhatta, R.; Schlecht, E.; König, S. Effects of Heat Stress across the Rural-Urban Interface on Phenotypic Trait Expressions of Dairy Cattle in a Tropical Savanna Region. Sustainability 2022, 14, 4590. [Google Scholar] [CrossRef]
- Velayudhan, S.M.; Brügemann, K.; Alam, S.; Yin, T.; Devaraj, C.; Sejian, V.; Schlecht, E.; König, S. Molecular, Physiological and Hematological Responses of Crossbred Dairy Cattle in a Tropical Savanna Climate. Biology 2022, 12, 26. [Google Scholar] [CrossRef]
- Kandeel, S.A.; Morin, D.E.; Calloway, C.D.; Constable, P.D. Association of California Mastitis Test Scores with Intramammary Infection Status in Lactating Dairy Cows Admitted to a Veterinary Teaching Hospital. J. Vet. Intern. Med. 2018, 32, 497–505. [Google Scholar] [CrossRef]
- Hoffmann, E.; Jose, M.; Nölke, N.; Möckel, T. Construction and Use of a Simple Index of Urbanisation in the Rural–Urban Interface of Bangalore, India. Sustainability 2017, 9, 2146. [Google Scholar] [CrossRef]
- National Research Council (NRC). A Guide to Environmental Research on Animals; National Research Council (NRC): Rockville, MD, USA, 1971. [Google Scholar]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.W.; Daly, M.J.; et al. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef]
- Wagner, P.; Yin, T.; Brügemann, K.; Engel, P.; Weimann, C.; Schlez, K.; König, S. Genome-Wide Associations for Microscopic Differential Somatic Cell Count and Specific Mastitis Pathogens in Holstein Cows in Compost-Bedded Pack and Cubicle Farming Systems. Animals 2021, 11, 1839. [Google Scholar] [CrossRef] [PubMed]
- Shabalina, T.; Yin, T.; May, K.; König, S. Proofs for Genotype by Environment Interactions Considering Pedigree and Genomic Data from Organic and Conventional Cow Reference Populations. J. Dairy Sci. 2021, 104, 4452–4466. [Google Scholar] [CrossRef]
- Misztal, I.; Lourenco, D.; Aguilar, I.; Legarra, A.; Vitezica, Z. Manual for BLUPF90 Family of Programs 2022. Available online: http://nce.ads.uga.edu/wiki/lib/exe/fetch.php?media=blupf90_all8.pdf (accessed on 28 November 2023).
- VanRaden, P.M. Efficient Methods to Compute Genomic Predictions. J. Dairy Sci. 2008, 91, 4414–4423. [Google Scholar] [CrossRef] [PubMed]
- Aguilar, I.; Misztal, I.; Tsuruta, S.; Legarra, A.; Wang, H. PREGSF90—POSTGSF90: Computational Tools for the Implementation of Single-Step Genomic Selection and Genome-Wide Association with Ungenotyped Individuals in BLUPF90 Programs. In Proceedings of the 10th World Congress of Genetics Applied to Livestock Production, Vancouver, BC, Canada, 17–22 August 2014. [Google Scholar]
- Deb, G.; Mufti, M.; Mostari, M.; Huque, K. Genetic Evaluation Of Bangladesh Livestock Research Institute Cattle Breed-1: Heritability And Genetic Correlation. Bangladesh J. Anim. Sci. 2008, 37, 25–33. [Google Scholar] [CrossRef]
- Roman, R.M.; Wilcox, C.J.; Martin, F.G. Estimates of Repeatability and Heritability of Productive and Reproductive Traits in a Herd of Jersey Cattle. Genet. Mol. Biol. 2000, 23, 113–119. [Google Scholar] [CrossRef]
- Liu, L.; Zhou, J.; Chen, C.J.; Zhang, J.; Wen, W.; Tian, J.; Zhang, Z.; Gu, Y. GWAS-Based Identification of New Loci for Milk Yield, Fat, and Protein in Holstein Cattle. Animals 2020, 10, 2048. [Google Scholar] [CrossRef] [PubMed]
- Costa, A.; Lopez-Villalobos, N.; Visentin, G.; Marchi, M.D.; Cassandro, M.; Penasa, M. Heritability and Repeatability of Milk Lactose and Its Relationships with Traditional Milk Traits, Somatic Cell Score and Freezing Point in Holstein Cows. Animal 2019, 13, 909–916. [Google Scholar] [CrossRef]
- Djedović, R.; Bogdanović, V.; Stanojević, D.; Beskorovajni, R.; Trivunović, S.; Petrović, M.; Samolovac, L. The Assessment of the Selection Effects on Milk Traits in Black-White Cattle. In Proceedings of the 23rd International Symposium “New Technologies in Contemporary Animal Production”, Novi Sad, Serbia, 19–21 June 2013. [Google Scholar]
- Schneider, H.; Segelke, D.; Tetens, J.; Thaller, G.; Bennewitz, J. A Genomic Assessment of the Correlation between Milk Production Traits and Claw and Udder Health Traits in Holstein Dairy Cattle. J. Dairy Sci. 2023, 106, 1190–1205. [Google Scholar] [CrossRef] [PubMed]
- Luttinen, A.; Juga, J. Genetic Relationships between Milk Yield, Somatic Cell Count, Mastitis, Milkability and Leakage in Finnish Dairy Cattle Population. Interbull Bull. 1997, 15, 78–83. [Google Scholar]
- Kawahara, T.; Gotoh, Y.; Yamaguchi, S.; Suzuki, M. Variance Component Estimates with Dominance Models for Milk Production in Holsteins of Japan Using Method R. Asian-Australas. J. Anim. Sci. 2006, 19, 769–774. [Google Scholar] [CrossRef]
- Moya, J.; Wilcox, C.J.; Bachman, K.C.; Martin, F.G. Genetic Trends in Milk Yield and Composition in a Subtropical Dairy Herd. Rev. Bras. Genet. 1985, 8, 509–521. [Google Scholar]
- Cho, C.I.; Alam, M.; Choi, T.J.; Choy, Y.H.; Choi, J.G.; Lee, S.S.; Cho, K.H. Models for Estimating Genetic Parameters of Milk Production Traits Using Random Regression Models in Korean Holstein Cattle. Asian-Australas. J. Anim. Sci. 2016, 29, 607–614. [Google Scholar] [CrossRef] [PubMed]
- Tiezzi, F.; Pretto, D.; Marchi, M.D.; Penasa, M.; Cassandro, M. Heritability and Repeatability of Milk Coagulation Properties Predicted by Mid-Infrared Spectroscopy during Routine Data Recording, and Their Relationships with Milk Yield and Quality Traits. Animal 2013, 7, 1592–1599. [Google Scholar] [CrossRef]
- Petrini, J.; Iung, L.; Rodriguez, M.; Salvian, M.; Pértille, F.; Rovadoscki, G.; Cassoli, L.; Coutinho, L.; Machado, P.; Wiggans, G.; et al. Genetic Parameters for Milk Fatty Acids, Milk Yield and Quality Traits of a Holstein Cattle Population Reared under Tropical Conditions. J. Anim. Breed. Genet. 2016, 133, 384–395. [Google Scholar] [CrossRef]
- Sneddon, N.; Lopez-Villalobos, N.; Davis, S.; Hickson, R.; Shalloo, L. Genetic Parameters for Milk Components including Lactose from Test Day Records in the New Zealand Dairy Herd. N. Z. J. Agric. Res. 2015, 58, 97–107. [Google Scholar] [CrossRef]
- Meena, H.R.; Mahapatra, R.K.; Sahoo, A. Variations in Nilk Composition of Hill and Pashmina Goats Under Temperate Climate of the Kumaon Himalaya. Indian J. Dairy Sci. 2009, 62, 25–30. [Google Scholar]
- Alrawi, A.A.; Pollak, E.J.; Laben, R.C. Genetic Analysis of California Mastitis Test Records. I. Coded Tests. J. Dairy Sci. 1979, 62, 1115–1124. [Google Scholar] [CrossRef]
- Pösö, J.; Mäntysaari, E.A. Relationships Between Clinical Mastitis, Somatic Cell Score, and Production for the First Three Lactations of Finnish Ayrshire. J. Dairy Sci. 1996, 79, 1284–1291. [Google Scholar] [CrossRef] [PubMed]
- Nash, D.L.; Rogers, G.W.; Cooper, J.B.; Hargrove, G.L.; Keown, J.F.; Hansen, L.B. Heritability of Clinical Mastitis Incidence and Relationships with Sire Transmitting Abilities for Somatic Cell Score, Udder Type Traits, Productive Life, and Protein Yield. J. Dairy Sci. 2000, 83, 2350–2360. [Google Scholar] [CrossRef] [PubMed]
- Carlén, E.; Strandberg, E.; Roth, A. Genetic Parameters for Clinical Mastitis, Somatic Cell Score, and Production in the First Three Lactations of Swedish Holstein Cows. J. Dairy Sci. 2004, 87, 3062–3070. [Google Scholar] [CrossRef] [PubMed]
- Zwald, N.R.; Weigel, K.A.; Chang, Y.M.; Welper, R.D.; Clay, J.S. Genetic Selection for Health Traits Using Producer-Recorded Data. I. Incidence Rates, Heritability Estimates, and Sire Breeding Values. J. Dairy Sci. 2004, 87, 4287–4294. [Google Scholar] [CrossRef] [PubMed]
- Gonyon, D.S.; Everson, D.O.; Christian, R.E. Heritability of Mastitis Score in Pacific Northwest Dairy Herds1. J. Dairy Sci. 1982, 65, 1269–1276. [Google Scholar] [CrossRef] [PubMed]
- Bouyai, D.; Duangjinda, M.; Pattarajinda, V.; Katawatin, S.; Sanitchon, J.; Bulakul, C.; Boonkum, W. Detection of Quantitative trait Loci for Clinical Mastitis in Crossbred Holsteins in the Tropics. Livest. Sci. 2012, 150, 22–30. [Google Scholar] [CrossRef]
- Bathla, S.; Rawat, P.; Baithalu, R.; Yadav, M.L.; Naru, J.; Tiwari, A.; Kumar, S.; Balhara, A.K.; Singh, S.; Chaudhary, S.; et al. Profiling of Urinary Proteins in Karan Fries Cows Reveals More than 1550 Proteins. J. Proteomics 2015, 127, 193–201. [Google Scholar] [CrossRef]
- Elangovan, M.; Ka, J.; Pak, B.; Choi, W.; Oh, S.-R.; Jin, S.-W.; Yoo, Y.J. Ubiquitin-Conjugating Enzyme V Variant 1 Enables Cellular Responses toward Fibroblast Growth Factor Signaling in Endothelium. FASEB J. 2022, 36, e22103. [Google Scholar] [CrossRef]
- Rani, P.; Onteru, S.K.; Singh, D. Genome-Wide Profiling and Analysis of microRNA Expression in Buffalo Milk Exosomes. Food Biosci. 2020, 38, 100769. [Google Scholar] [CrossRef]
- Rosen, J.M.; Wyszomierski, S.L.; Hadsell, D. Regulation of Milk Protein Gene Expression. Annu. Rev. Nutr. 1999, 19, 407–436. [Google Scholar] [CrossRef] [PubMed]
- Sigl, T.; Meyer, H.H.D.; Wiedemann, S. Gene Expression Analysis of Protein Synthesis Pathways in Bovine Mammary Epithelial Cells Purified from Milk during Lactation and Short-Term Restricted Feeding. J. Anim. Physiol. Anim. Nutr. 2014, 98, 84–95. [Google Scholar] [CrossRef] [PubMed]
- Doppler, W.; Welte, T.; Philipp, S. CCAAT/Enhancer-Binding Protein Isoforms γ and δ Are Expressed in Mammary Epithelial Cells and Bind to Multiple Sites in the β-Casein Gene Promoter. J. Biol. Chem. 1995, 270, 17962–17969. [Google Scholar] [CrossRef] [PubMed]
- Qanbari, S.; Pausch, H.; Jansen, S.; Somel, M.; Strom, T.M.; Fries, R.; Nielsen, R.; Simianer, H. Classic Selective Sweeps Revealed by Massive Sequencing in Cattle. PLOS Genet. 2014, 10, e1004148. [Google Scholar] [CrossRef] [PubMed]
- Vanvanhossou, S.F.U.; Yin, T.; Scheper, C.; Fries, R.; Dossa, L.H.; König, S. Unraveling Admixture, Inbreeding, and Recent Selection Signatures in West African Indigenous Cattle Populations in Benin. Front. Genet. 2021, 12, 657282. [Google Scholar] [CrossRef]
- Guo, H.J.; Tadi, P. Biochemistry, Ubiquitination; StatPearls Publishing: Treasure Island, FL, USA, 2022. [Google Scholar]
- Myung, J.; Kim, K.B.; Crews, C.M. The Ubiquitin-Proteasome Pathway and Proteasome Inhibitors. Med. Res. Rev. 2001, 21, 245–273. [Google Scholar] [CrossRef]
- Saravanan, K.A.; Panigrahi, M.; Kumar, H.; Parida, S.; Bhushan, B.; Gaur, G.K.; Dutt, T.; Mishra, B.P.; Singh, R.K. Genomic Scans for Selection Signatures Revealed Candidate Genes for Adaptation and Production Traits in a Variety of Cattle Breeds. Genomics 2021, 113, 955–963. [Google Scholar] [CrossRef]
- Singh, A.; Kumar, A.; Gondro, C.; Da Silva Romero, A.R.; Karthikeyan, A.; Mehrotra, A.; Pandey, A.K.; Dutt, T.; Mishra, B.P. Identification of Genes Affecting Milk Fat and Fatty Acid Composition in Vrindavani Crossbred Cattle Using 50 K SNP-Chip. Trop. Anim. Health Prod. 2021, 53, 347. [Google Scholar] [CrossRef]
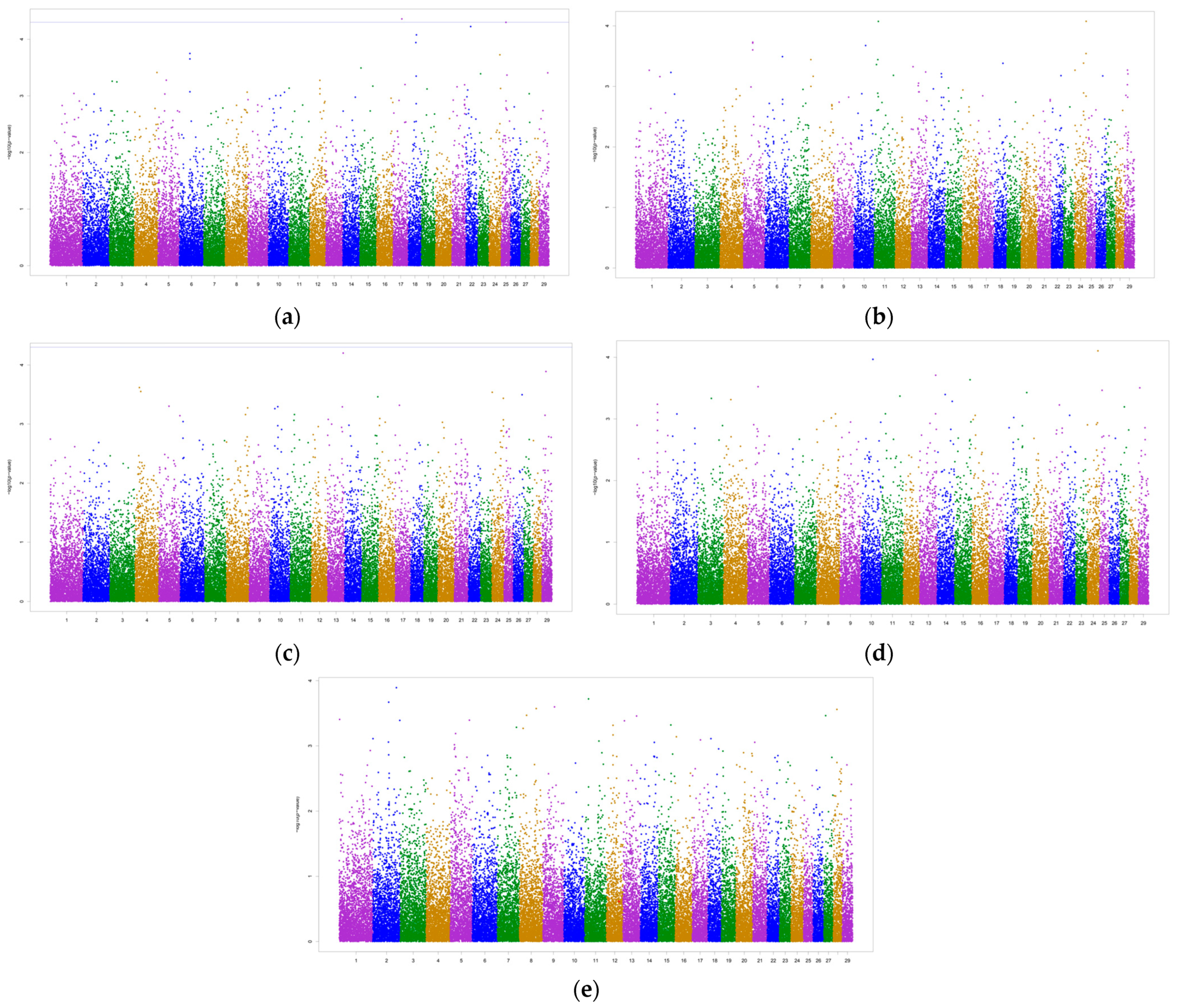

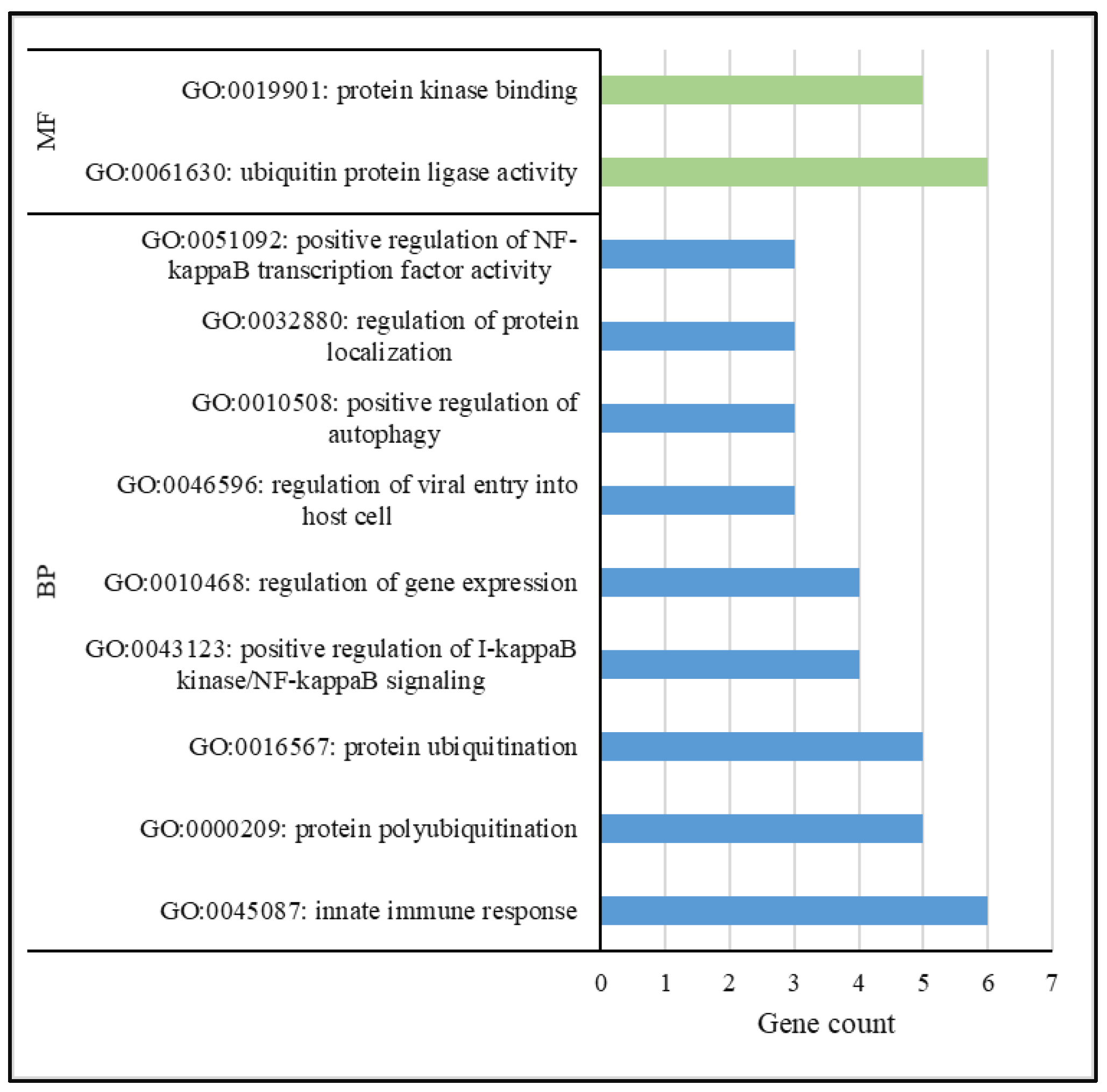
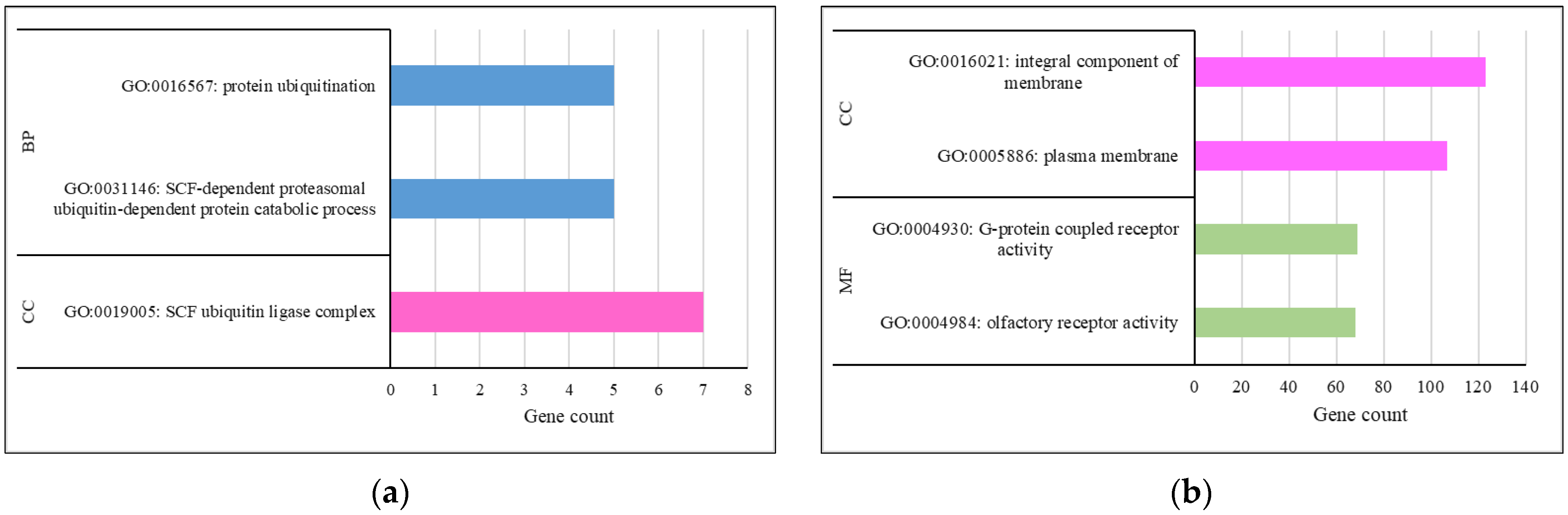

| Trait | Animals | Records (n) | h2 | σ2g | σ2pe | σ2e |
|---|---|---|---|---|---|---|
| Test-day milk yield | 125 | 527 | 0.25 (0.21) | 4.24 (3.59) | 5.54 (3.45) | 6.92 (0.49) |
| SNF | 126 | 496 | 0.13 (0.05) | 0.07 (0.03) | 0.15 × 10−4 (0.46 × 10−3) | 0.48 (0.04) |
| Lactose | 126 | 524 | 0.20 (0.16) | 0.024 (0.02) | 0.001 (0.02) | 0.096 (0.0068) |
| Density | 126 | 496 | 0.17 (0.23) | 1.42 (1.90) | 2.31 (1.89) | 4.40 (0.33) |
| Clinical mastitis | 126 | 267 | 0.48 (0.07) | 0.84 (0.18) | 0.13 × 10−4 (0.11 × 10−2) | 0.93 (0.11) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Velayudhan, S.M.; Yin, T.; Alam, S.; Brügemann, K.; Sejian, V.; Bhatta, R.; Schlecht, E.; König, S. Unraveling the Genomic Association for Milk Production Traits and Signatures of Selection of Cattle in a Harsh Tropical Environment. Biology 2023, 12, 1483. https://doi.org/10.3390/biology12121483
Velayudhan SM, Yin T, Alam S, Brügemann K, Sejian V, Bhatta R, Schlecht E, König S. Unraveling the Genomic Association for Milk Production Traits and Signatures of Selection of Cattle in a Harsh Tropical Environment. Biology. 2023; 12(12):1483. https://doi.org/10.3390/biology12121483
Chicago/Turabian StyleVelayudhan, Silpa Mullakkalparambil, Tong Yin, Shahin Alam, Kerstin Brügemann, Veerasamy Sejian, Raghavendra Bhatta, Eva Schlecht, and Sven König. 2023. "Unraveling the Genomic Association for Milk Production Traits and Signatures of Selection of Cattle in a Harsh Tropical Environment" Biology 12, no. 12: 1483. https://doi.org/10.3390/biology12121483
APA StyleVelayudhan, S. M., Yin, T., Alam, S., Brügemann, K., Sejian, V., Bhatta, R., Schlecht, E., & König, S. (2023). Unraveling the Genomic Association for Milk Production Traits and Signatures of Selection of Cattle in a Harsh Tropical Environment. Biology, 12(12), 1483. https://doi.org/10.3390/biology12121483








