The Diagnostic Landscape of Adult Neurogenetic Disorders
Abstract
:Simple Summary
Abstract
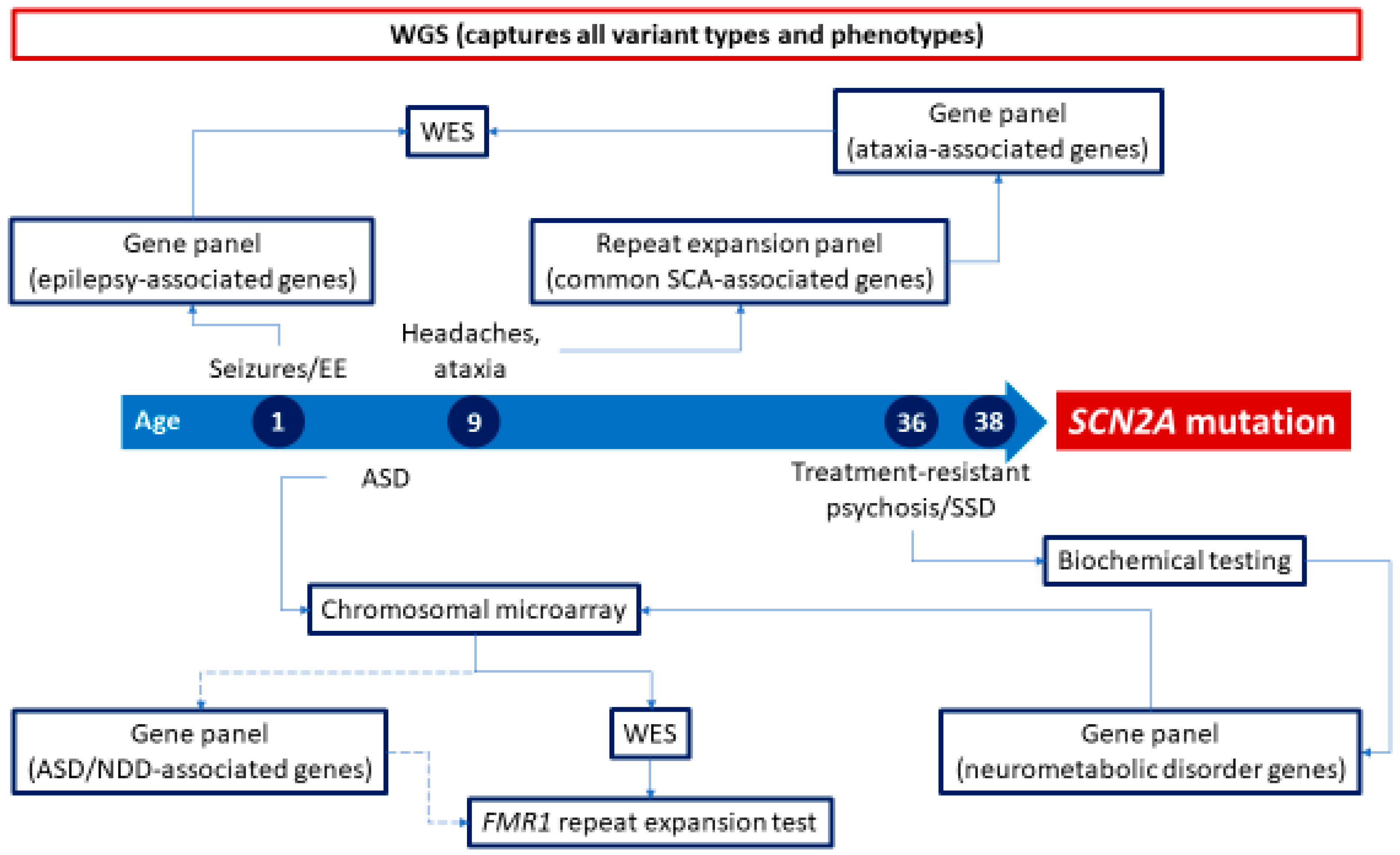
1. Introduction
2. Neurogenetic Syndromes
2.1. Movement Disorders: Parkinson’s Disease, Dystonia, Ataxia, Spastic Paraparesis
2.2. Epilepsy and Intellectual Disability
2.3. Neuromuscular Disorders
2.4. Cognitive Neurodegenerative Disease
2.5. Leukodystrophies and Other Diseases of White Matter
2.6. Neurometabolic Diseases
2.7. Episodic Neurologic Syndromes-Paroxysmal Movement Disorders, Episodic Ataxia, Hemiplegic Migraine
3. Current State and Challenges of Neurogenetic Testing
3.1. Chromosomal Microarray
3.2. Gene Panel Testing
3.3. Repeat Expansion Testing
3.4. Biochemical Testing
3.5. Whole-Exome Sequencing
4. The Future of Neurogenetic Testing
Whole-Genome Testing
5. Barriers to Neurogenetic Testing
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Delikurt, T.; Williamson, G.R.; Anastasiadou, V.; Skirton, H. A Systematic Review of Factors That Act as Barriers to Patient Referral to Genetic Services. Eur. J. Hum. Genet. 2015, 23, 739–745. [Google Scholar] [CrossRef] [PubMed]
- Penon-Portmann, M.; Chang, J.; Cheng, M.; Shieh, J.T. Genetics Workforce: Distribution of Genetics Services and Challenges to Health Care in California. Genet. Med. 2020, 22, 227–231. [Google Scholar] [CrossRef] [PubMed]
- GENE.00052 Whole Genome Sequencing, Whole Exome Sequencing, Gene Panels, and Molecular Profiling Medical Policy. Available online: https://www.anthem.com/dam/medpolicies/abc/active/policies/mp_pw_e000224.html (accessed on 13 August 2023).
- Whole Exome and Whole Genome Sequencing for Non-Cancer Indications Medical Coverage Policy. Available online: https://static.cigna.com/assets/chcp/pdf/coveragePolicies/medical/mm_0519_coveragepositioncriteria_exome_genome_sequence.pdf (accessed on 13 August 2023).
- Whole Exome and Whole Genome Sequencing UnitedHealthcare ® Commercial and Individual Exchange Medical Policy Whole Exome and Whole Genome Sequencing. 2023. Available online: https://www.uhcprovider.com/content/dam/provider/docs/public/policies/comm-medical-drug/whole-exome-and-whole-genome-sequencing.pdf (accessed on 13 August 2023).
- Reuter, C.M.; Kohler, J.N.; Bonner, D.; Zastrow, D.; Fernandez, L.; Dries, A.; Marwaha, S.; Davidson, J.; Brokamp, E.; Herzog, M.; et al. Yield of Whole Exome Sequencing in Undiagnosed Patients Facing Insurance Coverage Barriers to Genetic Testing. J. Genet. Couns. 2019, 28, 1107–1118. [Google Scholar] [CrossRef]
- Nakamura, K.; Kato, M.; Osaka, H.; Yamashita, S.; Nakagawa, E.; Haginoya, K.; Tohyama, J.; Okuda, M.; Wada, T.; Shimakawa, S.; et al. Clinical Spectrum of SCN2A Mutations Expanding to Ohtahara Syndrome. Neurology 2013, 81, 992–998. [Google Scholar] [CrossRef]
- Wolff, M.; Johannesen, K.M.; Hedrich, U.B.S.; Masnada, S.; Rubboli, G.; Gardella, E.; Lesca, G.; Ville, D.; Milh, M.; Villard, L.; et al. Genetic and Phenotypic Heterogeneity Suggest Therapeutic Implications in SCN2A-Related Disorders. Brain J. Neurol. 2017, 140, 1316–1336. [Google Scholar] [CrossRef] [PubMed]
- Sanders, S.J.; Murtha, M.T.; Gupta, A.R.; Murdoch, J.D.; Raubeson, M.J.; Willsey, A.J.; Ercan-Sencicek, A.G.; DiLullo, N.M.; Parikshak, N.N.; Stein, J.L.; et al. De Novo Mutations Revealed by Whole-Exome Sequencing Are Strongly Associated with Autism. Nature 2012, 485, 237–241. [Google Scholar] [CrossRef]
- Schwarz, N.; Hahn, A.; Bast, T.; Müller, S.; Löffler, H.; Maljevic, S.; Gaily, E.; Prehl, I.; Biskup, S.; Joensuu, T.; et al. Mutations in the Sodium Channel Gene SCN2A Cause Neonatal Epilepsy with Late-Onset Episodic Ataxia. J. Neurol. 2016, 263, 334–343. [Google Scholar] [CrossRef]
- Carroll, L.S.; Woolf, R.; Ibrahim, Y.; Williams, H.J.; Dwyer, S.; Walters, J.; Kirov, G.; O’Donovan, M.C.; Owen, M.J. Mutation Screening of SCN2A in Schizophrenia and Identification of a Novel Loss-of-Function Mutation. Psychiatr. Genet. 2016, 26, 60. [Google Scholar] [CrossRef]
- Suddaby, J.S.; Silver, J.; So, J. Understanding the Schizophrenia Phenotype in the First Patient with the Full SCN2A Phenotypic Spectrum. Psychiatr. Genet. 2019, 29, 91–94. [Google Scholar] [CrossRef]
- Morita, M.; Al-Chalabi, A.; Andersen, P.M.; Hosler, B.; Sapp, P.; Englund, E.; Mitchell, J.E.; Habgood, J.J.; de Belleroche, J.; Xi, J.; et al. A Locus on Chromosome 9p Confers Susceptibility to ALS and Frontotemporal Dementia. Neurology 2006, 66, 839–844. [Google Scholar] [CrossRef]
- Vance, C.; Al-Chalabi, A.; Ruddy, D.; Smith, B.N.; Hu, X.; Sreedharan, J.; Siddique, T.; Schelhaas, H.J.; Kusters, B.; Troost, D.; et al. Familial Amyotrophic Lateral Sclerosis with Frontotemporal Dementia Is Linked to a Locus on Chromosome 9p13.2–21.3. Brain 2006, 129, 868–876. [Google Scholar] [CrossRef] [PubMed]
- Wild, E.J.; Tabrizi, S.J. Huntington’s Disease Phenocopy Syndromes. Curr. Opin. Neurol. 2007, 20, 681–687. [Google Scholar] [CrossRef] [PubMed]
- Watts, G.D.J.; Wymer, J.; Kovach, M.J.; Mehta, S.G.; Mumm, S.; Darvish, D.; Pestronk, A.; Whyte, M.P.; Kimonis, V.E. Inclusion Body Myopathy Associated with Paget Disease of Bone and Frontotemporal Dementia Is Caused by Mutant Valosin-Containing Protein. Nat. Genet. 2004, 36, 377–381. [Google Scholar] [CrossRef] [PubMed]
- Al-Obeidi, E.; Al-Tahan, S.; Surampalli, A.; Goyal, N.; Wang, A.; Hermann, A.; Omizo, M.; Smith, C.; Mozaffar, T.; Kimonis, V. Genotype-Phenotype Study in Patients with VCP Valosin-Containing Protein Mutations Associated with Multisystem Proteinopathy. Clin. Genet. 2018, 93, 119–125. [Google Scholar] [CrossRef] [PubMed]
- Kazamel, M.; Sorenson, E.J.; McEvoy, K.M.; Jones, L.K., Jr.; Leep-Hunderfund, A.N.; Mauermann, M.L.; Milone, M. Clinical Spectrum of Valosin Containing Protein (VCP)-Opathy. Muscle Nerve 2016, 54, 94–99. [Google Scholar] [CrossRef] [PubMed]
- Schuermans, N.; Verdin, H.; Ghijsels, J.; Hellemans, M.; Debackere, E.; Bogaert, E.; Symoens, S.; Naesens, L.; Lecomte, E.; Crosiers, D.; et al. Exome Sequencing and Multigene Panel Testing in 1411 Patients With Adult-Onset Neurologic Disorders. Neurol. Genet. 2023, 9, e200071. [Google Scholar] [CrossRef]
- Wenger, A.M.; Guturu, H.; Bernstein, J.A.; Bejerano, G. Systematic Reanalysis of Clinical Exome Data Yields Additional Diagnoses: Implications for Providers. Genet. Med. 2017, 19, 209–214. [Google Scholar] [CrossRef] [PubMed]
- Ewans, L.J.; Schofield, D.; Shrestha, R.; Zhu, Y.; Gayevskiy, V.; Ying, K.; Walsh, C.; Lee, E.; Kirk, E.P.; Colley, A.; et al. Whole-Exome Sequencing Reanalysis at 12 Months Boosts Diagnosis and Is Cost-Effective When Applied Early in Mendelian Disorders. Genet. Med. 2018, 20, 1564–1574. [Google Scholar] [CrossRef]
- Costain, G.; Jobling, R.; Walker, S.; Reuter, M.S.; Snell, M.; Bowdin, S.; Cohn, R.D.; Dupuis, L.; Hewson, S.; Mercimek-Andrews, S.; et al. Periodic Reanalysis of Whole-Genome Sequencing Data Enhances the Diagnostic Advantage over Standard Clinical Genetic Testing. Eur. J. Hum. Genet. EJHG 2018, 26, 740–744. [Google Scholar] [CrossRef]
- Savatt, J.M.; Myers, S.M. Genetic Testing in Neurodevelopmental Disorders. Front. Pediatr. 2021, 9, 526779. [Google Scholar] [CrossRef]
- Srivastava, S.; Love-Nichols, J.A.; Dies, K.A.; Ledbetter, D.H.; Martin, C.L.; Chung, W.K.; Firth, H.V.; Frazier, T.; Hansen, R.L.; Prock, L.; et al. Meta-Analysis and Multidisciplinary Consensus Statement: Exome Sequencing Is a First-Tier Clinical Diagnostic Test for Individuals with Neurodevelopmental Disorders. Genet. Med. 2019, 21, 2413–2421. [Google Scholar] [CrossRef] [PubMed]
- Thevenon, J.; Duffourd, Y.; Masurel-Paulet, A.; Lefebvre, M.; Feillet, F.; El Chehadeh-Djebbar, S.; St-Onge, J.; Steinmetz, A.; Huet, F.; Chouchane, M.; et al. Diagnostic Odyssey in Severe Neurodevelopmental Disorders: Toward Clinical Whole-Exome Sequencing as a First-Line Diagnostic Test. Clin. Genet. 2016, 89, 700–707. [Google Scholar] [CrossRef] [PubMed]
- Gorcenco, S.; Ilinca, A.; Almasoudi, W.; Kafantari, E.; Lindgren, A.G.; Puschmann, A. New Generation Genetic Testing Entering the Clinic. Park. Relat. Disord. 2020, 73, 72–84. [Google Scholar] [CrossRef] [PubMed]
- Schormair, B.; Kemlink, D.; Mollenhauer, B.; Fiala, O.; Machetanz, G.; Roth, J.; Berutti, R.; Strom, T.M.; Haslinger, B.; Trenkwalder, C.; et al. Diagnostic Exome Sequencing in Early-Onset Parkinson’s Disease Confirms VPS13C as a Rare Cause of Autosomal-Recessive Parkinson’s Disease. Clin. Genet. 2018, 93, 603–612. [Google Scholar] [CrossRef]
- Gustavsson, E.K.; Trinh, J.; McKenzie, M.; Bortnick, S.; Petersen, M.S.; Farrer, M.J.; Aasly, J.O. Genetic Identification in Early Onset Parkinsonism among Norwegian Patients. Mov. Disord. Clin. Pract. 2017, 4, 499–508. [Google Scholar] [CrossRef]
- Trinh, J.; Lohmann, K.; Baumann, H.; Balck, A.; Borsche, M.; Brüggemann, N.; Dure, L.; Dean, M.; Volkmann, J.; Tunc, S.; et al. Utility and Implications of Exome Sequencing in Early-Onset Parkinson’s Disease. Mov. Disord. 2019, 34, 133–137. [Google Scholar] [CrossRef]
- Emelyanov, A.K.; Usenko, T.S.; Tesson, C.; Senkevich, K.A.; Nikolaev, M.A.; Miliukhina, I.V.; Kopytova, A.E.; Timofeeva, A.A.; Yakimovsky, A.F.; Lesage, S.; et al. Mutation Analysis of Parkinson’s Disease Genes in a Russian Data Set. Neurobiol. Aging 2018, 71, 267.e7–267.e10. [Google Scholar] [CrossRef]
- Youn, J.; Lee, C.; Oh, E.; Park, J.; Kim, J.S.; Kim, H.-T.; Cho, J.W.; Park, W.-Y.; Jang, W.; Ki, C.-S. Genetic Variants of PARK Genes in Korean Patients with Early-Onset Parkinson’s Disease. Neurobiol. Aging 2019, 75, 224.e9–224.e15. [Google Scholar] [CrossRef]
- Kumar, K.R.; Davis, R.L.; Tchan, M.C.; Wali, G.M.; Mahant, N.; Ng, K.; Kotschet, K.; Siow, S.-F.; Gu, J.; Walls, Z.; et al. Whole Genome Sequencing for the Genetic Diagnosis of Heterogenous Dystonia Phenotypes. Park. Relat. Disord. 2019, 69, 111–118. [Google Scholar] [CrossRef]
- Ruano, L.; Melo, C.; Silva, M.C.; Coutinho, P. The Global Epidemiology of Hereditary Ataxia and Spastic Paraplegia: A Systematic Review of Prevalence Studies. Neuroepidemiology 2014, 42, 174–183. [Google Scholar] [CrossRef]
- Davies, K.; Szmulewicz, D.J.; Corben, L.A.; Delatycki, M.; Lockhart, P.J. RFC1-Related Disease: Molecular and Clinical Insights. Neurol. Genet. 2022, 8, e200016. [Google Scholar] [CrossRef] [PubMed]
- Pellerin, D.; Danzi, M.C.; Wilke, C.; Renaud, M.; Fazal, S.; Dicaire, M.-J.; Scriba, C.K.; Ashton, C.; Yanick, C.; Beijer, D.; et al. Deep Intronic FGF14 GAA Repeat Expansion in Late-Onset Cerebellar Ataxia. N. Engl. J. Med. 2023, 388, 128–141. [Google Scholar] [CrossRef] [PubMed]
- Tesson, C.; Koht, J.; Stevanin, G. Delving into the Complexity of Hereditary Spastic Paraplegias: How Unexpected Phenotypes and Inheritance Modes Are Revolutionizing Their Nosology. Hum. Genet. 2015, 134, 511–538. [Google Scholar] [CrossRef] [PubMed]
- McKnight, D.; Bristow, S.L.; Truty, R.M.; Morales, A.; Stetler, M.; Westbrook, M.J.; Robinson, K.; Riethmaier, D.; Borlot, F.; Kellogg, M.; et al. Multigene Panel Testing in a Large Cohort of Adults With Epilepsy: Diagnostic Yield and Clinically Actionable Genetic Findings. Neurol. Genet. 2022, 8, e650. [Google Scholar] [CrossRef]
- Borlot, F.; de Almeida, B.I.; Combe, S.L.; Andrade, D.M.; Filloux, F.M.; Myers, K.A. Clinical Utility of Multigene Panel Testing in Adults with Epilepsy and Intellectual Disability. Epilepsia 2019, 60, 1661–1669. [Google Scholar] [CrossRef]
- Johannesen, K.M.; Nikanorova, N.; Marjanovic, D.; Pavbro, A.; Larsen, L.H.G.; Rubboli, G.; Møller, R.S. Utility of Genetic Testing for Therapeutic Decision-Making in Adults with Epilepsy. Epilepsia 2020, 61, 1234–1239. [Google Scholar] [CrossRef]
- Von Brauchitsch, S.; Haslinger, D.; Lindlar, S.; Thiele, H.; Bernsen, N.; Zahnert, F.; Reif, P.S.; Balcik, Y.; Au, P.Y.B.; Josephson, C.B.; et al. The Phenotypic and Genotypic Spectrum of Epilepsy and Intellectual Disability in Adults: Implications for Genetic Testing. Epilepsia Open 2023, 8, 497–508. [Google Scholar] [CrossRef]
- Sheidley, B.R.; Malinowski, J.; Bergner, A.L.; Bier, L.; Gloss, D.S.; Mu, W.; Mulhern, M.M.; Partack, E.J.; Poduri, A. Genetic Testing for the Epilepsies: A Systematic Review. Epilepsia 2022, 63, 375–387. [Google Scholar] [CrossRef]
- Ricos, M.G.; Hodgson, B.L.; Pippucci, T.; Saidin, A.; Ong, Y.S.; Heron, S.E.; Licchetta, L.; Bisulli, F.; Bayly, M.A.; Hughes, J.; et al. Mutations in the Mammalian Target of Rapamycin Pathway Regulators NPRL2 and NPRL3 Cause Focal Epilepsy. Ann. Neurol. 2016, 79, 120–131. [Google Scholar] [CrossRef]
- Dibbens, L.M.; de Vries, B.; Donatello, S.; Heron, S.E.; Hodgson, B.L.; Chintawar, S.; Crompton, D.E.; Hughes, J.N.; Bellows, S.T.; Klein, K.M.; et al. Mutations in DEPDC5 Cause Familial Focal Epilepsy with Variable Foci. Nat. Genet. 2013, 45, 546–551. [Google Scholar] [CrossRef]
- Whittaker, R.G.; Devine, H.E.; Gorman, G.S.; Schaefer, A.M.; Horvath, R.; Ng, Y.; Nesbitt, V.; Lax, N.Z.; McFarland, R.; Cunningham, M.O.; et al. Epilepsy in Adults with Mitochondrial Disease: A Cohort Study. Ann. Neurol. 2015, 78, 949–957. [Google Scholar] [CrossRef] [PubMed]
- Klein, C.J. Charcot-Marie-Tooth Disease and Other Hereditary Neuropathies. Contin. Lifelong Learn. Neurol. 2020, 26, 1224–1256. [Google Scholar] [CrossRef] [PubMed]
- Barreto, L.C.L.S.; Oliveira, F.S.; Nunes, P.S.; de França Costa, I.M.P.; Garcez, C.A.; Goes, G.M.; Neves, E.L.A.; de Souza Siqueira Quintans, J.; de Souza Araújo, A.A. Epidemiologic Study of Charcot-Marie-Tooth Disease: A Systematic Review. Neuroepidemiology 2016, 46, 157–165. [Google Scholar] [CrossRef] [PubMed]
- Magy, L.; Mathis, S.; Masson, G.L.; Goizet, C.; Tazir, M.; Vallat, J.-M. Updating the Classification of Inherited Neuropathies: Results of an International Survey. Neurology 2018, 90, e870–e876. [Google Scholar] [CrossRef] [PubMed]
- DiVincenzo, C.; Elzinga, C.D.; Medeiros, A.C.; Karbassi, I.; Jones, J.R.; Evans, M.C.; Braastad, C.D.; Bishop, C.M.; Jaremko, M.; Wang, Z.; et al. The Allelic Spectrum of Charcot-Marie-Tooth Disease in over 17,000 Individuals with Neuropathy. Mol. Genet. Genomic Med. 2014, 2, 522–529. [Google Scholar] [CrossRef]
- Weterman, M.A.; van Ruissen, F.; de Wissel, M.; Bordewijk, L.; Samijn, J.P.; van der Pol, W.L.; Meggouh, F.; Baas, F. Copy Number Variation Upstream of PMP22 in Charcot–Marie–Tooth Disease. Eur. J. Hum. Genet. 2010, 18, 421–428. [Google Scholar] [CrossRef]
- Pantera, H.; Moran, J.J.; Hung, H.A.; Pak, E.; Dutra, A.; Svaren, J. Regulation of the Neuropathy-Associated Pmp22 Gene by a Distal Super-Enhancer. Hum. Mol. Genet. 2018, 27, 2830–2839. [Google Scholar] [CrossRef]
- Foley, C.; Schofield, I.; Eglon, G.; Bailey, G.; Chinnery, P.F.; Horvath, R. Charcot–Marie–Tooth Disease in Northern England. J. Neurol. Neurosurg. Psychiatry 2012, 83, 572–573. [Google Scholar] [CrossRef]
- Lefter, S.; Hardiman, O.; Ryan, A.M. A Population-Based Epidemiologic Study of Adult Neuromuscular Disease in the Republic of Ireland. Neurology 2017, 88, 304–313. [Google Scholar] [CrossRef]
- Park, J.E.; Noh, S.-J.; Oh, M.; Cho, D.-Y.; Kim, S.Y.; Ki, C.-S. Frequency of Hereditary Neuropathy with Liability to Pressure Palsies (HNPP) Due to 17p11.2 Deletion in a Korean Newborn Population. Orphanet J. Rare Dis. 2018, 13, 40. [Google Scholar] [CrossRef]
- Senderek, J.; Lassuthova, P.; Kabzińska, D.; Abreu, L.; Baets, J.; Beetz, C.; Braathen, G.J.; Brenner, D.; Dalton, J.; Dankwa, L.; et al. The Genetic Landscape of Axonal Neuropathies in the Middle-Aged and Elderly: Focus on MME. Neurology 2020, 95, e3163–e3179. [Google Scholar] [CrossRef]
- Felice, K.J.; Whitaker, C.H.; Khorasanizadeh, S. Diagnostic Yield of Advanced Genetic Testing in Patients with Hereditary Neuropathies: A Retrospective Single-Site Study. Muscle Nerve 2021, 64, 454–461. [Google Scholar] [CrossRef]
- Higuchi, Y.; Takashima, H. Clinical Genetics of Charcot–Marie–Tooth Disease. J. Hum. Genet. 2023, 68, 199–214. [Google Scholar] [CrossRef]
- Liao, Q.; Zhang, Y.; He, J.; Huang, K. Global Prevalence of Myotonic Dystrophy: An Updated Systematic Review and Meta-Analysis. Neuroepidemiology 2022, 56, 163–173. [Google Scholar] [CrossRef] [PubMed]
- Chiò, A.; Logroscino, G.; Traynor, B.J.; Collins, J.; Simeone, J.C.; Goldstein, L.A.; White, L.A. Global Epidemiology of Amyotrophic Lateral Sclerosis: A Systematic Review of the Published Literature. Neuroepidemiology 2013, 41, 118–130. [Google Scholar] [CrossRef] [PubMed]
- Van Cauwenberghe, C.; Van Broeckhoven, C.; Sleegers, K. The Genetic Landscape of Alzheimer Disease: Clinical Implications and Perspectives. Genet. Med. 2016, 18, 421–430. [Google Scholar] [CrossRef] [PubMed]
- Onyike, C.U.; Diehl-Schmid, J. The Epidemiology of Frontotemporal Dementia. Int. Rev. Psychiatry 2013, 25, 130–137. [Google Scholar] [CrossRef]
- Rohrer, J.D.; Guerreiro, R.; Vandrovcova, J.; Uphill, J.; Reiman, D.; Beck, J.; Isaacs, A.M.; Authier, A.; Ferrari, R.; Fox, N.C.; et al. The Heritability and Genetics of Frontotemporal Lobar Degeneration. Neurology 2009, 73, 1451–1456. [Google Scholar] [CrossRef] [PubMed]
- DeJesus-Hernandez, M.; Mackenzie, I.R.; Boeve, B.F.; Boxer, A.L.; Baker, M.; Rutherford, N.J.; Nicholson, A.M.; Finch, N.A.; Gilmer, H.F.; Adamson, J.; et al. Expanded GGGGCC Hexanucleotide Repeat in Non-Coding Region of C9ORF72 Causes Chromosome 9p-Linked Frontotemporal Dementia and Amyotrophic Lateral Sclerosis. Neuron 2011, 72, 245–256. [Google Scholar] [CrossRef] [PubMed]
- Ladogana, A.; Puopolo, M.; Croes, E.A.; Budka, H.; Jarius, C.; Collins, S.; Klug, G.M.; Sutcliffe, T.; Giulivi, A.; Alperovitch, A.; et al. Mortality from Creutzfeldt–Jakob Disease and Related Disorders in Europe, Australia, and Canada. Neurology 2005, 64, 1586–1591. [Google Scholar] [CrossRef]
- Ayrignac, X.; Carra-Dalliere, C.; Menjot de Champfleur, N.; Denier, C.; Aubourg, P.; Bellesme, C.; Castelnovo, G.; Pelletier, J.; Audoin, B.; Kaphan, E.; et al. Adult-Onset Genetic Leukoencephalopathies: A MRI Pattern-Based Approach in a Comprehensive Study of 154 Patients. Brain J. Neurol. 2015, 138, 284–292. [Google Scholar] [CrossRef]
- Bonkowsky, J.L.; Nelson, C.; Kingston, J.L.; Filloux, F.M.; Mundorff, M.B.; Srivastava, R. The Burden of Inherited Leukodystrophies in Children. Neurology 2010, 75, 718–725. [Google Scholar] [CrossRef] [PubMed]
- Heim, P.; Claussen, M.; Hoffmann, B.; Conzelmann, E.; Gärtner, J.; Harzer, K.; Hunneman, D.H.; Köhler, W.; Kurlemann, G.; Kohlschütter, A. Leukodystrophy Incidence in Germany. Am. J. Med. Genet. 1997, 71, 475–478. [Google Scholar] [CrossRef]
- Köhler, W.; Curiel, J.; Vanderver, A. Adulthood Leukodystrophies. Nat. Rev. Neurol. 2018, 14, 94–105. [Google Scholar] [CrossRef] [PubMed]
- Vanderver, A.; Simons, C.; Helman, G.; Crawford, J.; Wolf, N.I.; Bernard, G.; Pizzino, A.; Schmidt, J.L.; Takanohashi, A.; Miller, D.; et al. Whole Exome Sequencing in Patients with White Matter Abnormalities. Ann. Neurol. 2016, 79, 1031–1037. [Google Scholar] [CrossRef] [PubMed]
- Manini, A.; Pantoni, L. Genetic Causes of Cerebral Small Vessel Diseases: A Practical Guide for Neurologists. Neurology 2023, 100, 766–783. [Google Scholar] [CrossRef]
- Fernández-Eulate, G.; Carreau, C.; Benoist, J.-F.; Lamari, F.; Rucheton, B.; Shor, N.; Nadjar, Y. Diagnostic Approach in Adult-Onset Neurometabolic Diseases. J. Neurol. Neurosurg. Psychiatry 2022, 93, 413–421. [Google Scholar] [CrossRef]
- Ferreira, E.A.; Buijs, M.J.N.; Wijngaard, R.; Daams, J.G.; Datema, M.R.; Engelen, M.; van Karnebeek, C.D.M.; Oud, M.M.; Vaz, F.M.; Wamelink, M.M.C.; et al. Inherited Metabolic Disorders in Adults: Systematic Review on Patient Characteristics and Diagnostic Yield of Broad Sequencing Techniques (Exome and Genome Sequencing). Front. Neurol. 2023, 14, 1206106. [Google Scholar] [CrossRef]
- Gorman, G.S.; Schaefer, A.M.; Ng, Y.; Gomez, N.; Blakely, E.L.; Alston, C.L.; Feeney, C.; Horvath, R.; Yu-Wai-Man, P.; Chinnery, P.F.; et al. Prevalence of Nuclear and Mitochondrial DNA Mutations Related to Adult Mitochondrial Disease. Ann. Neurol. 2015, 77, 753–759. [Google Scholar] [CrossRef]
- Van Rootselaar, A.-F.; van Westrum, S.S.; Tijssen, M.a.J.; Velis, D.N. The Paroxysmal Dyskinesias. Pract. Neurol. 2009, 9, 102–109. [Google Scholar] [CrossRef]
- Jen, J.C.; Graves, T.D.; Hess, E.J.; Hanna, M.G.; Griggs, R.C.; Baloh, R.W. The CINCH investigators. Primary Episodic Ataxias: Diagnosis, Pathogenesis and Treatment. Brain 2007, 130, 2484–2493. [Google Scholar] [CrossRef] [PubMed]
- Thomsen, L.L.; Eriksen, M.K.; Romer, S.F.; Andersen, I.; Ostergaard, E.; Keiding, N.; Olesen, J.; Russell, M. An Epidemiological Survey of Hemiplegic Migraine. Cephalalgia 2002, 22, 361–375. [Google Scholar] [CrossRef]
- Shao, L.; Akkari, Y.; Cooley, L.D.; Miller, D.T.; Seifert, B.A.; Wolff, D.J.; Mikhail, F.M. ACMG Laboratory Quality Assurance Committee. Chromosomal Microarray Analysis, Including Constitutional and Neoplastic Disease Applications, 2021 Revision: A Technical Standard of the American College of Medical Genetics and Genomics (ACMG). Genet. Med. 2021, 23, 1818–1829. [Google Scholar] [CrossRef] [PubMed]
- Bean, L.; Funke, B.; Carlston, C.M.; Gannon, J.L.; Kantarci, S.; Krock, B.L.; Zhang, S.; Bayrak-Toydemir, P. Diagnostic Gene Sequencing Panels: From Design to Report—A Technical Standard of the American College of Medical Genetics and Genomics (ACMG). Genet. Med. 2020, 22, 453–461. [Google Scholar] [CrossRef] [PubMed]
- Oda, M.; Maruyama, H.; Komure, O.; Morino, H.; Terasawa, H.; Izumi, Y.; Imamura, T.; Yasuda, M.; Ichikawa, K.; Ogawa, M.; et al. Possible Reduced Penetrance of Expansion of 44 to 47 CAG/CAA Repeats in the TATA-Binding Protein Gene in Spinocerebellar Ataxia Type 17. Arch. Neurol. 2004, 61, 209–212. [Google Scholar] [CrossRef]
- Richard, P.; Trollet, C.; Gidaro, T.; Demay, L.; Brochier, G.; Malfatti, E.; Tom, F.M.; Fardeau, M.; Lafor, P.; Romero, N.; et al. PABPN1 (GCN)11 as a Dominant Allele in Oculopharyngeal Muscular Dystrophy—Consequences in Clinical Diagnosis and Genetic Counselling. J. Neuromuscul. Dis. 2015, 2, 175–180. [Google Scholar] [CrossRef]
- Willemsen, R.; Levenga, J.; Oostra, B.A. CGG Repeat in the FMR1 Gene: Size Matters. Clin. Genet. 2011, 80, 214–225. [Google Scholar] [CrossRef]
- Murphy, N.A.; Arthur, K.C.; Tienari, P.J.; Houlden, H.; Chiò, A.; Traynor, B.J. Age-Related Penetrance of the C9orf72 Repeat Expansion. Sci. Rep. 2017, 7, 2116. [Google Scholar] [CrossRef]
- Bettencourt, C.; Lima, M. Machado-Joseph Disease: From First Descriptions to New Perspectives. Orphanet J. Rare Dis. 2011, 6, 35. [Google Scholar] [CrossRef]
- Maat-Kievit, A.; Losekoot, M.; Berg, H.V.D.B.-V.D.; Ommen, G.-J.V.; Niermeijer, M.; Breuning, M.; Tibben, A. New Problems in Testing for Huntington’s Disease: The Issue of Intermediate and Reduced Penetrance Alleles. J. Med. Genet. 2001, 38, e12. [Google Scholar] [CrossRef]
- Dolzhenko, E.; Bennett, M.F.; Richmond, P.A.; Trost, B.; Chen, S.; Van Vugt, J.J.F.A.; Nguyen, C.; Narzisi, G.; Gainullin, V.G.; Gross, A.M.; et al. ExpansionHunter Denovo: A Computational Method for Locating Known and Novel Repeat Expansions in Short-Read Sequencing Data. Genome Biol. 2020, 21, 102. [Google Scholar] [CrossRef] [PubMed]
- Rafehi, H.; Green, C.; Bozaoglu, K.; Gillies, G.; Delatycki, M.B.; Lockhart, P.J.; Scheffer, I.E.; Bahlo, M. Unexpected Diagnosis of Myotonic Dystrophy Type 2 Repeat Expansion by Genome Sequencing. Eur. J. Hum. Genet. 2023, 31, 122–124. [Google Scholar] [CrossRef] [PubMed]
- Cortese, A.; Simone, R.; Sullivan, R.; Vandrovcova, J.; Tariq, H.; Yau, W.Y.; Humphrey, J.; Jaunmuktane, Z.; Sivakumar, P.; Polke, J.; et al. Biallelic Expansion of an Intronic Repeat in RFC1 Is a Common Cause of Late-Onset Ataxia. Nat. Genet. 2019, 51, 649–658. [Google Scholar] [CrossRef] [PubMed]
- Syriani, D.A.; Wong, D.; Andani, S.; Gusmao, C.M.D.; Mao, Y.; Sanyoura, M.; Glotzer, G.; Lockhart, P.J.; Hassin-Baer, S.; Khurana, V.; et al. Prevalence of RFC1-Mediated Spinocerebellar Ataxia in a North American Ataxia Cohort. Neurol. Genet. 2020, 6, e440. [Google Scholar] [CrossRef] [PubMed]
- Rafehi, H.; Read, J.; Szmulewicz, D.J.; Davies, K.C.; Snell, P.; Fearnley, L.G.; Scott, L.; Thomsen, M.; Gillies, G.; Pope, K.; et al. An Intronic GAA Repeat Expansion in FGF14 Causes the Autosomal-Dominant Adult-Onset Ataxia SCA50/ATX-FGF14. Am. J. Hum. Genet. 2023, 110, 105–119. [Google Scholar] [CrossRef] [PubMed]
- Currò, R.; Salvalaggio, A.; Tozza, S.; Gemelli, C.; Dominik, N.; Galassi Deforie, V.; Magrinelli, F.; Castellani, F.; Vegezzi, E.; Businaro, P.; et al. RFC1 Expansions Are a Common Cause of Idiopathic Sensory Neuropathy. Brain 2021, 144, 1542–1550. [Google Scholar] [CrossRef] [PubMed]
- Tagliapietra, M.; Cardellini, D.; Ferrarini, M.; Testi, S.; Ferrari, S.; Monaco, S.; Cavallaro, T.; Fabrizi, G.M. RFC1 AAGGG Repeat Expansion Masquerading as Chronic Idiopathic Axonal Polyneuropathy. J. Neurol. 2021, 268, 4280–4290. [Google Scholar] [CrossRef]
- Miller, D.T.; Lee, K.; Abul-Husn, N.S.; Amendola, L.M.; Brothers, K.; Chung, W.K.; Gollob, M.H.; Gordon, A.S.; Harrison, S.M.; Hershberger, R.E.; et al. ACMG SF v3.2 List for Reporting of Secondary Findings in Clinical Exome and Genome Sequencing: A Policy Statement of the American College of Medical Genetics and Genomics (ACMG). Genet. Med. 2023, 25, 100866. [Google Scholar] [CrossRef]
- Anguela, X.M.; High, K.A. Entering the Modern Era of Gene Therapy. Annu. Rev. Med. 2019, 70, 273–288. [Google Scholar] [CrossRef]
- Xie, Y.-X.; Lv, W.-Q.; Chen, Y.-K.; Hong, S.; Yao, X.-P.; Chen, W.-J.; Zhao, M. Advances in Gene Therapy for Neurogenetic Diseases: A Brief Review. J. Mol. Med. 2022, 100, 385–394. [Google Scholar] [CrossRef]
- Hayeems, R.Z.; Dimmock, D.; Bick, D.; Belmont, J.W.; Green, R.C.; Lanpher, B.; Jobanputra, V.; Mendoza, R.; Kulkarni, S.; Grove, M.E.; et al. Clinical Utility of Genomic Sequencing: A Measurement Toolkit. NPJ Genomic Med. 2020, 5, 56. [Google Scholar] [CrossRef] [PubMed]
- Chung, C.C.Y.; Hue, S.P.Y.; Ng, N.Y.T.; Doong, P.H.L.; Chu, A.T.W.; Chung, B.H.Y. Meta-Analysis of the Diagnostic and Clinical Utility of Exome and Genome Sequencing in Pediatric and Adult Patients with Rare Diseases across Diverse Populations. Genet. Med. 2023, 25, 100896. [Google Scholar] [CrossRef] [PubMed]
- Schwarze, K.; Buchanan, J.; Taylor, J.C.; Wordsworth, S. Are Whole-Exome and Whole-Genome Sequencing Approaches Cost-Effective? A Systematic Review of the Literature. Genet. Med. 2018, 20, 1122–1130. [Google Scholar] [CrossRef] [PubMed]
- Smith, H.S.; Swint, J.M.; Lalani, S.R.; Yamal, J.M.; de Oliveira Otto, M.C.; Castellanos, S.; Taylor, A.; Lee, B.H.; Russell, H.V. Clinical Application of Genome and Exome Sequencing as a Diagnostic Tool for Pediatric Patients: A Scoping Review of the Literature. Genet. Med. 2019, 21, 3–16. [Google Scholar] [CrossRef]
- Alix, T.; Chéry, C.; Josse, T.; Bronowicki, J.-P.; Feillet, F.; Guéant-Rodriguez, R.-M.; Namour, F.; Guéant, J.-L.; Oussalah, A. Predictors of the Utility of Clinical Exome Sequencing as a First-Tier Genetic Test in Patients with Mendelian Phenotypes: Results from a Referral Center Study on 603 Consecutive Cases. Hum. Genomics 2023, 17, 5. [Google Scholar] [CrossRef]
- Shickh, S.; Gutierrez Salazar, M.; Zakoor, K.-R.; Lázaro, C.; Gu, J.; Goltz, J.; Kleinman, D.; Noor, A.; Khalouei, S.; Mighton, C.; et al. Exome and Genome Sequencing in Adults with Undiagnosed Disease: A Prospective Cohort Study. J. Med. Genet. 2021, 58, 275–283. [Google Scholar] [CrossRef]
- Shickh, S.; Mighton, C.; Uleryk, E.; Pechlivanoglou, P.; Bombard, Y. The Clinical Utility of Exome and Genome Sequencing across Clinical Indications: A Systematic Review. Hum. Genet. 2021, 140, 1403–1416. [Google Scholar] [CrossRef]
- Manickam, K.; McClain, M.R.; Demmer, L.A.; Biswas, S.; Kearney, H.M.; Malinowski, J.; Massingham, L.J.; Miller, D.; Yu, T.W.; Hisama, F.M.; et al. Exome and Genome Sequencing for Pediatric Patients with Congenital Anomalies or Intellectual Disability: An Evidence-Based Clinical Guideline of the American College of Medical Genetics and Genomics (ACMG). Genet. Med. 2021, 23, 2029–2037. [Google Scholar] [CrossRef]
- Stark, Z.; Tan, T.Y.; Chong, B.; Brett, G.R.; Yap, P.; Walsh, M.; Yeung, A.; Peters, H.; Mordaunt, D.; Cowie, S.; et al. A Prospective Evaluation of Whole-Exome Sequencing as a First-Tier Molecular Test in Infants with Suspected Monogenic Disorders. Genet. Med. 2016, 18, 1090–1096. [Google Scholar] [CrossRef]
- Khromykh, A.; Solomon, B.D. The Benefits of Whole-Genome Sequencing Now and in the Future. Mol. Syndromol. 2015, 6, 108–109. [Google Scholar] [CrossRef]
- Lionel, A.C.; Costain, G.; Monfared, N.; Walker, S.; Reuter, M.S.; Hosseini, S.M.; Thiruvahindrapuram, B.; Merico, D.; Jobling, R.; Nalpathamkalam, T.; et al. Improved Diagnostic Yield Compared with Targeted Gene Sequencing Panels Suggests a Role for Whole-Genome Sequencing as a First-Tier Genetic Test. Genet. Med. 2018, 20, 435–443. [Google Scholar] [CrossRef] [PubMed]
- Stavropoulos, D.J.; Merico, D.; Jobling, R.; Bowdin, S.; Monfared, N.; Thiruvahindrapuram, B.; Nalpathamkalam, T.; Pellecchia, G.; Yuen, R.K.C.; Szego, M.J.; et al. Whole-Genome Sequencing Expands Diagnostic Utility and Improves Clinical Management in Paediatric Medicine. NPJ Genomic Med. 2016, 1, 15012. [Google Scholar] [CrossRef] [PubMed]
- Wu, B.; Kang, W.; Wang, Y.; Zhuang, D.; Chen, L.; Li, L.; Su, Y.; Pan, X.; Wei, Q.; Tang, Z.; et al. Application of Full-Spectrum Rapid Clinical Genome Sequencing Improves Diagnostic Rate and Clinical Outcomes in Critically Ill Infants in the China Neonatal Genomes Project*. Crit. Care Med. 2021, 49, 1674. [Google Scholar] [CrossRef] [PubMed]
- Dolzhenko, E.; van Vugt, J.J.F.A.; Shaw, R.J.; Bekritsky, M.A.; Van Blitterswijk, M.; Narzisi, G.; Ajay, S.S.; Rajan, V.; Lajoie, B.R.; Johnson, N.H.; et al. Detection of Long Repeat Expansions from PCR-Free Whole-Genome Sequence Data. Genome Res. 2017, 27, 1895–1903. [Google Scholar] [CrossRef]
- Chen, X.; Shen, F.; Gonzaludo, N.; Malhotra, A.; Rogert, C.; Taft, R.J.; Bentley, D.R.; Eberle, M.A. Cyrius: Accurate CYP2D6 Genotyping Using Whole-Genome Sequencing Data. Pharmacogenom. J. 2021, 21, 251–261. [Google Scholar] [CrossRef]
- Splinter, K.; Adams, D.R.; Bacino, C.A.; Bellen, H.J.; Bernstein, J.A.; Cheatle-Jarvela, A.M.; Eng, C.M.; Esteves, C.; Gahl, W.A.; Hamid, R.; et al. Effect of Genetic Diagnosis on Patients with Previously Undiagnosed Disease. N. Engl. J. Med. 2018, 379, 2131–2139. [Google Scholar] [CrossRef] [PubMed]
- Trosman, J.R.; Weldon, C.B.; Douglas, M.P.; Kurian, A.W.; Kelley, R.K.; Deverka, P.A.; Phillips, K.A. Payer Coverage for Hereditary Cancer Panels: Barriers, Opportunities, and Implications for the Precision Medicine Initiative. JNCCN J. Natl. Compr. Cancer Netw. 2017, 15, 219–228. [Google Scholar] [CrossRef]
- Fogleman, A.J.; Zahnd, W.E.; Lipka, A.E.; Malhi, R.S.; Ganai, S.; Delfino, K.R.; Jenkins, W.D. Knowledge, Attitudes, and Perceived Barriers towards Genetic Testing across Three Rural Illinois Communities. J. Community Genet. 2019, 10, 417–423. [Google Scholar] [CrossRef]
- Gidding, S.S.; Sheldon, A.; Neben, C.L.; Williams, H.E.; Law, S.; Zhou, A.Y.; Wilemon, K.; Ahmed, C.D.; Kindt, I. Patient Acceptance of Genetic Testing for Familial Hypercholesterolemia in the CASCADE FH Registry. J. Clin. Lipidol. 2020, 14, 218–223. [Google Scholar] [CrossRef]
- Grant, P.; Langlois, S.; Lynd, L.D.; Austin, J.C.; Elliott, A.M. Out-of-pocket and Private Pay in Clinical Genetic Testing: A Scoping Review. Clin. Genet. 2021, 100, 504–521. [Google Scholar] [CrossRef]
- Steffen, L.E.; Du, R.; Gammon, A.; Mandelblatt, J.S.; Kohlmann, W.K.; Lee, J.H.; Buys, S.S.; Stroup, A.M.; Campo, R.A.; Flores, K.G.; et al. Genetic Testing in a Population-Based Sample of Breast and Ovarian Cancer Survivors from the REACH Randomized Trial: Cost Barriers and Moderators of Counseling Mode. Cancer Epidemiol. Biomark. Prev. 2017, 26, 1772–1780. [Google Scholar] [CrossRef] [PubMed]
- Regier, D.; Friedman, J.; Makela, N.; Ryan, M.; Marra, C. Valuing the Benefit of Diagnostic Testing for Genetic Causes of Idiopathic Developmental Disability: Willingness to Pay from Families of Affected Children. Clin. Genet. 2009, 75, 514–521. [Google Scholar] [CrossRef] [PubMed]
- Kohler, J.N.; Turbitt, E.; Lewis, K.L.; Wilfond, B.S.; Jamal, L.; Peay, H.L.; Biesecker, L.G.; Biesecker, B.B. Defining Personal Utility in Genomics: A Delphi Study. Clin. Genet. 2017, 92, 290–297. [Google Scholar] [CrossRef]
- Kohler, J.N.; Turbitt, E.; Biesecker, B.B. Personal Utility in Genomic Testing: A Systematic Literature Review. Eur. J. Hum. Genet. 2017, 25, 662–668. [Google Scholar] [CrossRef] [PubMed]
- Foster, M.W.; Mulvihill, J.J.; Sharp, R.R. Evaluating the Utility of Personal Genomic Information. Genet. Med. 2009, 11, 570–574. [Google Scholar] [CrossRef] [PubMed]
- Grosse, S.D.; Wordsworth, S.; Payne, K. Economic Methods for Valuing the Outcomes of Genetic Testing: Beyond Cost-Effectiveness Analysis. Genet. Med. 2008, 10, 648–654. [Google Scholar] [CrossRef]
- Grosse, S.D.; McBride, C.M.; Evans, J.P.; Khoury, M.J. Personal Utility and Genomic Information: Look before You Leap. Genet. Med. 2009, 11, 575–576. [Google Scholar] [CrossRef]
- Smith, H.S.; Morain, S.R.; Robinson, J.O.; Canfield, I.; Malek, J.; Rubanovich, C.K.; Bloss, C.S.; Ackerman, S.L.; Biesecker, B.; Brothers, K.B.; et al. Perceived Utility of Genomic Sequencing: Qualitative Analysis and Synthesis of a Conceptual Model to Inform Patient-Centered Instrument Development. Patient Patient-Centered Outcomes Res. 2021, 15, 317–328. [Google Scholar] [CrossRef]
- Snyder, C.; Brundage, M.; Rivera, Y.M.; Wu, A.W. A PRO-Cision Medicine Methods Toolkit to Address the Challenges of Personalizing Cancer Care Using Patient-Reported Outcomes: Introduction to the Supplement. Med. Care 2019, 57, S1. [Google Scholar] [CrossRef]
- Meienberg, J.; Bruggmann, R.; Oexle, K.; Matyas, G. Clinical Sequencing: Is WGS the Better WES? Hum. Genet. 2016, 135, 359–362. [Google Scholar] [CrossRef]
- Abyzov, A.; Urban, A.E.; Snyder, M.; Gerstein, M. CNVnator: An Approach to Discover, Genotype, and Characterize Typical and Atypical CNVs from Family and Population Genome Sequencing. Genome Res. 2011, 21, 974–984. [Google Scholar] [CrossRef] [PubMed]
- Colin, E.; Duffourd, Y.; Chevarin, M.; Tisserant, E.; Verdez, S.; Paccaud, J.; Bruel, A.-L.; Tran Mau-Them, F.; Denommé-Pichon, A.-S.; Thevenon, J.; et al. Stepwise Use of Genomics and Transcriptomics Technologies Increases Diagnostic Yield in Mendelian Disorders. Front. Cell Dev. Biol. 2023, 11, 1021920. [Google Scholar] [CrossRef] [PubMed]
- Cummings, B.B.; Marshall, J.L.; Tukiainen, T.; Lek, M.; Donkervoort, S.; Foley, A.R.; Bolduc, V.; Waddell, L.B.; Sandaradura, S.A.; O’Grady, G.L.; et al. Improving Genetic Diagnosis in Mendelian Disease with Transcriptome Sequencing. Sci. Transl. Med. 2017, 9, eaal5209. [Google Scholar] [CrossRef]
- Gonorazky, H.D.; Naumenko, S.; Ramani, A.K.; Nelakuditi, V.; Mashouri, P.; Wang, P.; Kao, D.; Ohri, K.; Viththiyapaskaran, S.; Tarnopolsky, M.A.; et al. Expanding the Boundaries of RNA Sequencing as a Diagnostic Tool for Rare Mendelian Disease. Am. J. Hum. Genet. 2019, 104, 1007. [Google Scholar] [CrossRef]
- Kadlubowska, M.K.; Schrauwen, I. Methods to Improve Molecular Diagnosis in Genomic Cold Cases in Pediatric Neurology. Genes 2022, 13, 333. [Google Scholar] [CrossRef] [PubMed]
- Lappalainen, T.; Scott, A.J.; Brandt, M.; Hall, I.M. Genomic Analysis in the Age of Human Genome Sequencing. Cell 2019, 177, 70–84. [Google Scholar] [CrossRef]
- Lee, H.; Huang, A.Y.; Wang, L.-K.; Yoon, A.J.; Renteria, G.; Eskin, A.; Signer, R.H.; Dorrani, N.; Nieves-Rodriguez, S.; Wan, J.; et al. Diagnostic Utility of Transcriptome Sequencing for Rare Mendelian Diseases. Genet. Med. 2020, 22, 490–499. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Li, J. A Deep Learning Framework Identifies Pathogenic Noncoding Somatic Mutations from Personal Prostate Cancer Genomes. Cancer Res. 2021, 80, 4644–4654. [Google Scholar] [CrossRef]
- Freeman, W.D.; Vatz, K.A.; Griggs, R.C.; Pedley, T. The Workforce Task Force Report: Clinical Implications for Neurology. Neurology 2013, 81, 479–486. [Google Scholar] [CrossRef]
- Lin, C.C.; Hill, C.E.; Kerber, K.A.; Burke, J.F.; Skolarus, L.E.; Esper, G.J.; de Havenon, A.; De Lott, L.B.; Callaghan, B.C. Patient Travel Distance to Neurologist Visits. Neurology 2023, 101, e1807–e1820. [Google Scholar] [CrossRef]
- Maiese, D.R.; Keehn, A.; Lyon, M.; Flannery, D.; Watson, M. Working Groups of the National Coordinating Center for Seven Regional Genetics Service Collaboratives. Current Conditions in Medical Genetics Practice. Genet. Med. 2019, 21, 1874–1877. [Google Scholar] [CrossRef] [PubMed]
- Jenkins, B.D.; Fischer, C.G.; Polito, C.A.; Maiese, D.R.; Keehn, A.S.; Lyon, M.; Edick, M.J.; Taylor, M.R.G.; Andersson, H.C.; Bodurtha, J.N.; et al. The 2019 US Medical Genetics Workforce: A Focus on Clinical Genetics. Genet. Med. 2021, 23, 1458–1464. [Google Scholar] [CrossRef] [PubMed]
- French, E.L.; Kader, L.; Young, E.E.; Fontes, J.D. Physician Perception of the Importance of Medical Genetics and Genomics in Medical Education and Clinical Practice. Med. Educ. Online 2023, 28, 2143920. [Google Scholar] [CrossRef] [PubMed]
- Hamilton, J.G.; Abdiwahab, E.; Edwards, H.M.; Fang, M.-L.; Jdayani, A.; Breslau, E.S. Primary Care Providers’ Cancer Genetic Testing-Related Knowledge, Attitudes, and Communication Behaviors: A Systematic Review and Research Agenda. J. Gen. Intern. Med. 2017, 32, 315–324. [Google Scholar] [CrossRef]
- Helman, G.; Bonkowsky, J.L.; Vanderver, A. Neurologist Comfort in the Use of Next-Generation Sequencing Diagnostics: Current State and Future Prospects. JAMA Neurol. 2016, 73, 621–622. [Google Scholar] [CrossRef]
- Hull, L.E.; Lynch, K.G.; Oslin, D.W. VA Primary Care and Mental Health Providers’ Comfort with Genetic Testing: Survey Results from the PRIME Care Study. J. Gen. Intern. Med. 2019, 34, 799–801. [Google Scholar] [CrossRef]
- Macklin, S.K.; Jackson, J.L.; Atwal, P.S.; Hines, S.L. Physician Interpretation of Variants of Uncertain Significance. Fam. Cancer 2019, 18, 121–126. [Google Scholar] [CrossRef]
| Clinical Presentation | First-Line Genetic Testing Strategy | Distinctive Clinical Features | Other Considerations |
|---|---|---|---|
| Parkinsonism | Gene panel | Early-onset < 40 years old | |
| Dystonia | Gene panel | Early age of onset, combined dystonia phenotype | |
| Chorea | Repeat Expansion Testing Panel +/− Sequencing Panel | Rule out Huntington disease and mimics | |
| Ataxia | Repeat Expansion Testing Panel +/− Sequencing Panel | Repeat expansion should be prioritized if pure SCA (most commonly SCA3, FXTAS); if complex, consider gene panel first | |
| Spastic Paraparesis | Gene panel | ||
| Epilepsy | Gene panel | Seizure onset in infancy, comorbid intellectual disability or developmental delay, pharmacoresistent epilepsy | |
| Neuropathy | Gene panel | Distal motor and/or sensory neuropathy | Consider sequence evaluation of PMP22 upstream regulatory elements |
| CANVAS | Single gene testing | Persistent cough | Novel repeat motifs identified, limited availability of testing |
| Muscle disorder | Gene panel | If findings strongly suggest a specific muscular dystrophy (e.g., DM, FSHD, OPMD) consider repeat expansion testing | |
| Motor Neuron Disease | Repeat Expansion Testing Panel +/− Sequencing Panel | Association with FTD | Repeat expansion should be prioritized, as most common diagnoses are ALS, SBMA, Friedreich ataxia |
| Dementia | Gene panel | Early-onset < 65 years old | If FTD, then prioritize repeat expansion (C9orf72) |
| Leukodystrophy | Gene panel | ||
| Cerebral ischemia | Gene panel | Recurrent stroke, early-onset < 50 years old, lack of typical vascular risk factors, symmetric imaging findings | |
| Episodic Neurological Syndrome | Gene panel |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Waung, M.W.; Ma, F.; Wheeler, A.G.; Zai, C.C.; So, J. The Diagnostic Landscape of Adult Neurogenetic Disorders. Biology 2023, 12, 1459. https://doi.org/10.3390/biology12121459
Waung MW, Ma F, Wheeler AG, Zai CC, So J. The Diagnostic Landscape of Adult Neurogenetic Disorders. Biology. 2023; 12(12):1459. https://doi.org/10.3390/biology12121459
Chicago/Turabian StyleWaung, Maggie W., Fion Ma, Allison G. Wheeler, Clement C. Zai, and Joyce So. 2023. "The Diagnostic Landscape of Adult Neurogenetic Disorders" Biology 12, no. 12: 1459. https://doi.org/10.3390/biology12121459
APA StyleWaung, M. W., Ma, F., Wheeler, A. G., Zai, C. C., & So, J. (2023). The Diagnostic Landscape of Adult Neurogenetic Disorders. Biology, 12(12), 1459. https://doi.org/10.3390/biology12121459






