Comparison of Traditional and Next-Generation Approaches for Uncovering Phytoplasma Diversity, with Discovery of New Groups, Subgroups and Potential Vectors
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
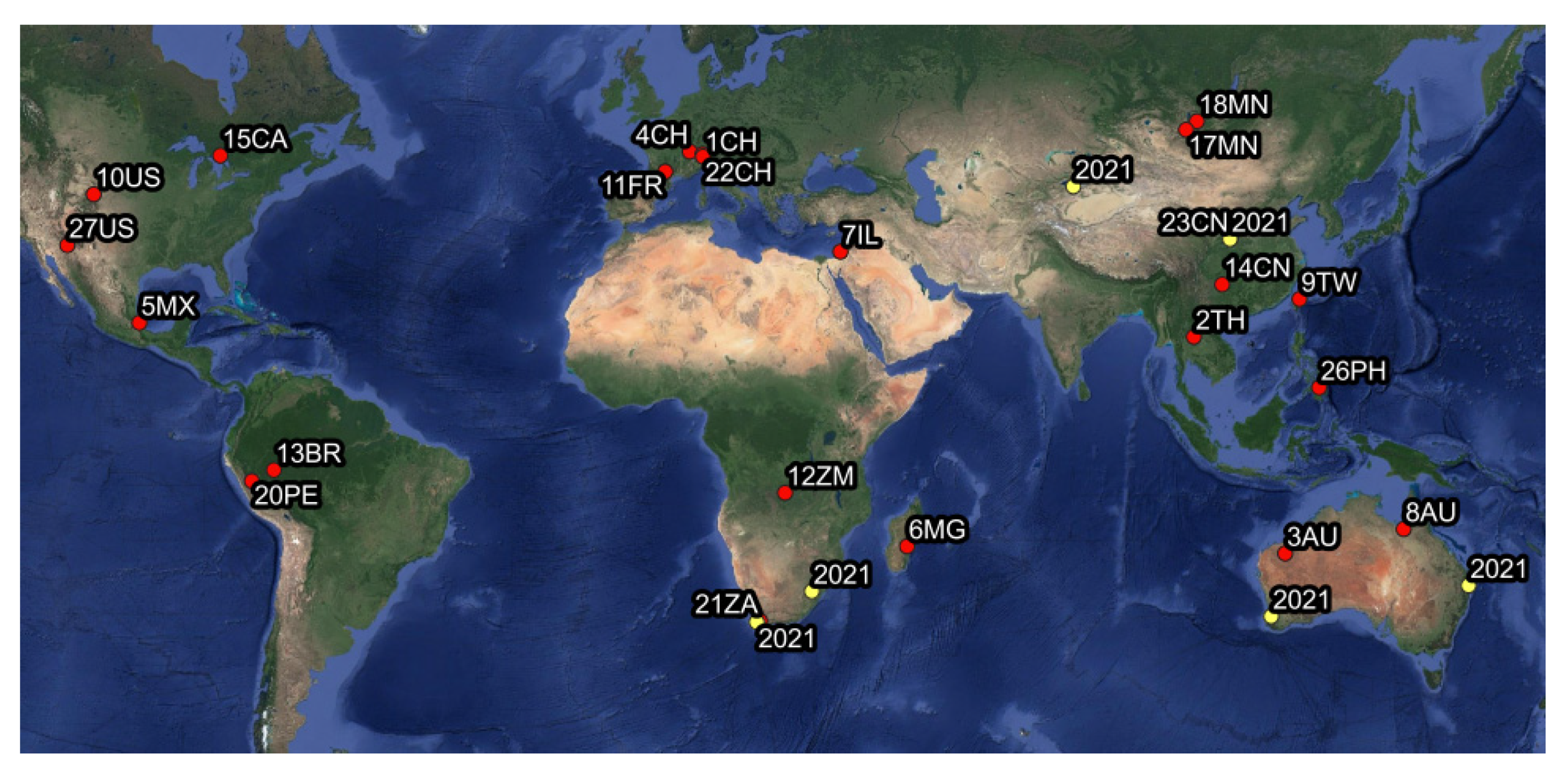
2.1. Leafhopper Collection
2.2. Phytoplasma Detection and Identification
2.2.1. qPCR
2.2.2. Anchored Hybrid Enrichment (AHE)
2.2.3. Nested PCR and Sanger DNA Sequencing
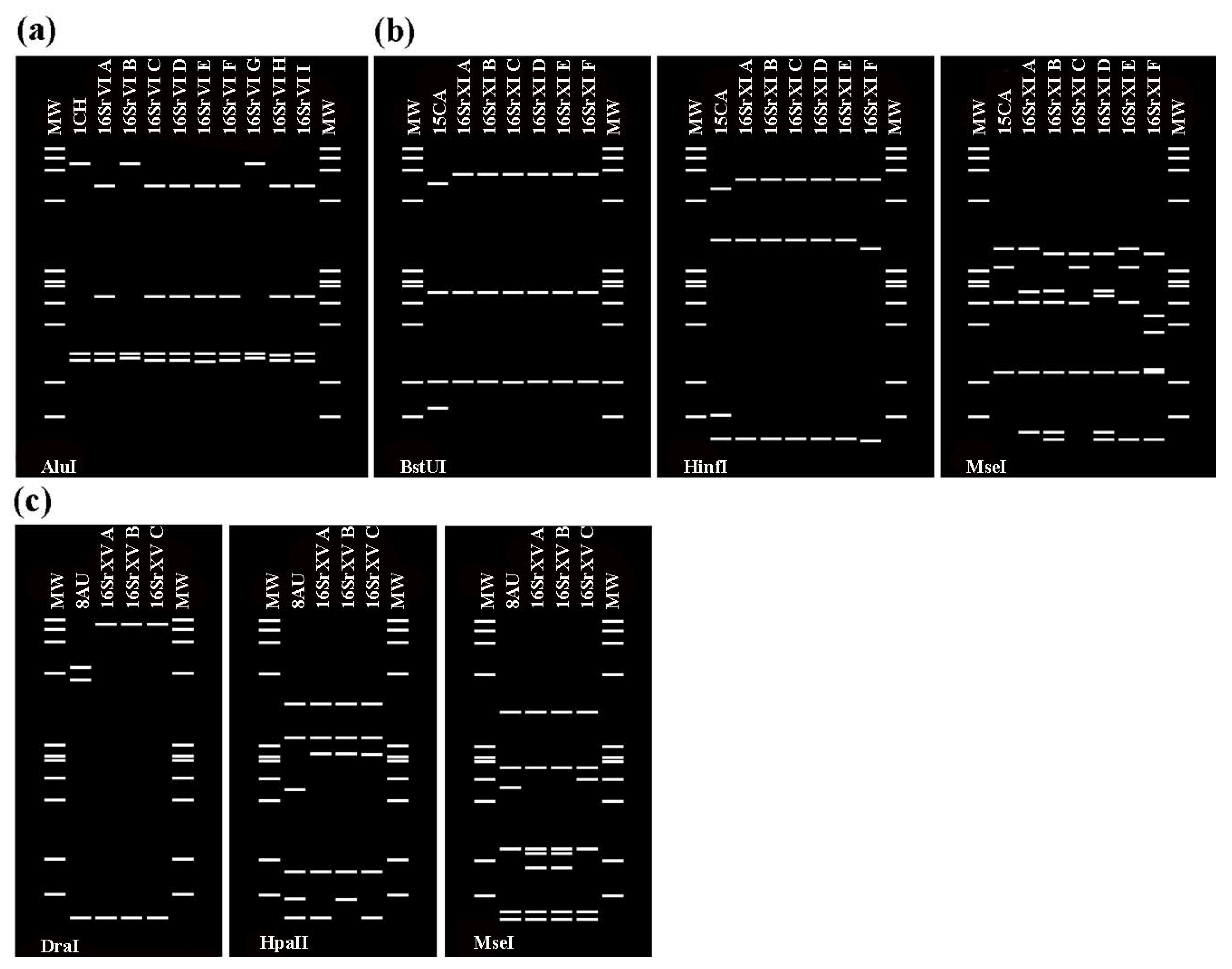
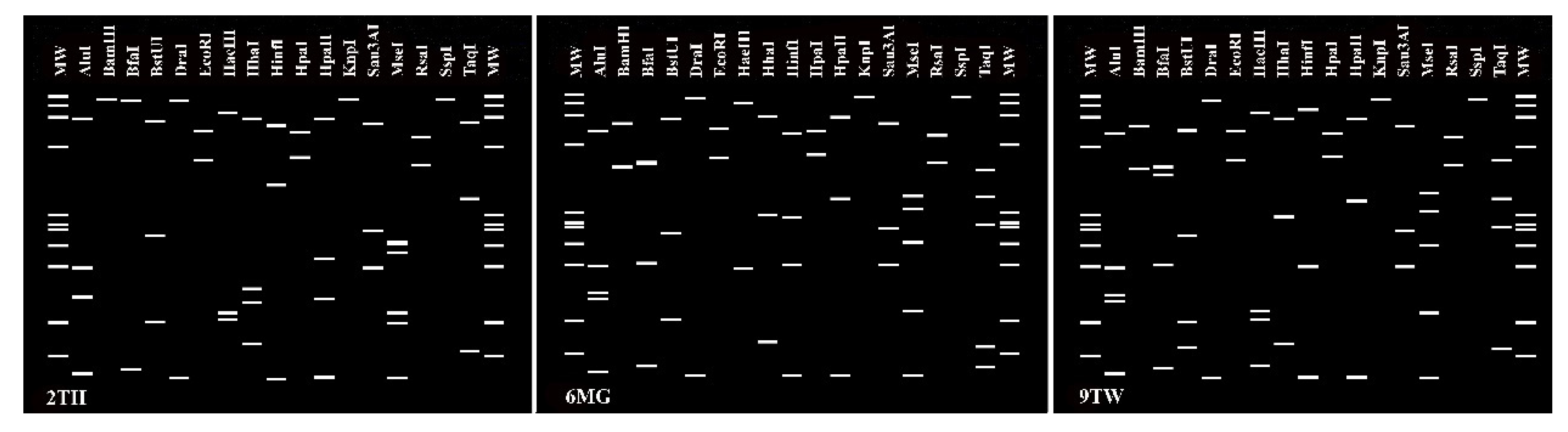
2.2.4. Virtual RFLP Analysis of 16S rRNA Genes
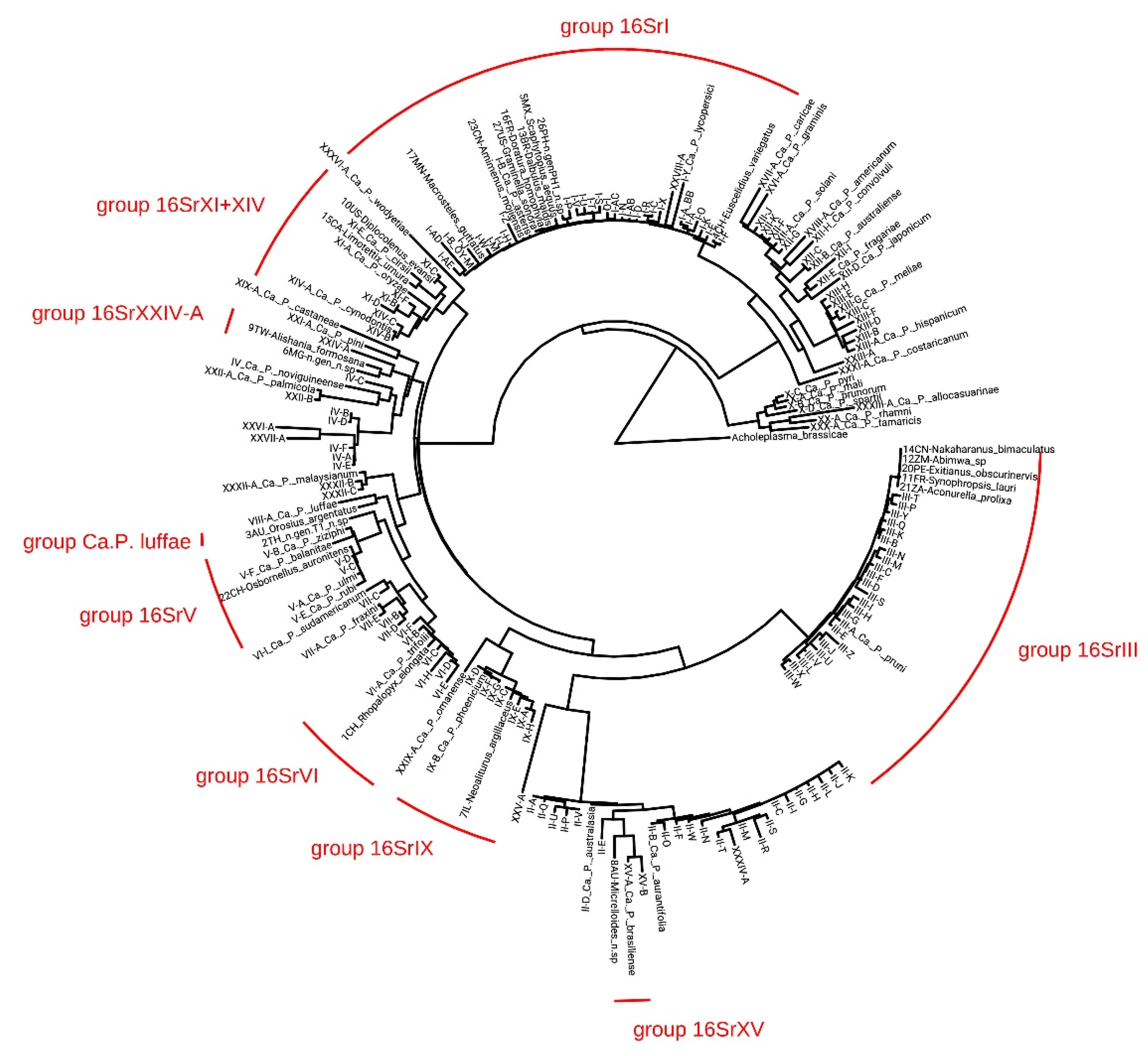
2.3. Phylogenetic Analysis
3. Results
3.1. Comparison of the Methods for the Detection and Sequencing of Phytoplasmas
3.1.1. qPCR Results
3.1.2. AHE Results
3.1.3. Nested PCR and Sanger Sequencing Results
3.1.4. Comparison among Methods of Detection and Sequencing
3.2. Characterization and Identification of Phytoplasma Strains
3.3. Insect-Phytoplasma Associations
4. Discussion
4.1. AHE Next Generation Sequencing as a Reliable Method for Future Phytoplasma Studies
4.2. Uncovering New Phytoplasma Strains and Host Associations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cao, Y.; Trivellone, V.; Dietrich, C.H. A Timetree for Phytoplasmas (Mollicutes) with New Insights on Patterns of Evolution and Diversification. Mol. Phylogenet. Evol. 2020, 149, 106826. [Google Scholar] [CrossRef]
- Moura, M.R.; Jetz, W. Shortfalls and Opportunities in Terrestrial Vertebrate Species Discovery. Nat. Ecol. Evol. 2021, 5, 631–639. [Google Scholar] [CrossRef]
- Trivellone, V.; Wei, W.; Filippin, L.; Dietrich, C.H. Screening Potential Insect Vectors in a Museum Biorepository Reveals Undiscovered Diversity of Plant Pathogens in Natural Areas. Ecol. Evol. 2021, 11, 6493–6503. [Google Scholar] [CrossRef]
- Wei, W.; Trivellone, V.; Dietrich, C.H.; Zhao, Y.; Bottner-Parker, K.D.; Ivanauskas, A. Identification of Phytoplasmas Representing Multiple New Genetic Lineages from Phloem-Feeding Leafhoppers Highlights the Diversity of Phytoplasmas and Their Potential Vectors. Pathogens 2021, 10, 352. [Google Scholar] [CrossRef]
- Trivellone, V.; Dietrich, C.H. Evolutionary Diversification in Insect Vector-Phytoplasma-Plant Associations. Ann. Entomol. Soc. Am. 2021, 114, 137–150. [Google Scholar] [CrossRef]
- Donkersley, P.; Silva, F.W.S.; Alves, M.S.; Carvalho, C.M.; Al-Sadi, A.M.; Elliot, S.L. Asymptomatic Phytoplasma Reveal a Novel and Troublesome Infection. Plant Pathol. Manag. Plant Dis. 2019, 1–20. [Google Scholar] [CrossRef]
- Zwolińska, A.; Krawczyk, K.; Borodynko-Filas, N.; Pospieszny, H. Non-Crop Sources of Rapeseed Phyllody Phytoplasma (‘Candidatus Phytoplasma Asteris’: 16SrI-B and 16SrI-(B/L)L), and Closely Related Strains. Crop Prot. 2019, 119, 59–68. [Google Scholar] [CrossRef]
- Trivellone, V. An Online Global Database of Hemiptera-Phytoplasma-Plant Biological Interactions. Biodivers. Data J. 2019, 7, e32910. [Google Scholar] [CrossRef]
- Lee, I.M.; Gundersen-Rindal, D.E.; Davis, R.E.; Bartoszyk, I.M. Revised Classification Scheme of Phytoplasmas Based on RFLP Analyses of 16S RRNA and Ribosomal Protein Gene Sequences. Int. J. Syst. Evol. Microbiol. 1998, 48, 1153–1169. [Google Scholar] [CrossRef]
- Lemmon, A.R.; Emme, S.A.; Lemmon, E.M. Anchored Hybrid Enrichment for Massively High-Throughput Phylogenomics. Syst. Biol. 2012, 61, 727–744. [Google Scholar] [CrossRef]
- Dietrich, C.H.; Allen, J.M.; Lemmon, A.R.; Lemmon, E.M.; Takiya, D.M.; Evangelista, O.; Walden, K.K.O.; Grady, P.G.S.; Johnson, K.P. Anchored Hybrid Enrichment-Based Phylogenomics of Leafhoppers and Treehoppers (Hemiptera: Cicadomorpha: Membracoidea). Insect Syst. Divers. 2017, 1, 57–72. [Google Scholar] [CrossRef]
- Cao, Y.; Dietrich, C.H.; Zahniser, J.N.; Dmitriev, D.A. Dense Sampling of Taxa and Characters Improves Phylogenetic Resolution among Deltocephaline Leafhoppers (Hemiptera: Cicadellidae: Deltocephalinae). Syst. Entomol. 2022, 47, 430–444. [Google Scholar] [CrossRef]
- Lee, I.-M.; Davis, R.E.; Gundersen-Rindal, D.E. Phytoplasma: Phytopathogenic Mollicutes. Annu. Rev. Microbiol. 2000, 54, 221–255. [Google Scholar] [CrossRef]
- Bertaccini, A.; Arocha-Rosete, Y.; Contaldo, N.; Duduk, B.; Fiore, N.; Montano, H.G.; Kube, M.; Kuo, C.-H.; Martini, M.; Oshima, K.; et al. Revision of the “Candidatus Phytoplasma” Species Description Guidelines. Int. J. Syst. Evol. Microbiol. 2022, 72, 005353. [Google Scholar] [CrossRef]
- Cao, Y.; Dietrich, C.H. Phylogenomics of Flavobacterial Insect Nutritional Endosymbionts with Implications for Auchenorrhyncha Phylogeny. Cladistics Int. J. Willi Hennig Soc. 2022, 38, 38–58. [Google Scholar] [CrossRef]
- Cao, Y.; Dietrich, C.H. Identification of Potential Host Plants of Sap-Sucking Insects (Hemiptera: Cicadellidae) Using Anchored Hybrid By-Catch Data. Insects 2021, 12, 964. [Google Scholar] [CrossRef]
- Angelini, E.; Luca Bianchi, G.; Filippin, L.; Morassutti, C.; Borgo, M. A New TaqMan Method for the Identification of Phytoplasmas Associated with Grapevine Yellows by Real-Time PCR Assay. J. Microbiol. Methods 2007, 68, 613–622. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 24 September 2021).
- Simpson, J.T.; Wong, K.; Jackman, S.D.; Schein, J.E.; Jones, S.J.M.; Birol, I. ABySS: A Parallel Assembler for Short Read Sequence Data. Genome Res. 2009, 19, 1117–1123. [Google Scholar] [CrossRef]
- Bai, X.; Zhang, J.; Ewing, A.; Miller, S.A.; Jancso Radek, A.; Shevchenko, D.V.; Tsukerman, K.; Walunas, T.; Lapidus, A.; Campbell, J.W.; et al. Living with Genome Instability: The Adaptation of Phytoplasmas to Diverse Environments of Their Insect and Plant Hosts. J. Bacteriol. 2006, 188, 3682–3696. [Google Scholar] [CrossRef]
- Johnson, M.G.; Gardner, E.M.; Liu, Y.; Medina, R.; Goffinet, B.; Shaw, A.J.; Zerega, N.J.C.; Wickett, N.J. HybPiper: Extracting Coding Sequence and Introns for Phylogenetics from High-Throughput Sequencing Reads Using Target Enrichment1. Appl. Plant Sci. 2016, 4, 1600016. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and Accurate Short Read Alignment with Burrows–Wheeler Transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Thorvaldsdóttir, H.; Robinson, J.T.; Mesirov, J.P. Integrative Genomics Viewer (IGV): High-Performance Genomics Data Visualization and Exploration. Brief. Bioinform. 2013, 14, 178–192. [Google Scholar] [CrossRef]
- Corpet, F. Multiple Sequence Alignment with Hierarchical Clustering. Nucleic Acids Res. 1988, 16, 10881–10890. [Google Scholar] [CrossRef]
- Bonfield, J.K.; Smith, K.F.; Staden, R. A New DNA Sequence Assembly Program. Nucleic Acids Res. 1995, 23, 4992–4999. [Google Scholar] [CrossRef]
- Zhao, Y.; Wei, W.; Lee, I.-M.; Shao, J.; Suo, X.; Davis, R.E. Construction of an Interactive Online Phytoplasma Classification Tool, IPhyClassifier, and Its Application in Analysis of the Peach X-Disease Phytoplasma Group (16SrIII). Int. J. Syst. Evol. Microbiol. 2009, 59, 2582–2593. [Google Scholar] [CrossRef]
- IRPCM Phytoplasma/Spiroplasma Working Team—Phytoplasma taxonomy group, ‘Candidatus Phytoplasma’, a Taxon for the Wall-Less, Non-Helical Prokaryotes That Colonize Plant Phloem and Insects. Int. J. Syst. Evol. Microbiol. 2004, 54, 1243–1255. [CrossRef]
- Kube, M.; Siewert, C.; Migdoll, A.M.; Duduk, B.; Holz, S.; Rabus, R.; Seemüller, E.; Mitrovic, J.; Müller, I.; Büttner, C.; et al. Analysis of the Complete Genomes of Acholeplasma brassicae, A. palmae and A. laidlawii and Their Comparison to the Obligate Parasites from ‘Candidatus Phytoplasma’. J. Mol. Microbiol. Biotechnol. 2014, 24, 19–36. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Nguyen, L.-T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Zhao, Y.; Davis, R.E. Criteria for Phytoplasma 16Sr Group/Subgroup Delineation and the Need of a Platform for Proper Registration of New Groups and Subgroups. Int. J. Syst. Evol. Microbiol. 2016, 66, 2121–2123. [Google Scholar] [CrossRef]
- de Oliveira, E.; de Sousa, S.M.; Landau, E.C. Transmission of Maize Bushy Stunt Phytoplasma by Dalbulus Maidis Leafhopper. Bull. Insectology 2011, 64, S153–S154. [Google Scholar]
- D’Amelio, R.; Berta, G.; Gamalero, E.; Massa, N.; Avidano, L.; Cantamessa, S.; D’Agostino, G.; Bosco, D.; Marzachì, C. Increased Plant Tolerance against Chrysanthemum Yellows Phytoplasma (‘Candidatus Phytoplasma Asteris’) Following Double Inoculation with Glomus Mosseae BEG12 and Pseudomonas Putida S1Pf1Rif. Plant Pathol. 2011, 60, 1014–1022. [Google Scholar] [CrossRef]
- Nault, L.R. Maize Bushy Stunt and Corn Stunt: A Comaprision of Disease Symptoms, Pathogen Host Range, and Vectors. Am. Phytopathol. Soc. 1980, 70, 659–662. [Google Scholar] [CrossRef]
- Orsagova, H.; Brezikova, M.; Schlesingerova, G. Presence of Phytoplasmas in Hemipterans in Czech Vineyards. Bull. Insectology 2011, 64, S119–S120. [Google Scholar]
- Trivellone, V.; Mitrović, M.; Dietrich, C.H.; Toševski, I. Osbornellus Auronitens (Hemiptera: Cicadellidae: Deltocephalinae), an Introduced Species New for the Palaearctic Region. Can. Entomol. 2017, 149, 551–559. [Google Scholar] [CrossRef]
- Trivellone, V.; Filippin, L.; Narduzzi-Wicht, B.; Angelini, E. A Regional-Scale Survey to Define the Known and Potential Vectors of Grapevine Yellow Phytoplasmas in Vineyards South of Swiss Alps. Eur. J. Plant Pathol. 2016, 145, 915–927. [Google Scholar] [CrossRef]
- Casati, P.; Jermini, M.; Quaglino, F.; Corbani, G.; Schaerer, S.; Passera, A.; Bianco, P.A.; Rigamonti, I.E. New Insights on Flavescence Dorée Phytoplasma Ecology in the Vineyard Agro-Ecosystem in Southern Switzerland. Ann. Appl. Biol. 2017, 171, 37–51. [Google Scholar] [CrossRef]
- Belgeri, E.; Rizzoli, A.; Jermini, M.; Angelini, E.; Filippin, L.; Rigamonti, I.E. First Report of Flavescence Dorée Phytoplasma Identification and Characterization in Three Species of Leafhoppers. J. Plant Pathol. 2022, 104, 375–379. [Google Scholar] [CrossRef]
- Pilkington, L.J.; Gurr, G.M.; Fletcher, M.J.; Nikandrow, A.; Elliott, E. Vector Status of Three Leafhopper Species for Australian Lucerne Yellows Phytoplasma. Aust. J. Entomol. 2004, 43, 366–373. [Google Scholar] [CrossRef]
- Rodrigues Jardim, B.; Kinoti, W.M.; Tran-Nguyen, L.T.T.; Gambley, C.; Rodoni, B.; Constable, F.E. ‘Candidatus Phytoplasma Stylosanthis’, a Novel Taxon with a Diverse Host Range in Australia, Characterised Using Multilocus Sequence Analysis of 16S RRNA, SecA, Tuf, and Rp Genes. Int. J. Syst. Evol. Microbiol. 2021, 71, ijsem004589. [Google Scholar] [CrossRef]
- Trivellone, V.; Achtziger, R.; Frund, E.; Funke, L.; Hollier, J.; Holzinger, W.; Huber, E.; Kunz, G.; Muhlethaler, R.; Nickel, H.; et al. Auchenorrinchi Ed Eterotteri (Hemiptera) Di Ecosistemi Naturali Di Rilevante Valore in Cantone Ticino (Svizzera). Boll. Della Soc. Ticinese Sci. Nat. 2021, 109, 183–197. [Google Scholar]
- Weintraub, P.G.; Orenstein, S. Potential Leafhopper Vectors of Phytoplasma in Carrots. Int. J. Trop. Insect Sci. 2004, 24, 228–235. [Google Scholar] [CrossRef]
- Weintraub, P.G.; Pivonia, S.; Rosner, A.; Gera, A. A New Disease in Limonium Latifolium Hybrids. II. Investigating Insect Vectors. HortScience 2004, 39, 1060–1061. [Google Scholar] [CrossRef]
- Martínez-Porchas, M.; Villalpando-Canchola, E.; Vargas-Albores, F. Significant Loss of Sensitivity and Specificity in the Taxonomic Classification Occurs When Short 16S RRNA Gene Sequences Are Used. Heliyon 2016, 2, e00170. [Google Scholar] [CrossRef]
- Chakravorty, S.; Helb, D.; Burday, M.; Connell, N.; Alland, D. A Detailed Analysis of 16S Ribosomal RNA Gene Segments for the Diagnosis of Pathogenic Bacteria. J. Microbiol. Methods 2007, 69, 330–339. [Google Scholar] [CrossRef]
- Janda, J.M.; Abbott, S.L. 16S RRNA Gene Sequencing for Bacterial Identification in the Diagnostic Laboratory: Pluses, Perils, and Pitfalls. J. Clin. Microbiol. 2007, 45, 2761–2764. [Google Scholar] [CrossRef]
- Martini, M.; Lee, I.-M.; Bottner, K.D.; Zhao, Y.; Botti, S.; Bertaccini, A.; Harrison, N.A.; Carraro, L.; Marcone, C.; Khan, A.J.; et al. Ribosomal Protein Gene-Based Phylogeny for Finer Differentiation and Classification of Phytoplasmas. Int. J. Syst. Evol. Microbiol. 2007, 57, 2037–2051. [Google Scholar] [CrossRef]
- Lee, I.M.; Bottner-Parker, K.D.; Zhao, Y.; Davis, R.E.; Harrison, N.A. Phylogenetic Analysis and Delineation of Phytoplasmas Based on SecY Gene Sequences. Int. J. Syst. Evol. Microbiol. 2010, 60, 2887–2897. [Google Scholar] [CrossRef]
- Trivellone, V.; Angelini, E.; Dmitriev, D.A.; Dietrich, C.H. Preliminary results on phylogenetic relatedness of potential and known auchenorrhyncha vectors of phytoplasmas. In Proceedings of the 15th International Auchenorrhyncha Congress and 10th International Workshop on Leafhoppers and Planthoppers of Economic Importance, Mendes, Brazil, 9 July 2017. [Google Scholar]
- Agosta, S.J. On Ecological Fitting, Plant-Insect Associations, Herbivore Host Shifts, and Host Plant Selection. Oikos 2006, 114, 556–565. [Google Scholar] [CrossRef]
- Davis, R.E.; Zhao, Y.; Dally, E.L.; Lee, I.-M.; Jomantiene, R.; Douglas, S.M. “Candidatus Phytoplasma Pruni”, a Novel Taxon Associated with X-Disease of Stone Fruits, Prunus Spp.: Multilocus Characterization Based on 16S RRNA, SecY, and Ribosomal Protein Genes. Int. J. Syst. Evol. Microbiol. 2013, 63, 766–776. [Google Scholar] [CrossRef]
- Pérez-López, E.; Wei, W.; Wang, J.; Davis, R.E.; Luna-Rodríguez, M.; Zhao, Y. Novel Phytoplasma Strains of X-Disease Group Unveil Genetic Markers That Distinguish North American and South American Geographic Lineages within Subgroups 16SrIII-J and 16SrIII-U. Ann. Appl. Biol. 2017, 171, 405–416. [Google Scholar] [CrossRef]
- De La Rue, S.J.; Hopkinson, R.; Foster, S.; Gibb, K.S. Phytoplasma Host Range and Symptom Expression in the Pasture Legume Stylosanthes. Field Crops Res. 2003, 84, 327–334. [Google Scholar] [CrossRef]
- Larsen, M.L.; Walter, G.H. Intraspecific Variation within Orosius Argentatus Evans (Hemiptera: Cicadellidae): Colour Polymorphisms, Morphometric Analyses and Host Associations. Aust. J. Entomol. 2007, 46, 207–216. [Google Scholar] [CrossRef]
- Liu, J.; Gopurenko, D.; Fletcher, M.J.; Johnson, A.C.; Gurr, G.M. Phytoplasmas–The “Crouching Tiger” Threat of Australian Plant Pathology. Front. Plant Sci. 2017, 8, 599. [Google Scholar] [CrossRef]
- Galetto, L.; Nardi, M.; Saracco, P.; Bressan, A.; Marzachì, C.; Bosco, D. Variation in Vector Competency Depends on Chrysanthemum Yellows Phytoplasma Distribution within Euscelidius Variegatus. Entomol. Exp. Appl. 2009, 131, 200–207. [Google Scholar] [CrossRef]
- Galetto, L.; Marzachì, C. Real-Time PCR Diagnosis and Quantification of Phytoplasmas. In Phytoplasmas: Genomes, Plant Hosts and Vectors; Weintraub, P.G., Jones, P., Eds.; CABI: Wallingford, CT, USA, 2009; pp. 1–18. ISBN 978-1-84593-530-6. [Google Scholar]
- Acs, Z.; Jović, J.; Ember, I.; Cvrković, T.; Nagy, Z.; Talaber, C.; Gergely, L.; Toševski, I.; Koelber, M. First Report of Maize Redness Disease in Hungary. Bull. Insectology 2011, 64, S229–S230. [Google Scholar]
- Lessio, F.; Picciau, L.; Gonella, E.; Mandrioli, M.; Tota, F.; Alma, A. The Mosaic Leafhopper Orientus Ishidae: Host Plants, Spatial Distribution, Infectivity, and Transmission of 16SrV Phytoplasmas to Vines. Bull. Insectology 2016, 69, 277–289. [Google Scholar]
- Mitrovic, M.; Toševski, I.; Krstic, O.; Cvrkovic, T.; Krnjajic, S.; Jovic, J. A Strain of Phytoplasma Related to 16SrII Group in Picris Hieracioides L. in Serbia. Bull. Insectology 2011, 64, S241–S242. [Google Scholar]
- Sforza, R.; Clair, D.; Daire, X.; Larrue, J.; Boudon-Padieu, E. The Role of Hyalesthes Obsoletus (Hemiptera: Cixiidae) in the Occurrence of Bois Noir of Grapevines in France. J. Phytopathol. 1998, 146, 549–556. [Google Scholar] [CrossRef]
- Trivellone, V.; Pinzauti, F.; Bagnoli, B. Reptalus Quinquecostatus (Dufour) (Auchenorrhyncha Cixiidae) as a Possible Vector of Stolbur-Phytoplasma in a Vineyard in Tuscany. Redia 2005, 88, 103–108. [Google Scholar]

| Code | Tribe | Species | qPCR | Cq 1 | AHE | len | nPCR-SS | len |
|---|---|---|---|---|---|---|---|---|
| 1CH | Cicadulini | Rhopalopyx elongata | phy | 22.08 | phy | 1530 | phy-part | 689 |
| 2TH | Athysanini | n. gen. T1 n. sp. | phy | 25.06 | phy | 1529 | phy | 1318 |
| 3AU | Opsiini | Orosius argentatus | phy | 23.23 | phy | 1528 | phy-part | 784 |
| 4CH | Athysanini | Euscelidius variegatus | phy | 26.41 | phy | 1527 | phy | 1368 |
| 5MX | Scaphytopiini | Scaphytopius aequus | phy | 20.28 | phy | 1527 | phy | 1378 |
| 6MG | Stenometopiini | Gen. sp. | phy | 27.20 | phy | 1525 | phy-part | 783 |
| 7IL | Opsiini | Neoaliturus argillaceus | phy | 21.38 | phy | 1525 | phy | 1366 |
| 8AU | Paralimnini | Micrelloides n. sp. | phy | 25.25 | phy | 1524 | phy | 1317 |
| 9TW | Opsiini | Alishania formosana | phy | 25.85 | phy | 1523 | phy | 1363 |
| 10US | Paralimnini | Diplocolenus evansi | phy | 20.57 | phy | 1522 | phy | 1367 |
| 11FR | Fieberiellini | Synophropsis lauri | phy | 28.29 | phy | 1522 | oBa-part | 812 |
| 12ZM | Selenocephalini | Abimwa sp. | phy | 27.65 | phy | 1522 | oBa | 1386 |
| 13BR | Macrostelini | Dalbulus maidis | phy | 23.75 | phy | 1397 | phy | 1381 |
| 14CN | Athysanini | Nakaharanus bimaculatus | oBa | 31.21 | phy | 1522 | oBa-part | 795 |
| 15CA | Limotettigini | Limotettix urnura | phy | 29.26 | phy | 1521 | oBa-part | 789 |
| 16FR | Chiasmini | Doratura homophyla | phy | 29.72 | phy | 1527 | oBa-part | 784 |
| 17MN | Macrostelini | Macrosteles guttatus | phy | 26.21 | phy | 1527 | phy | 1372 |
| 18MN | Paralimnini | Adarrus n. sp. | oBa | 32.74 | phy-part | 1235 2 | oBa | 1397 |
| 19CD | Scaphoideini | n. gen. ZA5 n. sp. 1 | oBa | 30.63 | phy-part | 1235 2 | oBa | 1372 |
| 20PE | Chiasmini | Exitianus obscurinervis | oBa | 31.82 | phy | 1518 | oBa | 1322 |
| 21ZA | Chiasmini | Aconurella prolixa | oBa | 38.09 | phy | 1464 | oBa-part | 959 |
| 22CH | Scaphoideini | Osbornellus auronitens | oBa | 31.27 | phy | 1527 | oBa | 1287 |
| 23CN | Scaphoideini | Amimenus mojiensis | phy | 27.80 | phy | 1526 | phy | 1374 |
| 24FR | Scaphoideini | Anoplotettix putoni | oBa | 32.73 | oBa | 1431 | oBa | 1224 |
| 25AU | Deltocephalini | n. gen. AU3 n. sp. 2 | oBa | 30.88 | phy-part | 910 2 | oBa | 1394 |
| 26PH | Scaphoideini | n. gen. PH1 n. sp. | phy | 29.88 | phy | 1527 | oBa | 1397 |
| 27US | Deltocephalini | Graminella sonora | phy | 21.13 | phy | 1527 | phy | 1383 |
| 28ZA | Opsiini | Neoaliturus angulatus | oBa | 30.99 | neg | 0 | oBa | 1288 |
| 29CN | Deltocephalini | Paramesodes sp. | phy | 28.27 | phy-part | 620 | phy-part | 873 |
| 30MX | Deltocephalini | Sanctanus fasciatus | oBa | 35.76 | oBa-part | 225 | oBa-part | 927 |
| 31PH | Scaphoideini | n. gen. PH1 n. sp. 2 | phy | 27.83 | neg | 0 | phy-part | 552 |
| 32BR | Scaphoideini | Scaphoidula dentata | oBa | 33.05 | oBa-part | 621 | oBa-part | 1208 |
| 33US | Limotettigini | Limotettix truncatus | oBa | 32.60 | neg | 0 | oBa | 1381 |
| 34AU | Deltocephalini | Horouta aristarche | oBa | 34.79 | oBa-part | 399 | oBa-part | 1090 |
| 35BR | Deltocephalini | Amplicephalus maculellus | phy | 28.71 | neg | 0 | oBa | 1277 |
| qPCR | nPCR-SS | AHE | ||
|---|---|---|---|---|
| phy | oBa | neg | ||
| phy | phy | 15 (43%) | 0 | 1 (3%) |
| phy | oBa | 5 (14%) | 0 | 1(3%) |
| oBa | phy | 0 | 0 | 0 |
| oBa | oBa | 7 (20%) | 4 (11%) | 2 (6%) |
| Code | Tribe | Species | 16Sr Group Subgroup | Comparison |
|---|---|---|---|---|
| 1CH | Cicadulini | Rhopalopyx elongata | 16SrVI-L | qPCR = AHE = nPCR-SS |
| 2TH | Athysanini | n. gen. T1 n. sp. | New group 1 | qPCR = AHE = nPCR-SS |
| 3AU | Opsiini | Orosius argentatus | 16SrXXXVII-A | qPCR = AHE = nPCR-SS |
| 4CH | Athysanini | Euscelidius variegatus | 16SrI-F | qPCR = AHE = nPCR-SS |
| 5MX | Scaphytopiini | Scaphytopius aequus | 16SrI-B variant | qPCR = AHE = nPCR-SS |
| 6MG | Stenometopiini | n. gen. n. sp. | New group 2 | qPCR = AHE = nPCR-SS |
| 7IL | Opsiini | Neoaliturus argillaceus | 16SrIX-J | qPCR = AHE = nPCR-SS |
| 8AU | Paralimnini | Micrelloides n. sp. | 16SrXV-D | qPCR = AHE = nPCR-SS |
| 9TW | Opsiini | Alishania formosana | New group 3 | qPCR = AHE = nPCR-SS |
| 10US | Paralimnini | Diplocolenus evansi | 16SrXI-C | qPCR = AHE = nPCR-SS |
| 11FR | Fieberiellini | Synophropsis lauri | 16SrIII-U variant | qPCR = AHE |
| 12ZM | Selenocephalini | Abimwa sp. | 16SrIII-U variant | qPCR = AHE |
| 13BR | Macrostelini | Dalbulus maidis | 16SrI-B | qPCR = AHE = nPCR-SS |
| 14CN | Athysanini | Nakaharanus bimaculatus | 16SrIII-U variant | AHE |
| 15CA | Limotettigini | Limotettix urnura | 16SrXI-G | qPCR = AHE |
| 16FR | Chiasmini | Doratura homophyla | 16SrI-B | qPCR = AHE |
| 17MN | Macrostelini | Macrosteles guttatus | 16SrI-B | qPCR = AHE = nPCR-SS |
| 20PE | Chiasmini | Exitianus obscurinervis | 16SrIII-U variant | AHE |
| 21ZA | Chiasmini | Aconurella prolixa | 16SrIII-U variant | AHE |
| 22CH | Scaphoideini | Osbornellus auronitens | 16SrV-C | AHE |
| 23CN | Scaphoideini | Amimenus mojiensis | 16SrI-B | qPCR = AHE = nPCR-SS |
| 26PH | Scaphoideini | n. gen. PH1 n. sp. | 16SrI-AO | qPCR = AHE |
| 27US | Deltocephalini | Graminella sonora | 16SrI-B | qPCR = AHE = nPCR-SS |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Trivellone, V.; Cao, Y.; Dietrich, C.H. Comparison of Traditional and Next-Generation Approaches for Uncovering Phytoplasma Diversity, with Discovery of New Groups, Subgroups and Potential Vectors. Biology 2022, 11, 977. https://doi.org/10.3390/biology11070977
Trivellone V, Cao Y, Dietrich CH. Comparison of Traditional and Next-Generation Approaches for Uncovering Phytoplasma Diversity, with Discovery of New Groups, Subgroups and Potential Vectors. Biology. 2022; 11(7):977. https://doi.org/10.3390/biology11070977
Chicago/Turabian StyleTrivellone, Valeria, Yanghui Cao, and Christopher H. Dietrich. 2022. "Comparison of Traditional and Next-Generation Approaches for Uncovering Phytoplasma Diversity, with Discovery of New Groups, Subgroups and Potential Vectors" Biology 11, no. 7: 977. https://doi.org/10.3390/biology11070977
APA StyleTrivellone, V., Cao, Y., & Dietrich, C. H. (2022). Comparison of Traditional and Next-Generation Approaches for Uncovering Phytoplasma Diversity, with Discovery of New Groups, Subgroups and Potential Vectors. Biology, 11(7), 977. https://doi.org/10.3390/biology11070977







