Phenolics from Chrozophora oblongifolia Aerial Parts as Inhibitors of α-Glucosidases and Advanced Glycation End Products: In-Vitro Assessment, Molecular Docking and Dynamics Studies
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Extraction and Isolation
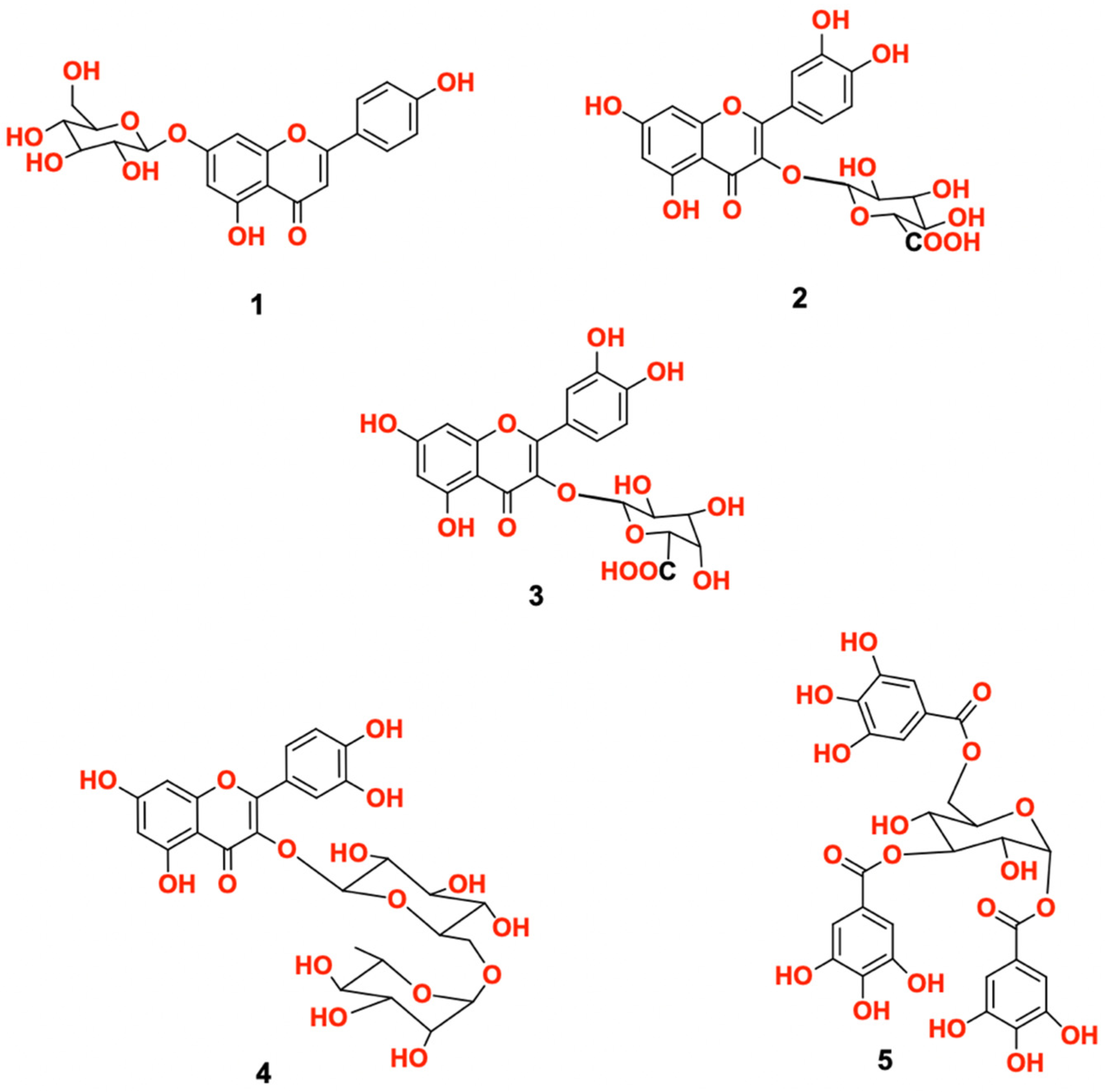
2.3. Characterization of Isolated Compounds
2.4. Biological Evaluation
2.4.1. Inhibition of α-Glucosidase
2.4.2. Inhibition of Pancreatic Lipase
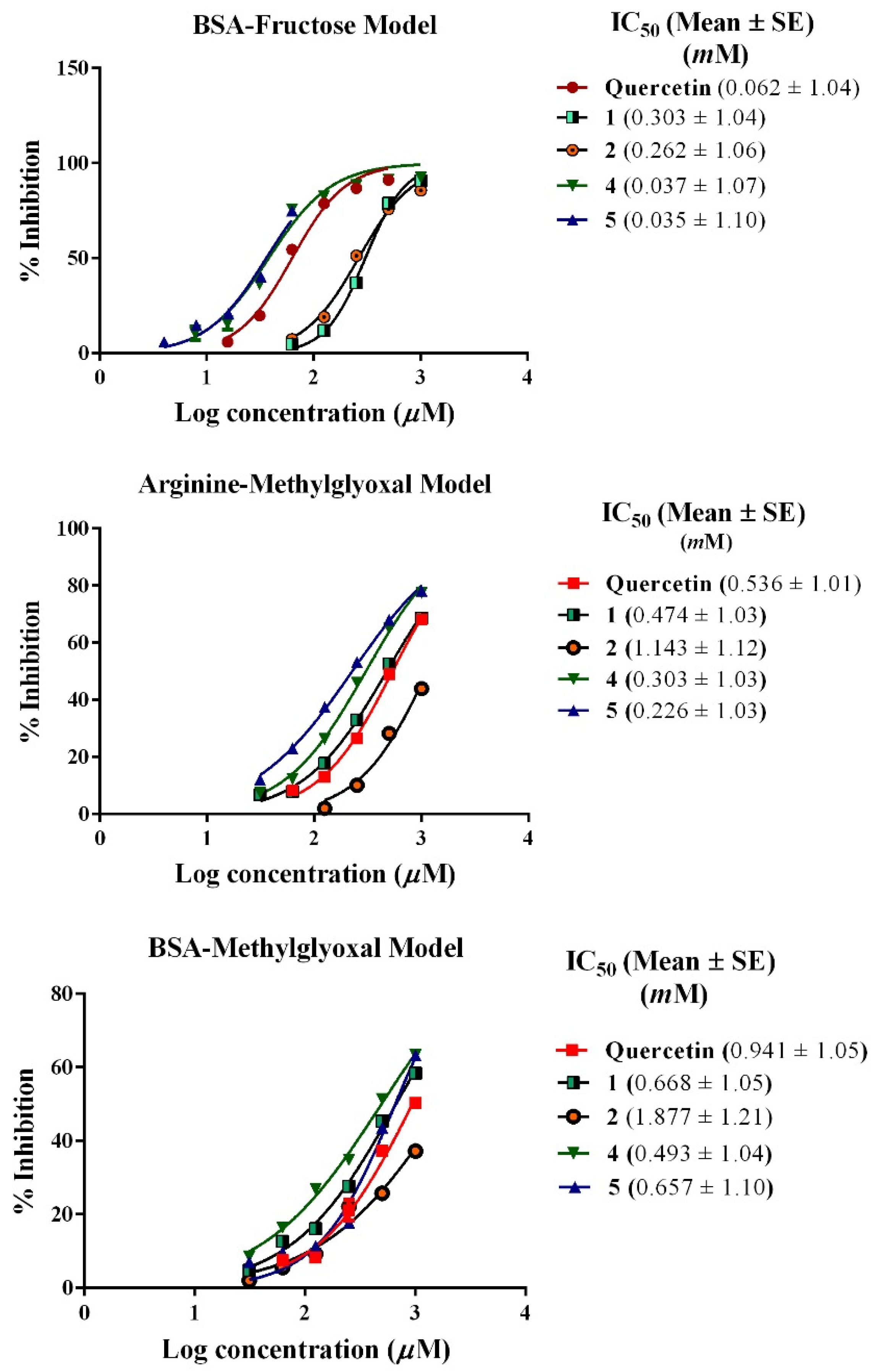
2.4.3. Inhibition of the Formation of Advanced Glycation End Products (AGEs)
BSA-Fructose Assay
Arginine-Methylglyoxal Assay
BSA-Methylglyoxal Assay
2.4.4. Antioxidant Activity
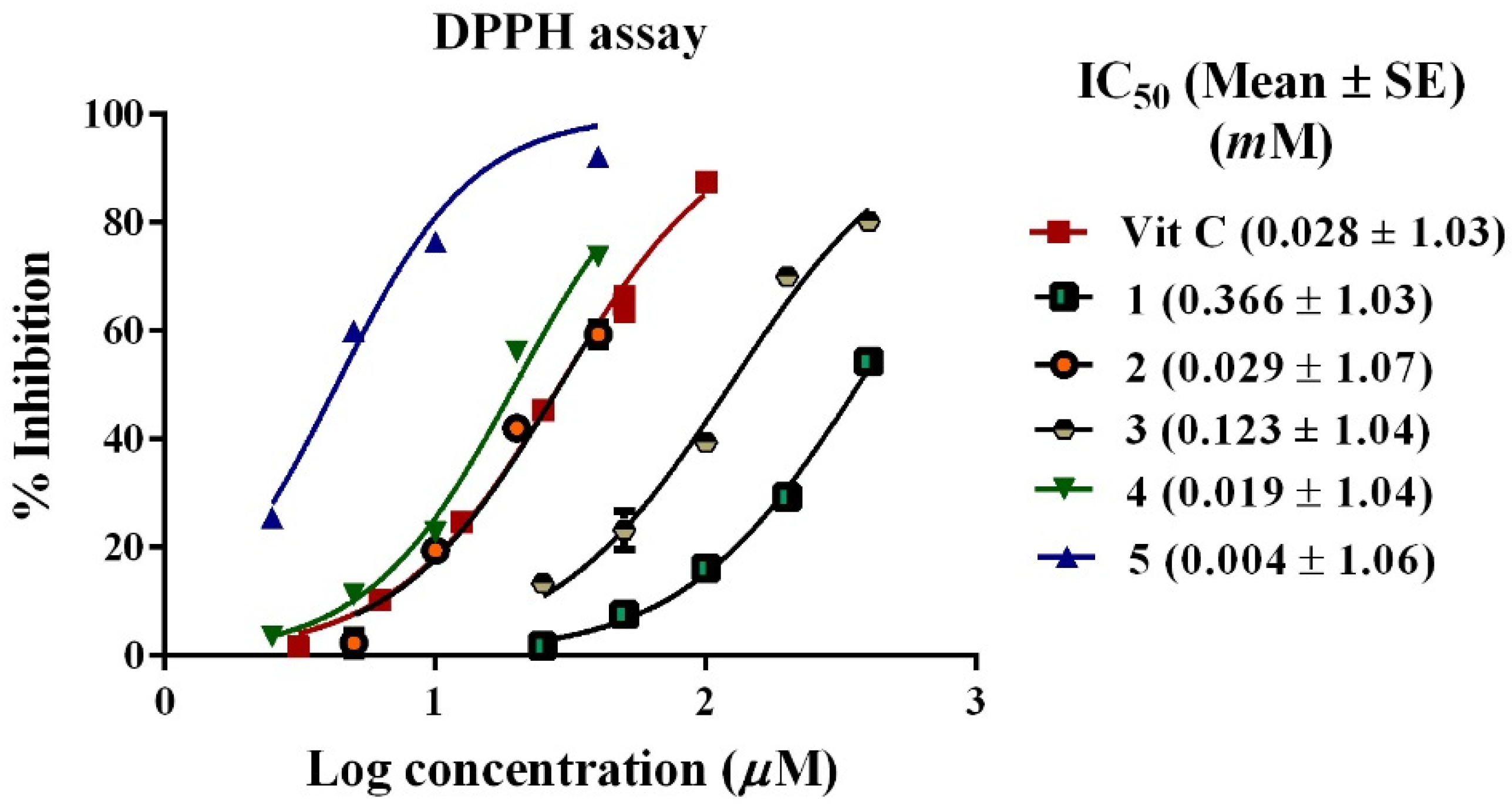
2,2-diphenyl-1-picrylhydrazyl (DPPH) Assay
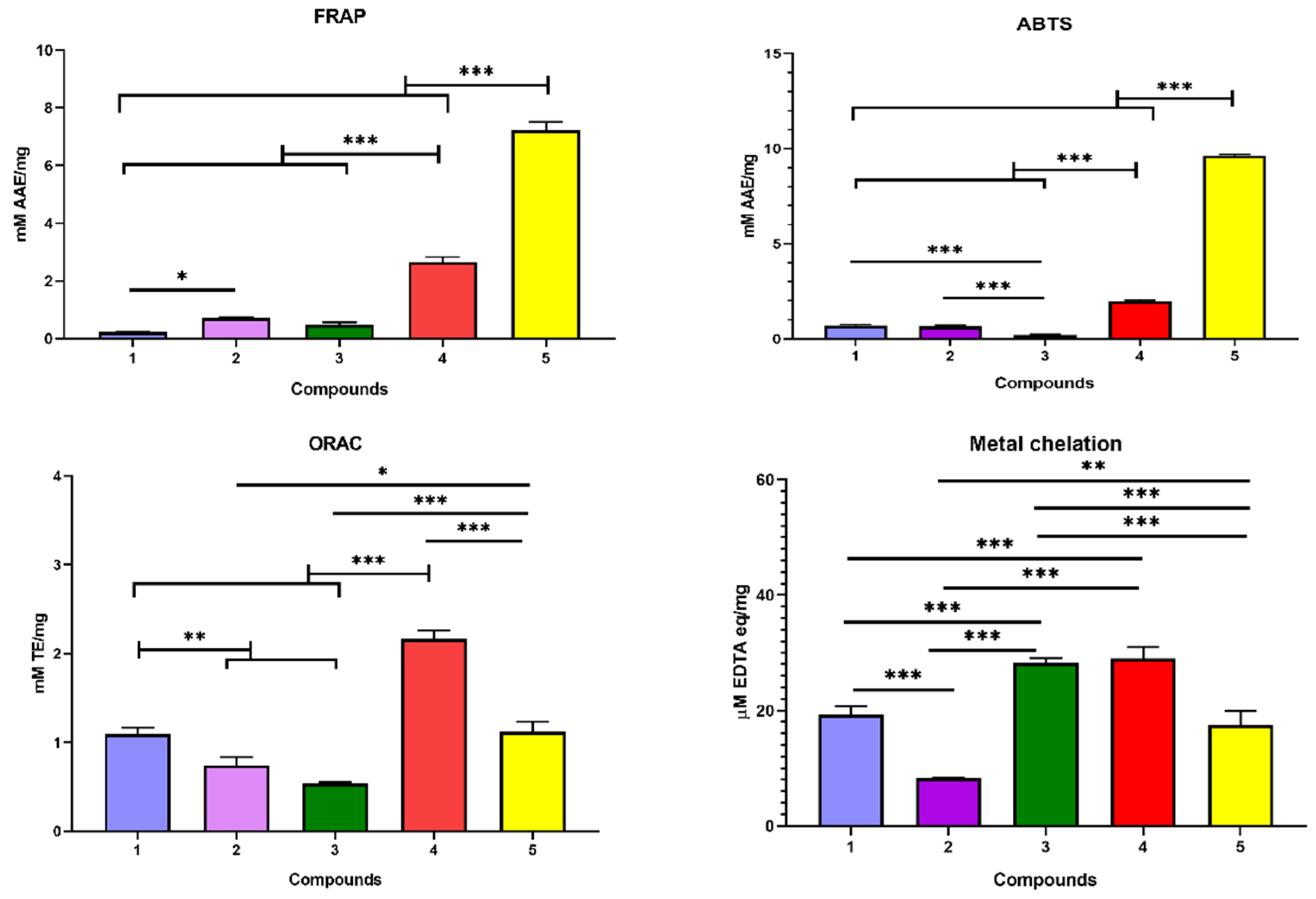
2,2′-azino-bis (3-ethylbenzothiazoline-6-sulfonic acid) (ABTS) Assay
Ferric Reducing Antioxidant Power (FRAP) Assay
Metal Chelation Assay
Oxygen Radical Absorbance Capacity (ORAC) Assay
2.4.5. Data Analysis
2.5. Ligand-Target Preparation and Molecular Docking Analysis
2.6. Molecular Dynamics (MD) Simulations
3. Results and Discussion
3.1. Inhibition of α-Glucosidase
3.2. Inhibition of Pancreatic Lipase
3.3. Inhibition of the Formation of Advanced Glycation End Products (AGEs)
3.4. Antioxidant Activity
3.5. Molecular Docking Analysis
3.6. Molecular Dynamics Simulation Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Justino, A.B.; Miranda, N.C.; Franco, R.R.; Martins, M.M.; da Silva, N.M.; Espindola, F.S. Annona muricata Linn. leaf as a source of antioxidant compounds with in vitro antidiabetic and inhibitory potential against α-amylase, α-glucosidase, lipase, non-enzymatic glycation and lipid peroxidation. Biomed. Pharmacother. 2018, 100, 83–92. [Google Scholar] [CrossRef]
- Vasarri, M.; Barletta, E.; Vinci, S.; Ramazzotti, M.; Francesconi, A.; Manetti, F.; Degl’Innocenti, D. Annona cherimola Miller Fruit as a Promising Candidate against Diabetic Complications: An In Vitro Study and Preliminary Clinical Results. Foods 2020, 9, 1350. [Google Scholar] [CrossRef] [PubMed]
- Glovaci, D.; Fan, W.; Wong, N.D. Epidemiology of diabetes mellitus and cardiovascular disease. Curr. Cardiol. Rep. 2019, 21, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Abdallah, H.M.; El-Bassossy, H.; Mohamed, G.A.; El-Halawany, A.M.; Alshali, K.Z.; Banjar, Z.M. Phenolics from Garcinia mangostana inhibit advanced glycation endproducts formation: Effect on amadori products, cross-linked structures and protein thiols. Molecules 2016, 21, 251. [Google Scholar] [CrossRef] [PubMed]
- Abdallah, H.M.; Zakaria, E.M.; El-Halawany, A.M.; Mohamed, G.A.; Safo, M.K.; El-Bassossy, H.M. Psiadia punctulata major flavonoids alleviate exaggerated vasoconstriction produced by advanced glycation end products. PLoS ONE 2019, 14, e0222101. [Google Scholar] [CrossRef] [PubMed]
- Song, F.; Schmidt, A.M. Glycation and insulin resistance: Novel mechanisms and unique targets? Arterioscler. Thromb. Vasc. Biol. 2012, 32, 1760–1765. [Google Scholar] [CrossRef]
- Shen, C.-Y.; Lu, C.-H.; Wu, C.-H.; Li, K.-J.; Kuo, Y.-M.; Hsieh, S.-C.; Yu, C.-L. The Development of Maillard Reaction, and Advanced Glycation End Product (AGE)-Receptor for AGE (RAGE) Signaling Inhibitors as Novel Therapeutic Strategies for Patients with AGE-Related Diseases. Molecules 2020, 25, 5591. [Google Scholar] [CrossRef]
- Wang, W.; Yagiz, Y.; Buran, T.J.; do Nascimento Nunes, C.; Gu, L. Phytochemicals from berries and grapes inhibited the formation of advanced glycation end-products by scavenging reactive carbonyls. Food Res. Int. 2011, 44, 2666–2673. [Google Scholar] [CrossRef]
- Pérez-Martínez, P.; Mikhailidis, D.P.; Athyros, V.G.; Bullo, M.; Couture, P.; Covas, M.I.; de Koning, L.; Delgado-Lista, J.; Diaz-Lopez, A.; Drevon, C.A. Lifestyle recommendations for the prevention and management of metabolic syndrome: An international panel recommendation. Nutr. Rev. 2017, 75, 307–326. [Google Scholar] [CrossRef]
- Payab, M.; Hasani-Ranjbar, S.; Shahbal, N.; Qorbani, M.; Aletaha, A.; Haghi-Aminjan, H.; Soltani, A.; Khatami, F.; Nikfar, S.; Hassani, S. Effect of the herbal medicines in obesity and metabolic syndrome: A systematic review and meta-analysis of clinical trials. Phytother. Res. 2020, 34, 526–545. [Google Scholar] [CrossRef]
- Abdel-Sattar, E.A.; Abdallah, H.M.; Khedr, A.; Abdel-Naim, A.B.; Shehata, I.A. Antihyperglycemic activity of Caralluma tuberculata in streptozotocin-induced diabetic rats. Food Chem. Toxicol. 2013, 59, 111–117. [Google Scholar] [CrossRef] [PubMed]
- Choucry, M.A.; Shalabi, A.A.; El Halawany, A.M.; El-Sakhawy, F.S.; Zaiter, A.; Morita, H.; Chaimbault, P.; Abdel-Sattar, E. New pregnane glycosides isolated from Caralluma hexagona lavranos as inhibitors of α-glucosidase, pancreatic lipase, and advanced glycation end products formation. ACS Omega 2021, 6, 18881–18889. [Google Scholar] [CrossRef]
- Clay, H.F.; Hubbard, J.C. Euphorbiaceae (Spurge Family). In The Hawai’i Garden; University of Hawaii Press: Honolulu, HI, USA, 2021; pp. 59–80. [Google Scholar]
- Hashim, O.; Abou-Zaid, M.; Abdel-Galil, F.; Saleh, N. The flavonoids of Egyptian Chrozophora species. Biochem. Syst. Ecol. 1990, 18, 151–152. [Google Scholar] [CrossRef]
- Usman, H.; Musa, Y.; Ahmadu, A.; Tijjani, M. Phytochemical and antimicrobial effects of Chrozophora senegalensis. Afr. J. Tradit. Complement. Altern. Med. 2007, 4, 488–494. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Tabussum, A.; Riaz, N.; Saleem, M.; Ashraf, M.; Ahmad, M.; Alam, U.; Jabeen, B.; Malik, A.; Jabbar, A. α-Glucosidase inhibitory constituents from Chrozophora plicata. Phytochem. Lett. 2013, 6, 614–619. [Google Scholar] [CrossRef]
- Abdallah, H.M.; Almowallad, F.M.; Esmat, A.; Shehata, I.A.; Abdel-Sattar, E.A. Anti-inflammatory activity of flavonoids from Chrozophora tinctoria. Phytochem. Lett. 2015, 13, 74–80. [Google Scholar] [CrossRef]
- Abdel-Naim, A.B.; Alghamdi, A.A.; Algandaby, M.M.; Al-Abbasi, F.A.; Al-Abd, A.M.; Eid, B.G.; Abdallah, H.M.; El-Halawany, A.M. Rutin isolated from Chrozophora tinctoria enhances bone cell proliferation and ossification markers. Oxid. Med. Cell. Longev. 2018, 2018, 5106469. [Google Scholar] [CrossRef]
- Abdallah, H.M.; El-Bassossy, H.M.; Mohamed, G.A.; El-Halawany, A.M.; Alshali, K.Z.; Banjar, Z.M. Phenolics from Garcinia mangostana alleviate exaggerated vasoconstriction in metabolic syndrome through direct vasodilatation and nitric oxide generation. BMC Complement. Altern. Med. 2016, 16, 359. [Google Scholar] [CrossRef]
- Abdallah, H.M.; El-Bassossy, H.M.; Mohamed, G.A.; El-Halawany, A.M.; Alshali, K.Z.; Banjar, Z.M. Mangostanaxanthones III and IV: Advanced glycation end-product inhibitors from the pericarp of Garcinia mangostana. J. Nat. Med. 2017, 71, 216–226. [Google Scholar] [CrossRef]
- Abdallah, H.M.; El-Bassossy, H.M.; El-Halawany, A.M.; Ahmed, T.A.; Mohamed, G.A.; Malebari, A.M.; Hassan, N.A. Self-Nanoemulsifying Drug Delivery System Loaded with Psiadia punctulata Major Metabolites for Hypertensive Emergencies: Effect on Hemodynamics and Cardiac Conductance. Front. Pharmacol. 2021, 12, 681070. [Google Scholar] [CrossRef]
- Gutiérrez-Grijalva, E.P.; Antunes-Ricardo, M.; Acosta-Estrada, B.A.; Gutiérrez-Uribe, J.A.; Heredia, J.B. Cellular antioxidant activity and in vitro inhibition of α-glucosidase, α-amylase and pancreatic lipase of oregano polyphenols under simulated gastrointestinal digestion. Food Res. Int. 2019, 116, 676–686. [Google Scholar] [CrossRef]
- Kordel, M.; Schmid, R.D. Inhibition of the lipase from Pseudomonas spec. ATCC 21808 by diethyl p-nitrophenylphosphate. Hints for one buried active site for lipolytic and esterolytic activity. Lipases Struct. Mech. Genet. Eng. 1991, 16, 385–387. [Google Scholar]
- Santos, J.S.; Brizola, V.R.A.; Granato, D. High-throughput assay comparison and standardization for metal chelating capacity screening: A proposal and application. Food Chem. 2017, 214, 515–522. [Google Scholar] [CrossRef]
- Liang, N.; Kitts, D.D. Antioxidant property of coffee components: Assessment of methods that define mechanisms of action. Molecules 2014, 19, 19180–19208. [Google Scholar] [CrossRef]
- Wadie, M.A.; Kishk, S.M.; Darwish, K.M.; Mostafa, S.M.; Elgawish, M.S. Simultaneous Determination of Losartan and Rosuvastatin in Rat Plasma Using Liquid Chromatography–Tandem Mass Spectrometric Technique for Application into Pharmacokinetic and Drug–Drug Interaction Studies. Chromatographia 2020, 83, 1477–1494. [Google Scholar] [CrossRef]
- Malebari, A.; Ibrahim, T.; Salem, I.; Salama, I.; Khayyat, A.; Mostafa, S.; El-Sabbagh, O.; Darwish, K. The Anticancer Activity for the Bumetanide-Based Analogs via Targeting the Tumor-Associated Membrane Bound Human Carbonic Anhydrase-IX Enzyme. Pharmaceuticals 2020, 13, 252. [Google Scholar] [CrossRef]
- El Raey, M.A.; El-Hagrassi, A.M.; Osman, A.F.; Darwish, K.M.; Emam, M. Acalypha wilkesiana flowers: Phenolic profiling, cytotoxic activity of their biosynthesized silver nanoparticles and molecular docking study for its constituents as Topoisomerase-I inhibitors. Biocatal. Agric. Biotechnol. 2019, 20, 101243. [Google Scholar] [CrossRef]
- Liang, J.; Edelsbrunner, H.; Woodward, C. Anatomy of protein pockets and cavities: Measurement of binding site geometry and implications for ligand design. Protein Sci. 1998, 7, 1884–1897. [Google Scholar] [CrossRef]
- Kitchen, D.B.; Decornez, H.; Furr, J.R.; Bajorath, J. Docking and scoring in virtual screening for drug discovery: Methods and applications. Nat. Rev. Drug Discov. 2004, 3, 935–949. [Google Scholar] [CrossRef]
- Wojciechowski, M.; Lesyng, B. Generalized Born Model: Analysis, Refinement, and Applications to Proteins. J. Phys. Chem. B 2004, 108, 18368–18376. [Google Scholar] [CrossRef]
- Labute, P. The generalized Born/volume integral implicit solvent model: Estimation of the free energy of hydration using London dispersion instead of atomic surface area. J. Comput. Chem. 2008, 29, 1693–1698. [Google Scholar] [CrossRef]
- Schrödinger, L.; DeLano, W. The PyMOL Molecular Graphics System, 2.0.6; Schrödinger, LLC: New York, NY, USA, 2016. [Google Scholar]
- Elhady, S.S.; Abdelhameed, R.F.A.; Malatani, R.T.; Alahdal, A.M.; Bogari, H.A.; Almalki, A.J.; Mohammad, K.A.; Ahmed, S.A.; Khedr, A.I.M.; Darwish, K.M. Molecular Docking and Dynamics Simulation Study of Hyrtios erectus Isolated Scalarane Sesterterpenes as Potential SARS-CoV-2 Dual Target Inhibitors. Biology 2021, 10, 389. [Google Scholar] [CrossRef]
- Vanommeslaeghe, K.; Hatcher, E.; Acharya, C.; Kundu, S.; Zhong, S.; Shim, J.; Darian, E.; Guvench, O.; Lopes, P.; Vorobyov, I.; et al. CHARMM general force field: A force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. J. Comput. Chem. 2010, 31, 671–690. [Google Scholar] [CrossRef]
- Saleh, A.H.; Abdelwaly, A.; Darwish, K.M.; Eissa, A.; Chittiboyina, A.; Helal, M.A. Deciphering the molecular basis of the kappa opioid receptor selectivity: A Molecular Dynamics study. J. Mol. Graph. Model. 2021, 106, 107940. [Google Scholar] [CrossRef]
- Ross, G.A.; Rustenburg, A.S.; Grinaway, P.B.; Fass, J.; Chodera, J.D. Biomolecular Simulations under Realistic Macroscopic Salt Conditions. J. Phys. Chem. B 2018, 122, 5466–5486. [Google Scholar] [CrossRef]
- Darden, T.; York, D.; Pedersen, L. Particle mesh Ewald: An N⋅log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993, 98, 10089–10092. [Google Scholar] [CrossRef]
- Hess, B.; Bekker, H.; Berendsen, H.J.C.; Fraaije, J.G.E.M. LINCS: A linear constraint solver for molecular simulations. J. Comput. Chem. 1997, 18, 1463–1472. [Google Scholar] [CrossRef]
- Páll, S.; Hess, B. A flexible algorithm for calculating pair interactions on SIMD architectures. Comput. Phys. Comm. 2013, 184, 2641–2650. [Google Scholar] [CrossRef]
- Kumari, R.; Kumar, R.; Lynn, A. g_mmpbsa—A GROMACS Tool for High-Throughput MM-PBSA Calculations. J. Chem. Inf. Model. 2014, 54, 1951–1962. [Google Scholar] [CrossRef]
- Schalkwijk, C.; Stehouwer, C. Methylglyoxal, a highly reactive dicarbonyl compound, in diabetes, its vascular complications, and other age-related diseases. Physiol. Rev. 2020, 100, 407–461. [Google Scholar] [CrossRef]
- Justino, A.B.; Franco, R.R.; Silva, H.C.; Saraiva, A.L.; Sousa, R.M.; Espindola, F.S. B procyanidins of Annona crassiflora fruit peel inhibited glycation, lipid peroxidation and protein-bound carbonyls, with protective effects on glycated catalase. Sci. Rep. 2019, 9, 19183. [Google Scholar] [CrossRef]
- Ravichandran, G.; Lakshmanan, D.K.; Murugesan, S.; Elangovan, A.; Rajasekaran, N.S.; Thilagar, S. Attenuation of protein glycation by functional polyphenolics of dragon fruit (Hylocereus polyrhizus); an in vitro and in silico evaluation. Food Res. Int. 2021, 140, 110081. [Google Scholar] [CrossRef]
- Allaman, I.; Bélanger, M.; Magistretti, P.J. Methylglyoxal, the dark side of glycolysis. Front. Neurosci. 2015, 9, 23. [Google Scholar] [CrossRef]
- Albano, C.; Negro, C.; Tommasi, N.; Gerardi, C.; Mita, G.; Miceli, A.; De Bellis, L.; Blando, F. Betalains, phenols and antioxidant capacity in Cactus Pear [Opuntia ficus-indica (L.) Mill.] fruits from Apulia (South Italy) Genotypes. Antioxidant 2015, 4, 269–280. [Google Scholar] [CrossRef]
- Opitz, S.E.; Smrke, S.; Goodman, B.A.; Keller, M.; Schenker, S.; Yeretzian, C. Antioxidant generation during coffee roasting: A comparison and interpretation from three complementary assays. Foods 2014, 3, 586–604. [Google Scholar] [CrossRef]
- Suleria, H.A.; Barrow, C.J.; Dunshea, F.R. Screening and characterization of phenolic compounds and their antioxidant capacity in different fruit peels. Foods 2020, 9, 1206. [Google Scholar] [CrossRef]
- Escribano, J.; Cabanes, J.; Jiménez-Atiénzar, M.; Ibañez-Tremolada, M.; Gómez-Pando, L.R.; García-Carmona, F.; Gandía-Herrero, F. Characterization of betalains, saponins and antioxidant power in differently colored quinoa (Chenopodium quinoa) varieties. Food Chem. 2017, 234, 285–294. [Google Scholar] [CrossRef]
- Mridula, S.; Masroor, W.S.; Xavier, M.; Hui, T.W.; Chan, H.K.; Chirara, K.; Nwabueze, O.P. Antioxidant and anti-advanced glycation end products formation properties of palmatine. J. Pharm. Pharmacogn. Res. 2021, 9, 366–378. [Google Scholar]
- Ren, L.; Qin, X.; Cao, X.; Wang, L.; Bai, F.; Bai, G.; Shen, Y. Structural insight into substrate specificity of human intestinal maltase-glucoamylase. Protein Cell 2011, 2, 827–836. [Google Scholar] [CrossRef]
- Sim, L.; Quezada-Calvillo, R.; Sterchi, E.E.; Nichols, B.L.; Rose, D.R. Human intestinal maltase-glucoamylase: Crystal structure of the N-terminal catalytic subunit and basis of inhibition and substrate specificity. J. Mol. Biol. 2008, 375, 782–792. [Google Scholar] [CrossRef]
- Sim, L.; Willemsma, C.; Mohan, S.; Naim, H.Y.; Pinto, B.M.; Rose, D.R. Structural basis for substrate selectivity in human maltase-glucoamylase and sucrase-isomaltase N-terminal domains. J. Biol. Chem. 2010, 285, 17763–17770. [Google Scholar] [CrossRef] [PubMed]
- Lovering, A.L.; Lee, S.S.; Kim, Y.W.; Withers, S.G.; Strynadka, N.C. Mechanistic and structural analysis of a family 31 alpha-glycosidase and its glycosyl-enzyme intermediate. J. Biol. Chem. 2005, 280, 2105–2115. [Google Scholar] [CrossRef] [PubMed]
- Ernst, H.A.; Lo Leggio, L.; Willemoës, M.; Leonard, G.; Blum, P.; Larsen, S. Structure of the Sulfolobus solfataricus alpha-glucosidase: Implications for domain conservation and substrate recognition in GH31. J. Mol. Biol. 2006, 358, 1106–1124. [Google Scholar] [CrossRef]
- Roig-Zamboni, V.; Cobucci-Ponzano, B.; Iacono, R.; Ferrara, M.C.; Germany, S.; Bourne, Y.; Parenti, G.; Moracci, M.; Sulzenbacher, G. Structure of human lysosomal acid α-glucosidase–a guide for the treatment of Pompe disease. Nat. Commun. 2017, 8, 1111. [Google Scholar] [CrossRef]
- Nagy, M.I.; Darwish, K.M.; Kishk, S.M.; Tantawy, M.A.; Nasr, A.M.; Qushawy, M.; Swidan, S.A.; Mostafa, S.M.; Salama, I. Design, Synthesis, Anticancer Activity, and Solid Lipid Nanoparticle Formulation of Indole- and Benzimidazole-Based Compounds as Pro-Apoptotic Agents Targeting Bcl-2 Protein. Pharmaceuticals 2021, 14, 113. [Google Scholar] [CrossRef]
- Kontoyianni, M.; McClellan, L.M.; Sokol, G.S. Evaluation of Docking Performance: Comparative Data on Docking Algorithms. J. Med. Chem. 2004, 47, 558–565. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Wang, Y.; Han, L.; Fu, X.; Wang, S.; Li, W.; Han, W. Targeting N-Terminal Human Maltase-Glucoamylase to Unravel Possible Inhibitors Using Molecular Docking, Molecular Dynamics Simulations, and Adaptive Steered Molecular Dynamics Simulations. Front. Chem. 2021, 9, 635. [Google Scholar] [CrossRef]
- Cavasotto, C.N. Binding Free Energy Calculation Using Quantum Mechanics Aimed for Drug Lead Optimization. Methods Mol. Biol. 2020, 2114, 257–268. [Google Scholar] [CrossRef]

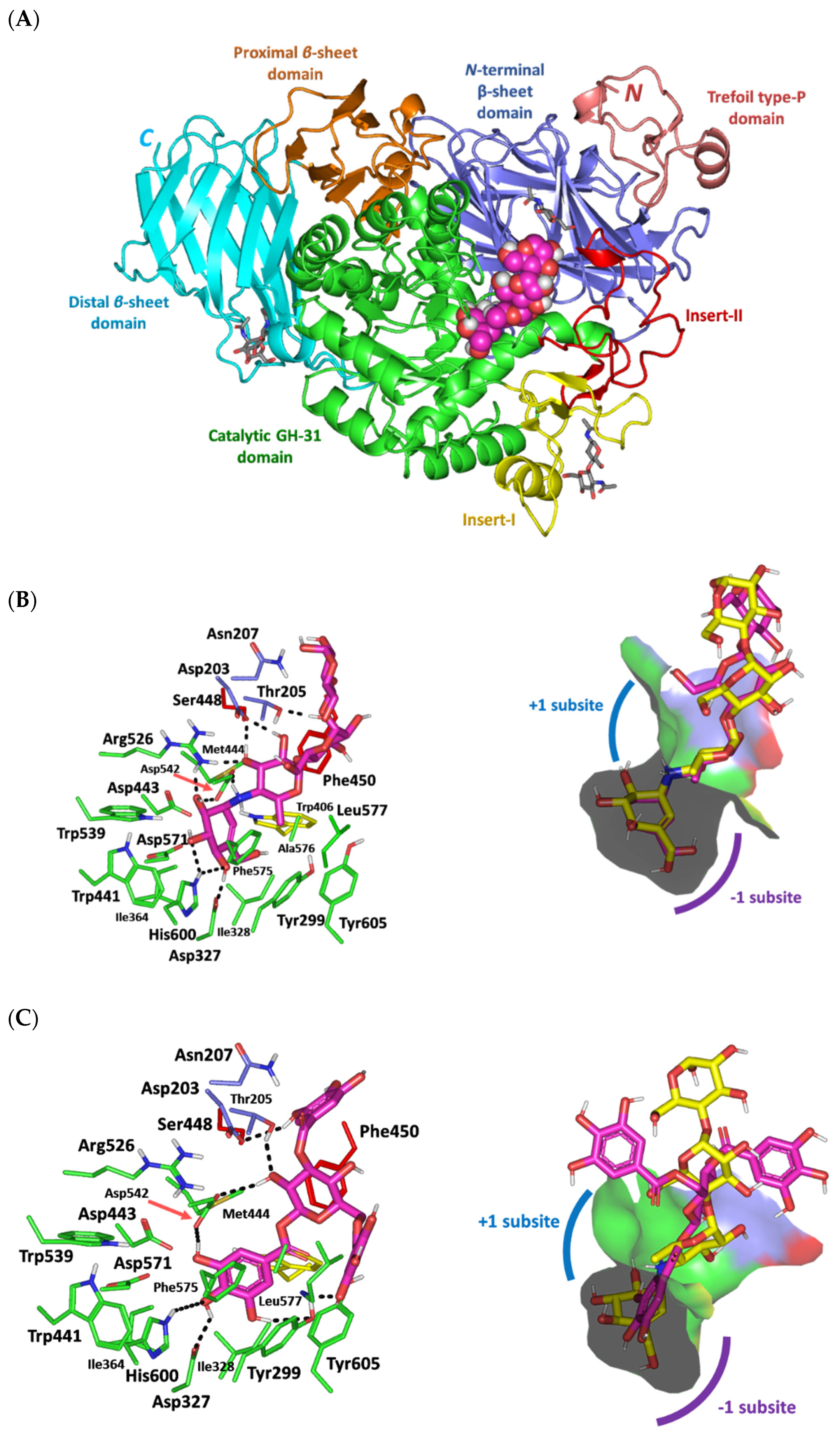
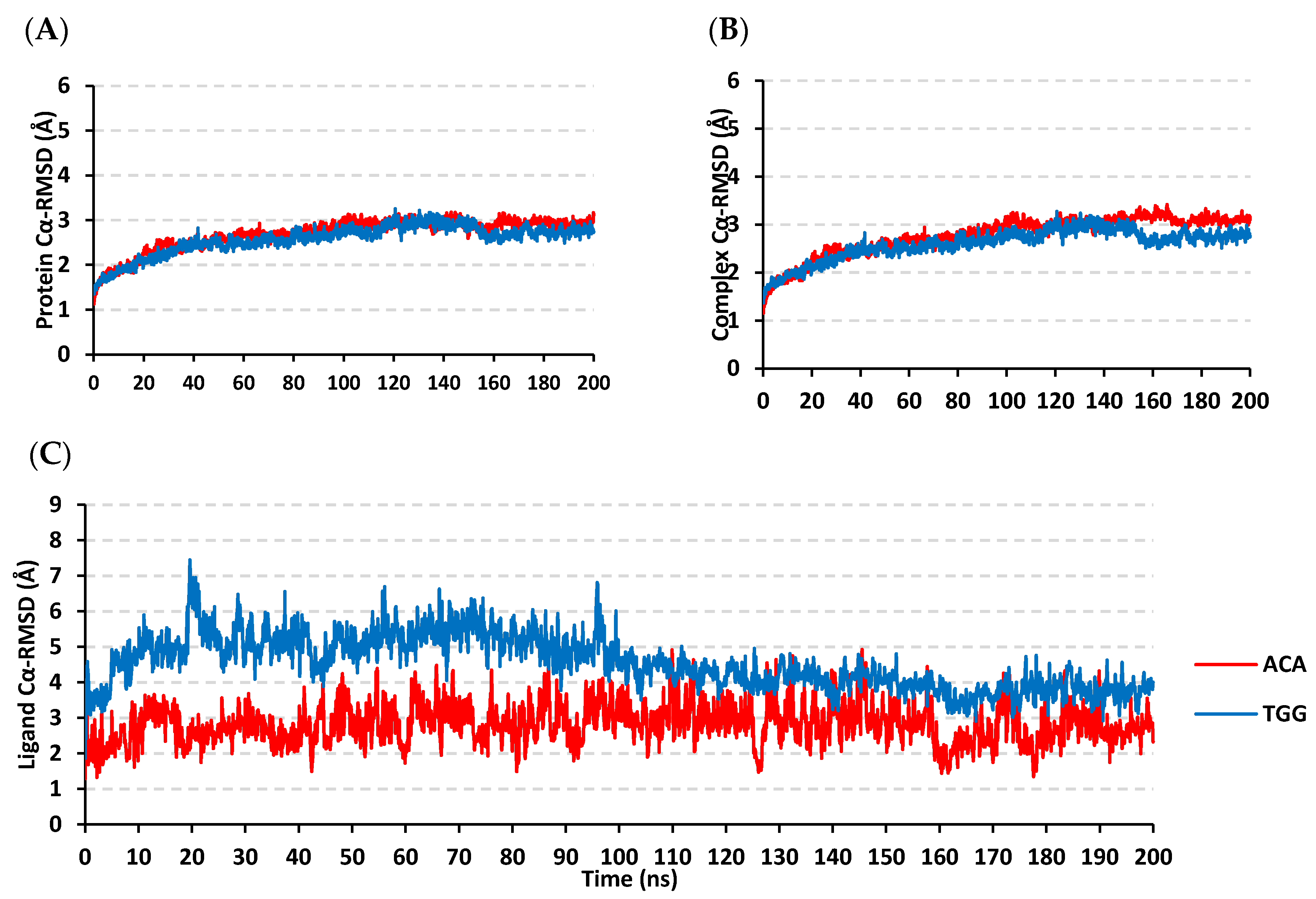
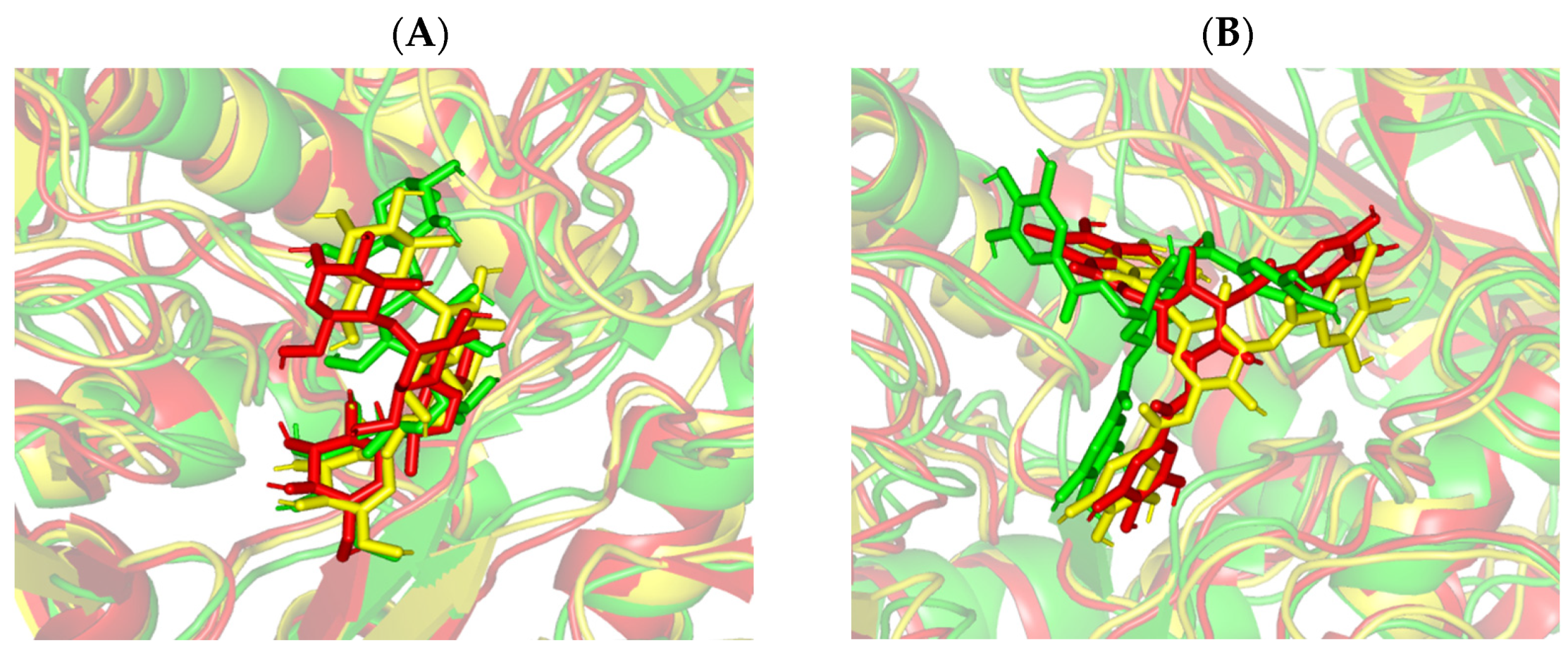
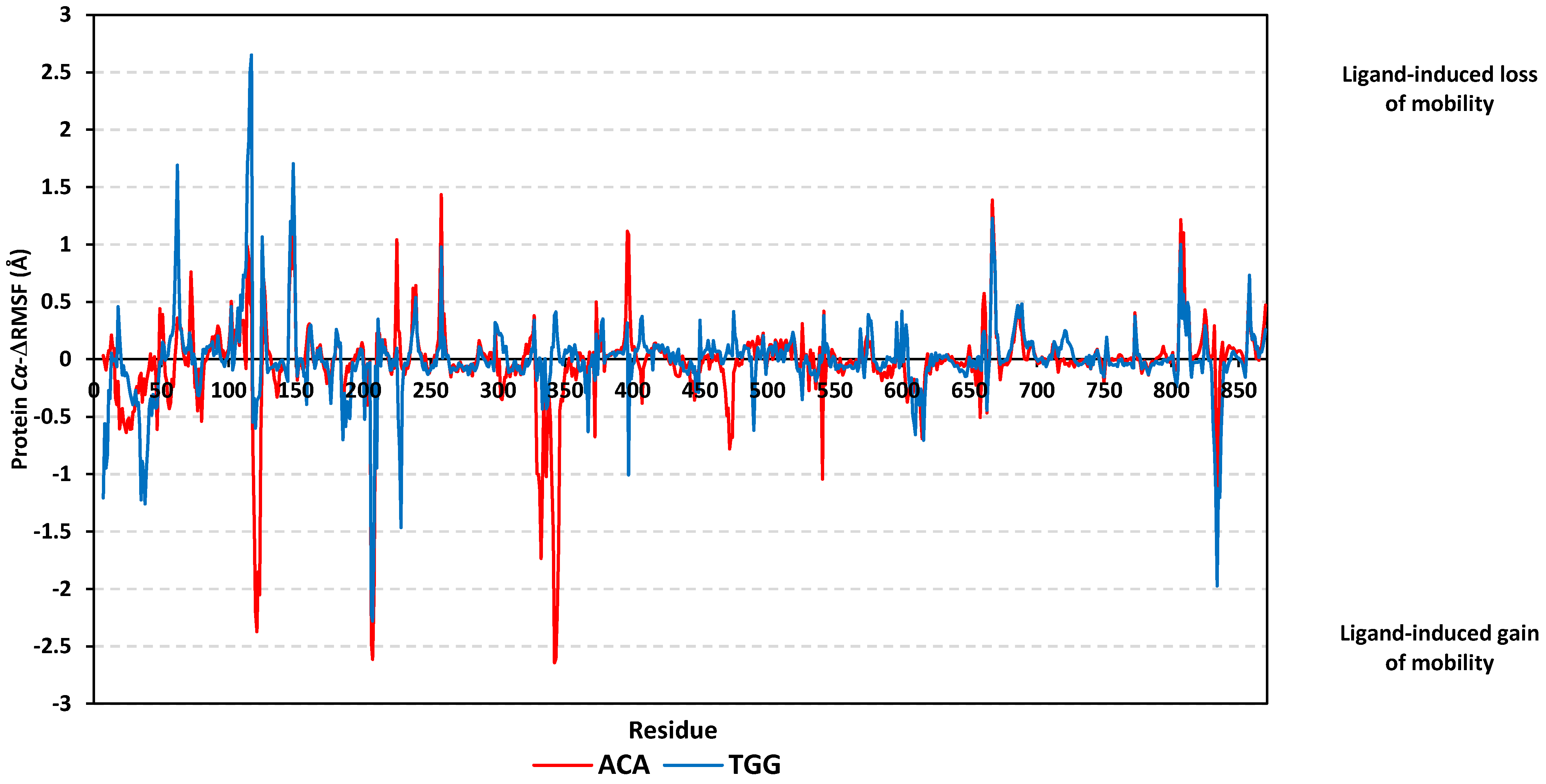

| Ligand | Docking Energy (Kcal/mol) | H-Bond Interactions [Length (Å); Angle (°); Binding Residues] | Hydrophobic Interactions | π-Interactions |
|---|---|---|---|---|
| Acarbose | −6.8 | 1.9 Å; 159°; Asp203 (sidechain CO−/6-deoxyglucosyl 3′-OH) 2.0 Å; 160°; Asp203 (sidechain CO−/6-deoxyglucosyl 4′-OH) 2.1 Å; 164°; Thr205 (sidechain OH/+3 maltosyl 6′-OH) 1.9 Å; 175°; Asp327 (sidechain CO−/valienamine 4′-OH) 2.0 Å; 168°; Arg526 (sidechain =NHH/6-deoxyglucosyl 3′-OH) 2.0 Å; 146°; Arg526 (sidechain =NHH/valienamine 6′-OH) 1.9 Å; 142°; Asp542 (sidechain CO−/glycosidic linker NH) 1.7 Å; 155°; Asp542 (sidechain C=O/valienamine 6′-OH) 2.3 Å; 142°; His600 (sidechain NH/valienamine 4′-OH) 2.3 Å; 138°; His600 (sidechain NH/valienamine 5′-OH) | Tyr299, Ile328, Ile364, Trp406, Trp441, Phe450, Trp539, Phe575, Ala576, Leu577, Tyr605 | - |
| 1,3,6-Trigalloyl glucose | −7.3 | 2.5 Å; 145°; Asp203 (sidechain CO−/C3-galloyl 3′-OH) 3.2 Å; 124°; Thr205 (sidechain OH−/sugar C2-OH) 2.5 Å; 154°; Tyr299 (sidechain OH/C1-galloyl 6′-OH) 2.3 Å; 127°; Asp327 (sidechain CO −/C1-galloyl 4′-OH) 2.5 Å; 154°; Asp542 (sidechain CO −/sugar C2-OH) 2.5 Å; 137°; Asp542 (sidechain C=O/C1-galloyl 3′-OH) 2.0 Å; 157°; Leu577 (mainchain NH/C6-galloyl 3′-OH) 3.2 Å; 144°; His600 (sidechain NH/C1-galloyl 4′-OH) | Tyr299, Ile328, Ile364, Trp406, Trp441, Phe450, Trp539, Phe575, Ala576, Leu577, Tyr605 | Tyr299 (H-π) Phe575 (H-π) |
| Canonical Domains Forming Substrate-Binding Site | Comprising Residues | Acarbose | 1,3,6- Trigalloylglucose |
|---|---|---|---|
| N-terminal β-sheet domain | Arg202 | −0.3 | −0.1 |
| Asp203 | −0.4 | −0.0 | |
| Thr204 | −0.1 | −0.1 | |
| Thr205 | −0.0 | −0.0 | |
| Pro206 | −2.5 | −2.2 | |
| Asn207 | −0.3 | −0.1 | |
| Asn209 | −0.6 | −0.1 | |
| Thr211 | 0.2 | 0.4 | |
| Tyr214 | 0.1 | 0.2 | |
| Catalytic GH-31 domain | Arg298 | 0.3 | 0.3 |
| Tyr299 | 0.0 | 0.3 | |
| Asp327 | 0.4 | 0.3 | |
| Ile328 | −0.2 | 0.0 | |
| Ile364 | −0.0 | −0.0 | |
| Trp441 | −0.1 | −0.1 | |
| Asp443 | −0.3 | −0.0 | |
| Met444 | −0.2 | −0.0 | |
| Ser448 | −0.2 | −0.1 | |
| Arg526 | 0.3 | −0.4 | |
| Trp539 | 0.1 | 0.0 | |
| Gly541 | −1.0 | 0.0 | |
| Asp542 | 0.4 | 0.4 | |
| Asp571 | 0.0 | −0.1 | |
| Phe575 | 0.1 | 0.4 | |
| Ala576 | 0.1 | 0.3 | |
| Leu577 | 0.2 | 0.3 | |
| Arg598 | −0.0 | −0.1 | |
| His600 | 0.3 | 0.4 | |
| Gly602 | −0.1 | 0.2 | |
| Gln603 | −0.3 | 0.3 | |
| Phe605 | −0.3 | −0.2 | |
| Catalytic insert-I loop | Val405 | −0.1 | 0.2 |
| Trp406 | −0.1 | 0.4 | |
| Catalytic insert-II loop | Ser448 | −0.2 | −0.1 |
| Phe450 | −0.1 | 0.3 | |
| Leu473 | −0.7 | 0.7 | |
| Asp474 | −0.7 | 0.3 |
| Energy (kJ/mol ± SD) | Ligand-Protein Complex | |
|---|---|---|
| Acarbose | 1,3,6-trigalloylglucose | |
| ΔGvan der Waals | −52.6 ± 10.9 | −147.6 ± 20.2 |
| ΔGElectrostatic | −280.1 ± 32.5 | −165.2 ± 38.7 |
| ΔGSolvation; Polar | 311.6 ± 29.7 | 267.6 ± 56.3 |
| ΔGSolvation; non-polar; SASA | −19.5 ± 0.8 | −24.5 ± 3.5 |
| ΔGTotal binding | −40.7 ± 22.8 | −69.6 ± 24.9 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abdallah, H.M.; Kashegari, A.T.; Shalabi, A.A.; Darwish, K.M.; El-Halawany, A.M.; Algandaby, M.M.; Ibrahim, S.R.M.; Mohamed, G.A.; Abdel-Naim, A.B.; Koshak, A.E.; et al. Phenolics from Chrozophora oblongifolia Aerial Parts as Inhibitors of α-Glucosidases and Advanced Glycation End Products: In-Vitro Assessment, Molecular Docking and Dynamics Studies. Biology 2022, 11, 762. https://doi.org/10.3390/biology11050762
Abdallah HM, Kashegari AT, Shalabi AA, Darwish KM, El-Halawany AM, Algandaby MM, Ibrahim SRM, Mohamed GA, Abdel-Naim AB, Koshak AE, et al. Phenolics from Chrozophora oblongifolia Aerial Parts as Inhibitors of α-Glucosidases and Advanced Glycation End Products: In-Vitro Assessment, Molecular Docking and Dynamics Studies. Biology. 2022; 11(5):762. https://doi.org/10.3390/biology11050762
Chicago/Turabian StyleAbdallah, Hossam M., Albraa T. Kashegari, Akram A. Shalabi, Khaled M. Darwish, Ali M. El-Halawany, Mardi M. Algandaby, Sabrin R. M. Ibrahim, Gamal A. Mohamed, Ashraf B. Abdel-Naim, Abdulrahman E. Koshak, and et al. 2022. "Phenolics from Chrozophora oblongifolia Aerial Parts as Inhibitors of α-Glucosidases and Advanced Glycation End Products: In-Vitro Assessment, Molecular Docking and Dynamics Studies" Biology 11, no. 5: 762. https://doi.org/10.3390/biology11050762
APA StyleAbdallah, H. M., Kashegari, A. T., Shalabi, A. A., Darwish, K. M., El-Halawany, A. M., Algandaby, M. M., Ibrahim, S. R. M., Mohamed, G. A., Abdel-Naim, A. B., Koshak, A. E., Proksch, P., & Elhady, S. S. (2022). Phenolics from Chrozophora oblongifolia Aerial Parts as Inhibitors of α-Glucosidases and Advanced Glycation End Products: In-Vitro Assessment, Molecular Docking and Dynamics Studies. Biology, 11(5), 762. https://doi.org/10.3390/biology11050762













