Non-Coding Transcriptome Provides Novel Insights into the Escherichia coli F17 Susceptibility of Sheep Lamb
Abstract
:Simple Summary
Abstract
1. Introduction
2. Material and Methods
2.1. Sample Collection
2.2. RNA Extraction and Sequencing
2.3. Analysis of miRNA and circRNA Expression
2.4. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) Functional Analyses
2.5. Identification of Potential circRNA/miRNA Biomarkers for E. coli F17 Infection Using Machine Learning Methods
2.6. Acquisition of lncRNA and mRNA Expression Dataset
2.7. ceRNA Network Construction
2.8. Validation of Sequencing Data
3. Results
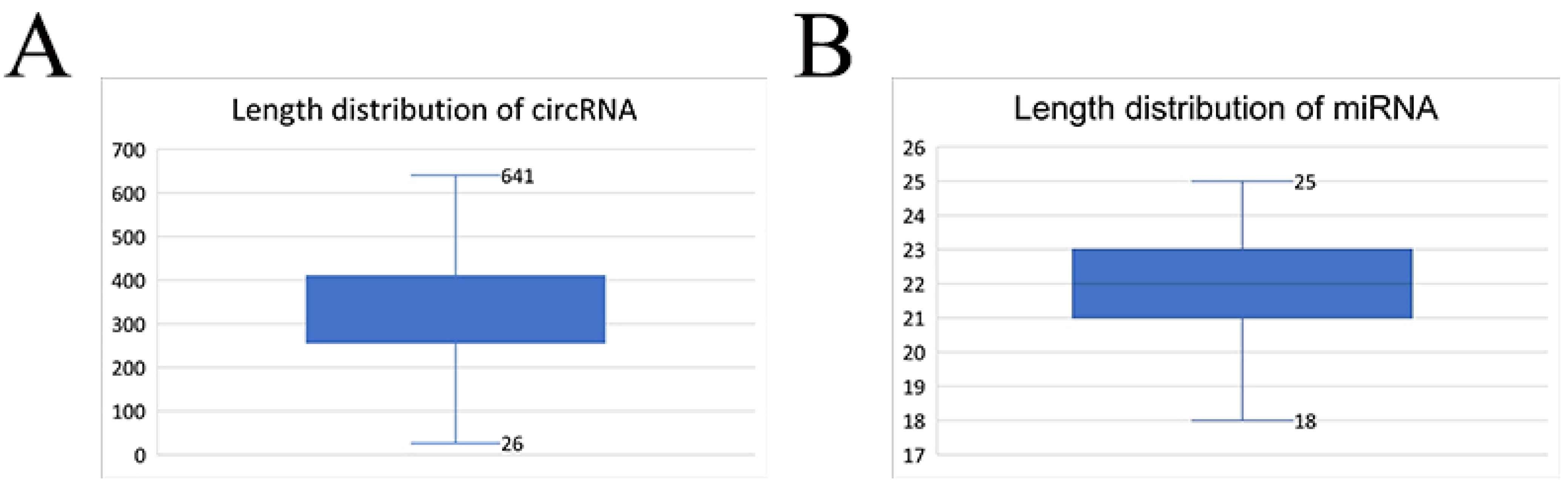
3.1. Overview of the Sequencing Data
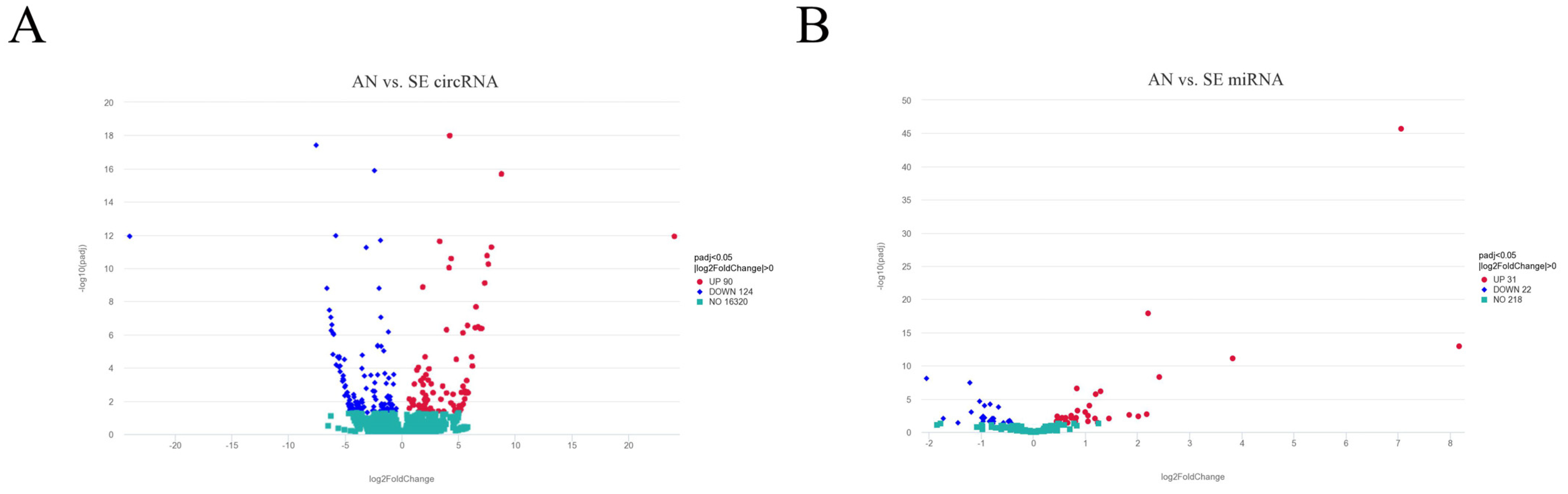
3.2. Differentially Expressed circRNAs and miRNAs
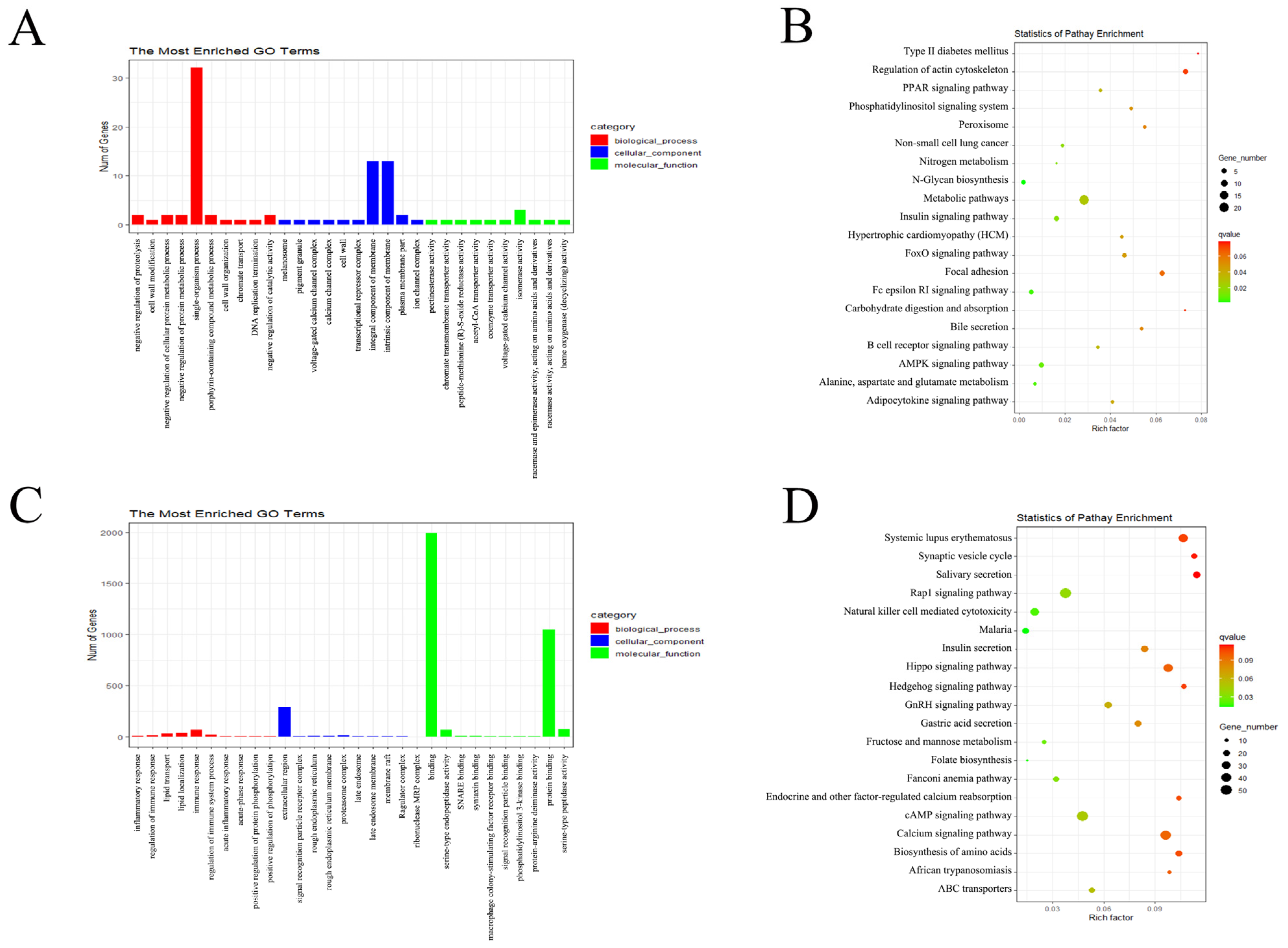
3.3. Functional Analysis
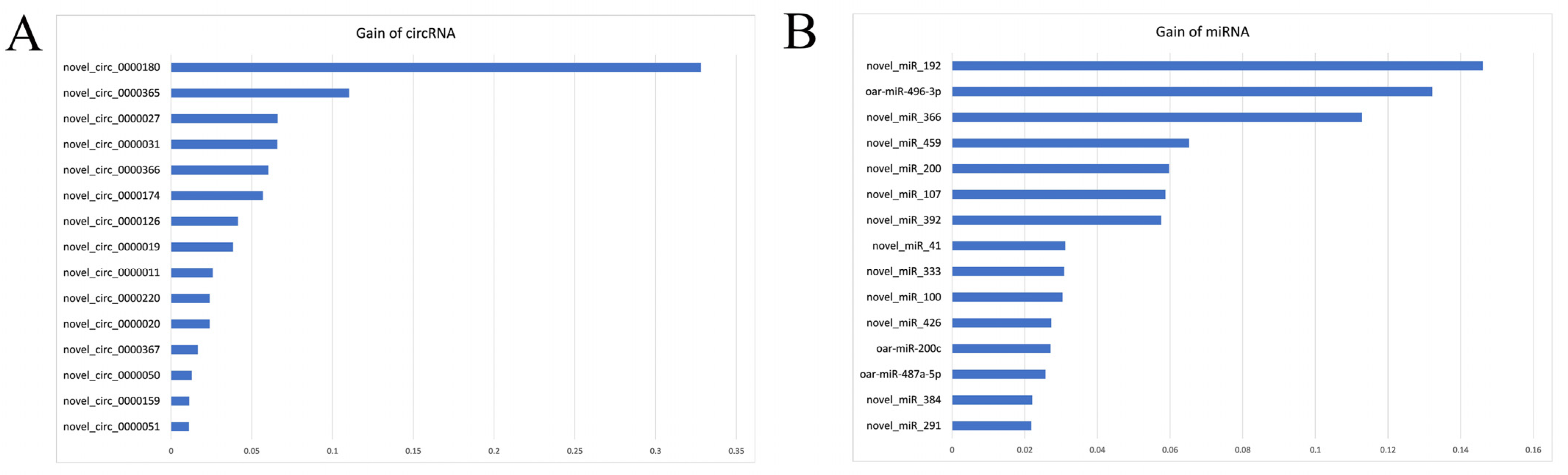
3.4. Potential circRNA/miRNA Biomarkers for E. coli F17 Infection
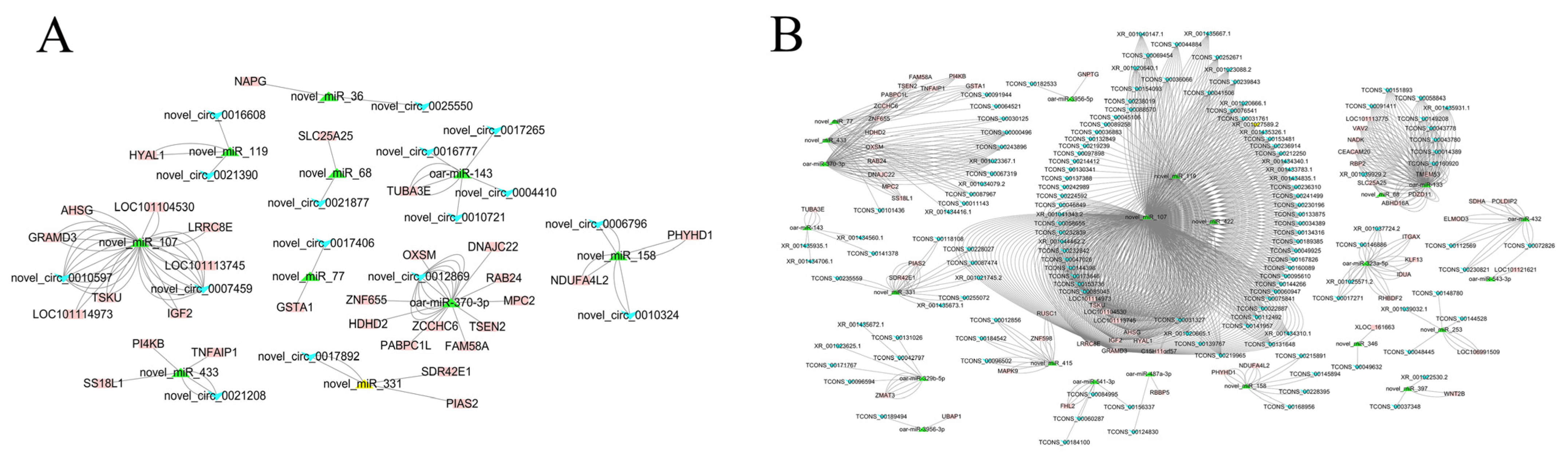
3.5. ceRNA Network
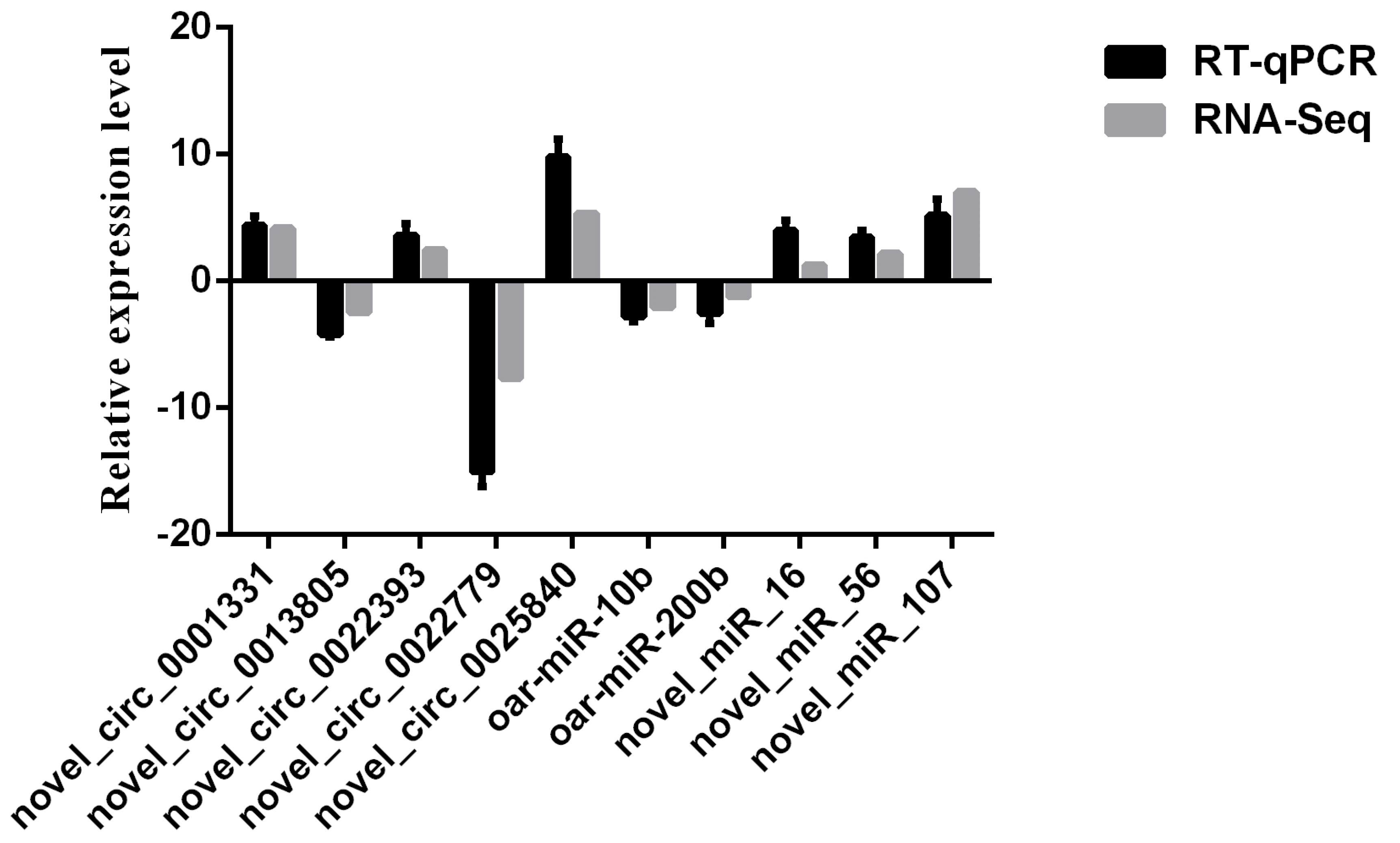
3.6. Validation of Sequencing Data
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Kaper, J.B.; Nataro, J.P.; Mobley, H.L.T. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2004, 2, 123–140. [Google Scholar] [CrossRef] [PubMed]
- Levine, M.M. Escherichia coli that Cause Diarrhea: Enterotoxigenic, Enteropathogenic, Enteroinvasive, Enterohemorrhagic, and Enteroadherent. J. Infect. Dis. 1987, 155, 377–389. [Google Scholar] [CrossRef] [PubMed]
- Bandyopadhyay, S.; Mahanti, A.; Lodh, C.; Samanta, I.; Biswas, T.K.; Dutta, T.K.; Baruah, K.K.; Bhattacharya, D. The prevalence and drug resistance profile of Shiga-toxin producing (STEC), enteropathogenic (EPEC) and enterotoxigenic (ETEC) Escherichia coli in free ranging diarrheic and non-diarrheic yaks of West Kameng, Arunachal Pradesh, India. Vet. Arh. 2015, 85, 501–510. [Google Scholar]
- Cheng, D.R.; Zhu, S.Y.; Su, Z.R.; Zuo, W.Y.; Lu, H. Prevalence of the E. coli type three secretion system 2 (ETT2) locus among enterotoxigenic E. coli ( ETEC), shigatoxin-producing E. coli (STEC) from weaned piglets. Afr. J. Microbiol. Res. 2011, 5, 4697–4701. [Google Scholar] [CrossRef]
- Ogundare, S.T.; Fasanmi, O.G.; Fasina, F.O. Risk Factors for Prevalence of Enterotoxigenic Escherichia coli (ETEC) in Diarrheic and Non-diarrheic Neonatal and Weaner Pigs, South Africa. Biomed. Environ. Sci. 2018, 31, 149–154. [Google Scholar] [CrossRef]
- Subekti, D.S.; Lesmana, M.; Tjaniadi, P.; Machpud, N.; Sriwati; Sukarma; Daniel, J.C.; Alexander, W.K.; Campbell, J.R.; Corwin, A.L.; et al. Prevalence of enterotoxigenic Escherichia coli (ETEC) in hospitalized acute diarrhea patients in Denpasar, Bali, Indonesia. Diagn. Microbiol. Infect. Dis. 2003, 47, 399–405. [Google Scholar] [CrossRef]
- Isidean, S.D.; Riddle, M.S.; Savarino, S.J.; Porter, C.K. A systematic review of ETEC epidemiology focusing on colonization factor and toxin expression. Vaccine 2011, 29, 6167–6178. [Google Scholar] [CrossRef]
- Kolenda, R.; Burdukiewicz, M.; Schierack, P. A systematic review and meta-analysis of the epidemiology of pathogenic Escherichia coli of calves and the role of calves as reservoirs for human pathogenic E. coli. Front. Cell. Infect. Microbiol. 2015, 5, 23. [Google Scholar] [CrossRef]
- Weiner, M.; Dacko, J.; Osek, J. Correlation between the presence of F5, F6, F17, F18, F41 fimbriae and the toxicity profile in Escherichia coli strains isolated from piglets with diarrhea. Med. Weter. 2004, 60, 1342–1346. [Google Scholar]
- Cid, D.; Sanz, R.; Marin, I.; De Greve, H.; Ruiz-Santa-Quiteria, J.A.; Amils, R.; De la Fuente, R. Characterization of nonenterotoxigenic Escherichia coli strains producing F17 fimbriae isolated from diarrheic lambs and goat kids. J. Clin. Microbiol. 1999, 37, 1370–1375. [Google Scholar] [CrossRef] [Green Version]
- Bertagna, F.; Treglia, G.; Orlando, E.; Dognini, L.; Giovanella, L.; Sadeghi, R.; Giubbini, R. Prevalence and clinical significance of incidental F18-FDG breast uptake: A systematic review and meta-analysis. Jpn. J. Radiol. 2014, 32, 59–68. [Google Scholar] [CrossRef]
- Kwon, D.; Choi, C.; Jung, T.; Chung, H.K.; Kim, J.P.; Bae, S.S.; Cho, W.S.; Kim, J.; Chae, C. Genotypic prevalence of the fimbrial adhesins (F4 F5, F6, F41 and F18) and toxins (LT, STa. STb and Stx2e) in Escherichia coli isolated from postweaning pigs with diarrhoea or oedema disease in Korea. Vet. Rec. 2002, 150, 35–37. [Google Scholar] [CrossRef] [PubMed]
- Ryu, J.H.; Kim, S.; Park, J.; Choi, K.S. Characterization of virulence genes in Escherichia coli strains isolated from pre-weaned calves in the Republic of Korea. Acta Vet. Scand. 2020, 62, 45. [Google Scholar] [CrossRef] [PubMed]
- Siuce, J.; Maturrano, L.; Wheeler, J.C.; Rosadio, R. Diarrheagenic Escherichia coli isolates from neonatal alpacas mainly display F17 fimbriae adhesion gene. Trop. Anim. Health Prod. 2020, 52, 3917–3921. [Google Scholar] [CrossRef] [PubMed]
- Bihannic, M.; Ghanbarpour, R.; Auvray, F.; Cavalie, L.; Chatre, P.; Boury, M.; Brugere, H.; Madec, J.Y.; Oswald, E. Identification and detection of three new F17 fimbrial variants in Escherichia coli strains isolated from cattle. Vet. Res. 2014, 45, 76. [Google Scholar] [CrossRef] [Green Version]
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef]
- Capel, B.; Swain, A.; Nicolis, S.; Hacker, A.; Walter, M.; Koopman, P.; Goodfellow, P.; Lovell-Badge, R. Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell 1993, 73, 1019–1030. [Google Scholar] [CrossRef]
- Cai, Y.F.; Wan, J. Competing Endogenous RNA Regulations in Neurodegenerative Disorders: Current Challenges and Emerging Insights. Front. Mol. Neurosci. 2018, 11, 370. [Google Scholar] [CrossRef] [Green Version]
- Jin, W.; Ibeagha-Awemu, E.M.; Liang, G.; Beaudoin, F.; Zhao, X.; Guan, L.L. Transcriptome microRNA profiling of bovine mammary epithelial cells challenged with Escherichia coli or Staphylococcus aureus bacteria reveals pathogen directed microRNA expression profiles. BMC Genom. 2014, 15, 181. [Google Scholar] [CrossRef] [Green Version]
- Naeem, A.; Zhong, K.; Moisa, S.J.; Drackley, J.K.; Moyes, K.M.; Loor, J.J. Bioinformatics analysis of microRNA and putative target genes in bovine mammary tissue infected with Streptococcus uberis. J. Dairy Sci. 2012, 95, 6397–6408. [Google Scholar] [CrossRef] [Green Version]
- Julie, A.H.; Yoo, D.; Liu, H.C. Characterization of the microRNAome in porcine reproductive and respiratory syndrome virus infected macrophages. PLoS ONE 2013, 8, e82054. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Q.; Guo, X.K.; Gao, L.; Huang, C.; Li, N.; Jia, X.; Liu, W.; Feng, W.H. MicroRNA-23 inhibits PRRSV replication by directly targeting PRRSV RNA and possibly by upregulating type I interferons. Virology 2014, 450–451, 182–195. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; You, Z.; Wang, M.; Yuan, Y.; Liu, C.; Yang, N.; Zhang, H.; Lian, L. Genome-wide analysis of circular RNAs involved in Marek’s disease tumourigenesis in chickens. RNA Biol. 2020, 17, 517–527. [Google Scholar] [CrossRef]
- Tian, F.; Luo, J.; Zhang, H.; Chang, S.; Song, J. MiRNA expression signatures induced by Marek's disease virus infection in chickens. Genomics 2012, 99, 152–159. [Google Scholar] [CrossRef] [Green Version]
- Salmena, L.; Poliseno, L.; Tay, Y.; Kats, L.; Pandolfi, P.P. A ceRNA hypothesis: The Rosetta Stone of a hidden RNA language? Cell 2011, 146, 353–358. [Google Scholar] [CrossRef] [Green Version]
- Yang, R.C.; Chen, J.Q.; Xu, B.J.; Yang, B.; Fu, J.Y.; Xiao, S.Y.; Tan, C.; Chen, H.C.; Wang, X.R. circ_2858 Helps Blood-Brain Barrier Disruption by Increasing VEGFA via Sponging miR-93-5p during Escherichia coli Meningitis. Mol. Ther. Nucleic Acids 2020, 22, 708–721. [Google Scholar] [CrossRef]
- Xu, B.J.; Yang, R.C.; Fu, J.Y.; Yang, B.; Chen, J.Q.; Tan, C.; Chen, H.C.; Wang, X.R. LncRSPH9-4 Facilitates Meningitic Escherichia coli-Caused Blood-Brain Barrier Disruption via miR-17-5p/MMP3 Axis. Int. J. Mol. Sci. 2021, 22, 6343. [Google Scholar] [CrossRef]
- Dai, C.H.; Wang, F.; Wang, S.Q.; Wu, Z.C.; Wu, S.L.; Bao, W.B. miR-215 Targeting Novel Genes EREG, NIPAL1 and PTPRU Regulates the Resistance to E.coli F18 in Piglets. Genes 2020, 11, 1050. [Google Scholar] [CrossRef]
- Sun, L.; Wu, S.; Dai, C.-H.; Sun, S.-Y.; Zhu, G.-Q.; Wu, S.-L.; Bao, W.-B. Insight into the molecular mechanism of miR-192 regulating Escherichia coli resistance in piglets. Biosci. Rep. 2018, 38, BSR20171160. [Google Scholar] [CrossRef] [Green Version]
- Jin, C.; Bao, J.; Wang, Y.; Chen, W.; Wu, T.; Wang, L.; Lv, X.; Gao, W.; Wang, B.; Zhu, G.; et al. Changes in long non-coding RNA expression profiles related to the antagonistic effects of Escherichia coli F17 on lamb spleens. Sci. Rep. 2018, 8, 16514. [Google Scholar] [CrossRef] [Green Version]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [Green Version]
- Friedlander, M.R.; Mackowiak, S.D.; Li, N.; Chen, W.; Rajewsky, N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 2012, 40, 37–52. [Google Scholar] [CrossRef]
- Wen, M.; Shen, Y.; Shi, S.; Tang, T. miREvo: An integrative microRNA evolutionary analysis platform for next-generation sequencing experiments. BMC Bioinform. 2012, 13, 140. [Google Scholar] [CrossRef] [Green Version]
- Memczak, S.; Jens, M.; Elefsinioti, A.; Torti, F.; Krueger, J.; Rybak, A.; Maier, L.; Mackowiak, S.D.; Gregersen, L.H.; Munschauer, M.; et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013, 495, 333–338. [Google Scholar] [CrossRef]
- Gao, Y.; Wang, J.; Zhao, F. CIRI: An efficient and unbiased algorithm for de novo circular RNA identification. Genome Biol. 2015, 16, 4. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Feng, Z.; Wang, X.; Wang, X.; Zhang, X. DEGseq: An R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 2010, 26, 136–138. [Google Scholar] [CrossRef]
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol. 2010, 11, R14. [Google Scholar] [CrossRef] [Green Version]
- Xie, C.; Mao, X.; Huang, J.; Ding, Y.; Wu, J.; Dong, S.; Kong, L.; Gao, G.; Li, C.Y.; Wei, L. KOBAS 2.0: A web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011, 39, W316–W322. [Google Scholar] [CrossRef] [Green Version]
- Benjamini, Y.; Drai, D.; Elmer, G.; Kafkafi, N.; Golani, I. Controlling the false discovery rate in behavior genetics research. Behav. Brain Res. 2001, 125, 279–284. [Google Scholar] [CrossRef] [Green Version]
- Wiener, A.L.a.M. Classification and Regression by randomForest. R News 2002, 2, 18–22. [Google Scholar]
- Chen, T.; He, T.; Michael, B.; Vadim, K.; Tang, Y.; Hyunsu, C.; Chen, K.; Rory, M.; Ignacio, C.; Zhou, T.; et al. xgboost: Extreme Gradient Boosting. R Package Version 1.5.0.1. 2001. Available online: https://CRAN.R-project.org/package=xgboost (accessed on 5 October 2021).
- Chen, W.; Alexandre, P.A.; Ribeiro, G.; Fukumasu, H.; Sun, W.; Reverter, A.; Li, Y. Identification of Predictor Genes for Feed Efficiency in Beef Cattle by Applying Machine Learning Methods to Multi-Tissue Transcriptome Data. Front. Genet. 2021, 12, 619857. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [Green Version]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [Green Version]
- Sun, L.; Luo, H.T.; Bu, D.C.; Zhao, G.G.; Yu, K.T.; Zhang, C.H.; Liu, Y.N.; Chen, R.S.; Zhao, Y. Utilizing sequence intrinsic composition to classify protein-coding and long non-coding transcripts. Nucleic Acids Res. 2013, 41, e166. [Google Scholar] [CrossRef]
- Kang, Y.J.; Yang, D.C.; Kong, L.; Hou, M.; Meng, Y.Q.; Wei, L.P.; Gao, G. CPC2: A fast and accurate coding potential calculator based on sequence intrinsic features. Nucleic Acids Res. 2017, 45, W12–W16. [Google Scholar] [CrossRef] [Green Version]
- El-Gebali, S.; Mistry, J.; Bateman, A.; Eddy, S.R.; Luciani, A.; Potter, S.C.; Qureshi, M.; Richardson, L.J.; Salazar, G.A.; Smart, A.; et al. The Pfam protein families database in 2019. Nucleic Acids Res. 2019, 47, D427–D432. [Google Scholar] [CrossRef]
- Enright, A.J.; John, B.; Gaul, U.; Tuschl, T.; Sander, C.; Marks, D.S. MicroRNA targets in Drosophila. Genome Biol. 2003, 5, R1. [Google Scholar] [CrossRef] [Green Version]
- Rehmsmeier, M.; Steffen, P.; Hochsmann, M.; Giegerich, R. Fast and effective prediction of microRNA/target duplexes. RNA 2004, 10, 1507–1517. [Google Scholar] [CrossRef] [Green Version]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(-Delta Delta C) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Jin, C.Y.; Bao, J.J.; Wang, Y.; Chen, W.H.; Zou, S.X.; Wu, T.Y.; Wang, L.H.; Lv, X.Y.; Gao, W.; Wang, B.Z.; et al. Changes in circRNA expression profiles related to the antagonistic effects of Escherichia coli F17 in lamb spleens. Sci. Rep. UK 2018, 8, 14524. [Google Scholar] [CrossRef]
- Zhao, X.W.; Huang, D.W.; Zhu, H.L.; Pan, X.C.; Wang, X.X.; Qi, Y.X.; Cheng, G.L.; Zhao, H.L.; Yang, Y.X. Alterations of the circular RNA profile in the jejunum of neonatal calves in response to colostrum and milk feeding. J. Dairy Sci. 2019, 102, 7038–7048. [Google Scholar] [CrossRef]
- Chen, J.N.; Wang, H.W.; Jin, L.; Wang, L.Y.; Huang, X.; Chen, W.W.; Yan, M.M.; Liu, G.L. Profile analysis of circRNAs induced by porcine endemic diarrhea virus infection in porcine intestinal epithelial cells. Virology 2019, 527, 169–179. [Google Scholar] [CrossRef]
- Ebbesen, K.K.; Hansen, T.B.; Kjems, J. Insights into circular RNA biology. RNA Biol. 2017, 14, 1035–1045. [Google Scholar] [CrossRef]
- Altirriba, J.; Gasa, R.; Casas, S.; Ramirez-Bajo, M.J.; Ros, S.; Gutierrez-Dalmau, A.; de Villa, M.C.R.; Barbera, A.; Gomis, R. The role of transmembrane protein 27 (TMEM27) in islet physiology and its potential use as a beta cell mass biomarker. Diabetologia 2010, 53, 1406–1414. [Google Scholar] [CrossRef] [Green Version]
- Di Zanni, E.; Gradogna, A.; Picco, C.; Scholz-Starke, J.; Boccaccio, A. TMEM16E/ANO5 mutations related to bone dysplasia or muscular dystrophy cause opposite effects on lipid scrambling. Hum. Mutat. 2020, 41, 1157–1170. [Google Scholar] [CrossRef] [Green Version]
- Tan, W.X.; Sun, G.; Shangguan, M.Y.; Gui, Z.; Bao, Y.; Li, Y.F.; Jia, Z.H. Novel role of lncRNA CHRF in cisplatin resistance of ovarian cancer is mediated by miR-10b induced EMT and STAT3 signaling. Sci. Rep. UK 2020, 10, 14768. [Google Scholar] [CrossRef]
- Wimmer, M.; Zauner, R.; Ablinger, M.; Pinon-Hofbauer, J.; Guttmann-Gruber, C.; Reisenberger, M.; Lettner, T.; Niklas, N.; Proell, J.; Sajinovic, M.; et al. A cancer stem cell-like phenotype is associated with miR-10b expression in aggressive squamous cell carcinomas. Cell Commun. Signal. 2020, 18, 61. [Google Scholar] [CrossRef] [Green Version]
- Gabriely, G.; Yi, M.; Narayan, R.S.; Niers, J.M.; Wurdinger, T.; Imitola, J.; Ligon, K.L.; Kesari, S.; Esau, C.; Stephens, R.M.; et al. Human Glioma Growth Is Controlled by MicroRNA-10b. Cancer Res. 2011, 71, 3563–3572. [Google Scholar] [CrossRef] [Green Version]
- Zhen, L.M.; Li, J.; Zhang, M.R.; Yang, K. MiR-10b decreases sensitivity of glioblastoma cells to radiation by targeting AKT. J. Biol. Res.-Thessalon. 2016, 23, 14. [Google Scholar] [CrossRef] [Green Version]
- Prager, I.; Watzl, C. Mechanisms of natural killer cell-mediated cellular cytotoxicity. J. Leukoc. Biol. 2019, 105, 1319–1329. [Google Scholar] [CrossRef]
- Shi, M.; Li, Z.Y.; Zhang, L.M.; Wu, X.Y.; Xiang, S.H.; Wang, Y.G.; Zhang, Y.Q. Hsa_circ_0007456 regulates the natural killer cell-mediated cytotoxicity toward hepatocellular carcinoma via the miR-6852-3p/ICAM-1 axis. Cell Death Dis. 2021, 12, 94. [Google Scholar] [CrossRef]
- Saxberg, A.D.; Martinez, M.; Fendley, G.A.; Zoghbi, M.E. Production of a human mitochondrial ABC transporter in E. coli. Protein Expr. Purif. 2021, 178, 105778. [Google Scholar] [CrossRef]
- Zhou, Y.; Ojeda-May, P.; Nagaraju, M.; Kim, B.; Pu, J.Z. Mapping Free Energy Pathways for ATP Hydrolysis in the E. coli ABC Transporter HlyB by the String Method. Molecules 2018, 23, 2652. [Google Scholar] [CrossRef] [Green Version]
- Verma, P.; Shakya, M. Machine learning model for predicting Major Depressive Disorder using RNA-Seq data: Optimization of classification approach. Cogn. Neurodynamics 2021, 1–11. [Google Scholar] [CrossRef]
- Lee, J.; Geng, S.; Li, S.; Li, L.W. Single Cell RNA-Seq and Machine Learning Reveal Novel Subpopulations in Low-Grade Inflammatory Monocytes with Unique Regulatory Circuits. Front. Immunol. 2021, 12, 266. [Google Scholar] [CrossRef]
- Yu, Z.Z.; Wang, Z.; Yu, X.C.; Zhang, Z. RNA-Seq-Based Breast Cancer Subtypes Classification Using Machine Learning Approaches. Comput. Intell. Neurosci. 2020, 2020, 13. [Google Scholar] [CrossRef]
- Estevez, O.; Anibarro, L.; Garet, E.; Pallares, A.; Barcia, L.; Calvino, L.; Maueia, C.; Mussa, T.; Fdez-Riverola, F.; Glez-Pena, D.; et al. An RNA-seq Based Machine Learning Approach Identifies Latent Tuberculosis Patients with an Active Tuberculosis Profile. Front. Immunol. 2020, 11, 1470. [Google Scholar] [CrossRef]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef] [Green Version]
- Meng, F.; Wehbe-Janek, H.; Henson, R.; Smith, H.; Patel, T. Epigenetic regulation of microRNA-370 by interleukin-6 in malignant human cholangiocytes. Oncogene 2008, 27, 378–386. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Zeng, J.; Zhou, M.; Li, B.; Zhang, Y.; Huang, T.; Wang, L.; Jia, J.; Chen, C. The tumor suppressive role of miRNA-370 by targeting FoxM1 in acute myeloid leukemia. Mol. Cancer 2012, 11, 56. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, K.W.; Chu, T.H.; Gong, N.R.; Chiang, W.F.; Yang, C.C.; Liu, C.J.; Wu, C.H.; Lin, S.C. miR-370 modulates insulin receptor substrate-1 expression and inhibits the tumor phenotypes of oral carcinoma. Oral Dis. 2013, 19, 611–619. [Google Scholar] [CrossRef]
- Xu, W.P.; Yi, M.; Li, Q.Q.; Zhou, W.P.; Cong, W.M.; Yang, Y.; Ning, B.F.; Yin, C.; Huang, Z.W.; Wang, J.; et al. Perturbation of MicroRNA-370/Lin-28 homolog A/nuclear factor kappa B regulatory circuit contributes to the development of hepatocellular carcinoma. Hepatology 2013, 58, 1977–1991. [Google Scholar] [CrossRef]
- Ye, L.; Su, X.; Wu, Z.; Zheng, X.; Wang, J.; Zi, C.; Zhu, G.; Wu, S.; Bao, W. Analysis of differential miRNA expression in the duodenum of Escherichia coli F18-sensitive and -resistant weaned piglets. PLoS ONE 2012, 7, e43741. [Google Scholar] [CrossRef]
- Chen, Y.; Zhao, Y.; Chen, W.; Xie, L.; Zhao, Z.A.; Yang, J.; Chen, Y.; Lei, W.; Shen, Z. MicroRNA-133 overexpression promotes the therapeutic efficacy of mesenchymal stem cells on acute myocardial infarction. Stem Cell Res. Ther. 2017, 8, 268. [Google Scholar] [CrossRef]
- Zhang, G.; Wang, J.; Zheng, R.; Song, B.; Huang, L.; Liu, Y.; Hao, Y.; Bai, X. MiR-133 Targets YES1 and Inhibits the Growth of Triple-Negative Breast Cancer Cells. Technol. Cancer Res. Treat. 2020, 19, 1533033820927011. [Google Scholar] [CrossRef]
- Almeida Junior, L.D.; Quaglio, A.E.V.; de Almeida Costa, C.A.R.; Di Stasi, L.C. Intestinal anti-inflammatory activity of Ground Cherry (Physalis angulata L.) standardized CO2 phytopharmaceutical preparation. World J. Gastroenterol. 2017, 23, 4369–4380. [Google Scholar] [CrossRef]
| Sample | Raw Reads | Clean Reads | Mapping Rate (%) | Error Rate (%) | Q20 (%) | Q30 (%) | GC Content (%) |
|---|---|---|---|---|---|---|---|
| AN1 | 86,448,964 | 85,310,470 | 98.68 | 0.03 | 97.55 | 93.27 | 51.32 |
| AN2 | 82,985,976 | 82,314,372 | 99.19 | 0.03 | 97.00 | 91.76 | 46.32 |
| AN3 | 81,095,934 | 79,701,960 | 98.28 | 0.03 | 97.49 | 93.16 | 51.61 |
| AN4 | 94,502,330 | 93,722,960 | 99.18 | 0.03 | 97.25 | 92.49 | 48.07 |
| AN5 | 84,496,940 | 83,246,004 | 98.52 | 0.03 | 97.45 | 93.06 | 50.25 |
| AN6 | 83,613,850 | 82,012,052 | 98.08 | 0.03 | 97.49 | 93.19 | 54.43 |
| SE1 | 82,325,980 | 81,420,394 | 98.90 | 0.03 | 97.31 | 92.67 | 52.18 |
| SE2 | 83,101,628 | 81,439,640 | 98.00 | 0.03 | 97.39 | 93.00 | 48.07 |
| SE3 | 83,731,304 | 82,241,834 | 98.22 | 0.03 | 97.45 | 93.09 | 49.79 |
| SE4 | 80,794,124 | 79,478,658 | 98.37 | 0.03 | 96.90 | 91.99 | 56.07 |
| SE5 | 92,174,900 | 90,902,860 | 98.62 | 0.03 | 97.35 | 92.99 | 49.55 |
| SE6 | 84,577,884 | 83,190,218 | 98.36 | 0.03 | 96.54 | 91.14 | 49.89 |
| Sample Name | Raw Reads | Clean Reads | Clean Bases | Error Rate (%) | Q20 (%) | Q30 (%) | GC Content (%) |
|---|---|---|---|---|---|---|---|
| AN1 | 16,273,383 | 16,059,313 | 98.68 | 0.01 | 99.49 | 98.16 | 49.67 |
| AN2 | 13,434,545 | 13,274,363 | 98.81 | 0.01 | 99.50 | 98.35 | 48.56 |
| AN3 | 14,558,297 | 14,136,725 | 97.10 | 0.01 | 99.06 | 96.65 | 48.87 |
| AN4 | 11,883,680 | 11,545,885 | 97.16 | 0.01 | 99.10 | 96.96 | 49.54 |
| AN5 | 15,402,425 | 15,008,710 | 97.44 | 0.01 | 99.04 | 96.79 | 49.09 |
| AN6 | 11,625,621 | 10,918,820 | 93.92 | 0.01 | 99.30 | 97.26 | 50.01 |
| SE1 | 18,148,953 | 17,949,815 | 98.90 | 0.01 | 99.49 | 98.30 | 48.85 |
| SE2 | 13,392,060 | 13,198,054 | 98.55 | 0.01 | 99.32 | 97.92 | 49.46 |
| SE3 | 10,839,760 | 10,594,527 | 97.74 | 0.01 | 99.34 | 97.74 | 49.80 |
| SE4 | 13,718,249 | 13,297,138 | 96.93 | 0.01 | 99.02 | 97.04 | 49.09 |
| SE5 | 12,498,474 | 11,906,416 | 95.26 | 0.01 | 98.97 | 96.90 | 50.58 |
| SE6 | 11,896,114 | 11,605,384 | 97.56 | 0.01 | 99.33 | 97.73 | 48.81 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, W.; Lv, X.; Zhang, W.; Hu, T.; Cao, X.; Ren, Z.; Getachew, T.; Mwacharo, J.M.; Haile, A.; Sun, W. Non-Coding Transcriptome Provides Novel Insights into the Escherichia coli F17 Susceptibility of Sheep Lamb. Biology 2022, 11, 348. https://doi.org/10.3390/biology11030348
Chen W, Lv X, Zhang W, Hu T, Cao X, Ren Z, Getachew T, Mwacharo JM, Haile A, Sun W. Non-Coding Transcriptome Provides Novel Insights into the Escherichia coli F17 Susceptibility of Sheep Lamb. Biology. 2022; 11(3):348. https://doi.org/10.3390/biology11030348
Chicago/Turabian StyleChen, Weihao, Xiaoyang Lv, Weibo Zhang, Tingyan Hu, Xiukai Cao, Ziming Ren, Tesfaye Getachew, Joram M. Mwacharo, Aynalem Haile, and Wei Sun. 2022. "Non-Coding Transcriptome Provides Novel Insights into the Escherichia coli F17 Susceptibility of Sheep Lamb" Biology 11, no. 3: 348. https://doi.org/10.3390/biology11030348
APA StyleChen, W., Lv, X., Zhang, W., Hu, T., Cao, X., Ren, Z., Getachew, T., Mwacharo, J. M., Haile, A., & Sun, W. (2022). Non-Coding Transcriptome Provides Novel Insights into the Escherichia coli F17 Susceptibility of Sheep Lamb. Biology, 11(3), 348. https://doi.org/10.3390/biology11030348






