Genome-Wide Association Study Revealed the Effect of rs312715211 in ZNF652 Gene on Abdominal Fat Percentage of Chickens
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Population and Sample Collection
2.2. Genotyping and Quality Control
2.3. Genome-Wide Association Study
2.4. Fixation Indices (FST) and Heterozygosity (π)
2.5. Dual-Luciferase Reporter Assay
2.6. RNA-Seq and Weighted Gene Correlation Network Analysis (WGCNA)
2.7. Kyoto Encyclopedia of Genes and Genomes (KEGG)
2.8. Statistical Analysis
3. Results
3.1. Phenotypic Statistics and Heritability Evaluation
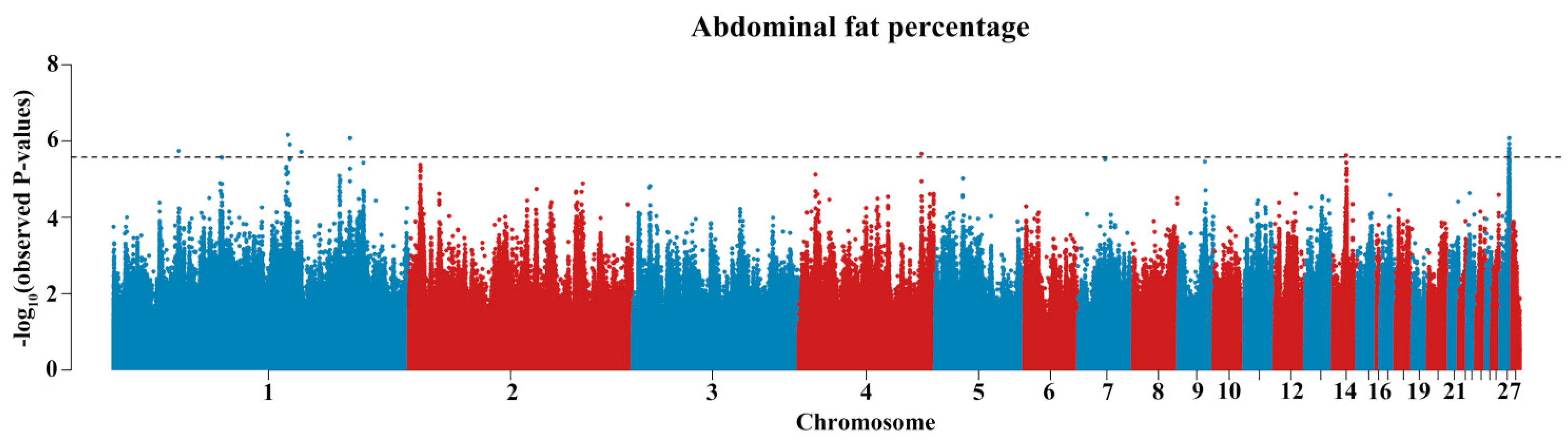
3.2. GWAS Identified the Effective Variants and Candidate Genes
3.3. rs312715211 in the Intron Region of ZNF652 Was the Primary Variant Associated with AFP
3.4. rs312715211 Can Affect the Activity of ZNF652 Promoter
3.5. rs312715211 Mutation Can Increase AFP and AFW and Decrease ECW
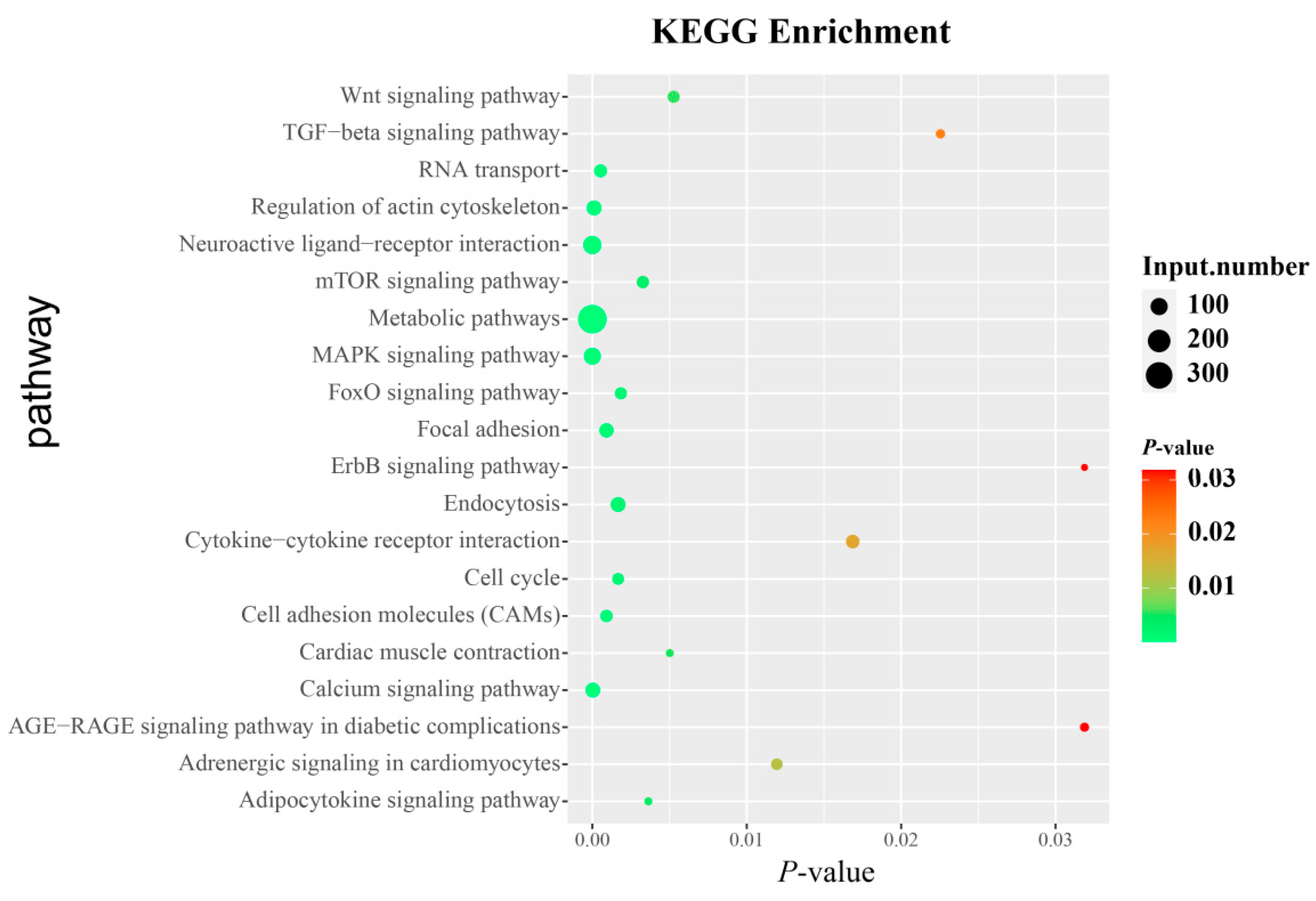
3.6. Identification of Candidate Genes and Pathways Related to AFP
3.7. ZNF652 May Regulate Abdominal Fat and BW through MAPK/FoxO Signaling Pathways
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AFW | Abdominal fat weight |
| AFP | Abdominal fat percentage |
| ECW | Eviscerated carcass weight |
| IMF | Intramuscular fat |
| GWAS | Genome-wide association study |
| LMM | Linear mixed model |
| PCA | Principal component analysis |
| LD | Linkage disequilibrium |
| SNP | Single nucleotide polymorphism |
| FST | Fixation indices |
| Heterozygosity | π |
| WGCNA | RNA-seq and weighted gene correlation network analysis |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
References
- Zerehdaran, S.A.L.J.; Vereijken, A.J.; Van Arendonk, J.A.M.; Van der Waaijt, E.H. Estimation of genetic parameters for fat deposition and carcass traits in broilers. Poult Sci. 2004, 83, 521–525. [Google Scholar] [CrossRef] [PubMed]
- Moreira, G.C.M.; Godoy, T.F.; Boschiero, C.; Gheyas, A.; Gasparin, G.; Andrade, S.C.S.; Paduan, M.; Montenegro, H.; Burt, D.W.; Ledur, M.C.; et al. Variant discovery in a QTL region on chromosome 3 associated with fatness in chickens. Anim. Genet. 2015, 46, 141–147. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Cheng, B.H.; Yang, L.L.; Wang, Z.P.; Zhang, H.L.; Xu, S.S.; Wang, S.Z.; Wang, Y.X.; Zhang, H.; Li, H. Identification of a potential functional single nucleotide polymorphism for fatness and growth traits in the 3′-untranslated region of the PCSK1 gene in chickens. J. Anim. Sci. 2017, 95, 4776–4786. [Google Scholar] [CrossRef] [PubMed]
- Chao, X.; Guo, L.; Wang, Q.; Huang, W.; Liu, M.; Luan, K.; Jiang, J.; Lin, S.; Nie, Q.; Luo, W.; et al. miR-429-3p/LPIN1 axis promotes chicken abdominal fat deposition via PPARγ pathway. Front. Cell Dev. Biol. 2020, 8, 595637. [Google Scholar] [CrossRef]
- Huang, H.Y.; Liu, R.R.; Zhao, G.P.; Li, Q.H.; Zheng, M.Q.; Zhang, J.J.; Li, S.F.; Liang, Z.; Wen, J. Integrated analysis of microRNA and mRNA expression profiles in abdominal adipose tissues in chickens. Sci. Rep. 2015, 5, 16132. [Google Scholar] [CrossRef] [PubMed]
- Chabault, M.; Baéza, E.; Gigaud, V.; Chartrin, P.; Chapuis, H.; Boulay, M.; Arnould, C.; D’Abbadie, F.; Berri, C.; Bihan-Duval, L. Analysis of a slow-growing line reveals wide genetic variability of carcass and meat quality-related traits. BMC Genet. 2012, 13, 90. [Google Scholar] [CrossRef]
- Demeure, O.; Duclos, M.J.; Bacciu, N.; Le Mignon, G.; Filangi, O.; Pitel, F.; Boland, A.; Lagarrigue, S.; Cogburn, L.A.; Simon, J.; et al. Genome-wide interval mapping using SNPs identifies new QTL for growth, body composition and several physiological variables in an F2 intercross between fat and lean chicken lines. Genet. Sel. Evol. 2013, 45, 36. [Google Scholar] [CrossRef]
- Leenstra, F.R.; Pit, R. Fat deposition in a broiler sire strain. 4. Performance of broiler progeny of four differently selected sire lines. Poult. Sci. 1988, 67, 10–15. [Google Scholar] [CrossRef]
- Baéza, E.; Le Bihan-Duval, E. Chicken lines divergent for low or high abdominal fat deposition: A relevant model to study the regulation of energy metabolism. Animal 2013, 7, 965–973. [Google Scholar] [CrossRef]
- Leng, L.; Zhang, H.; Dong, J.Q.; Wang, Z.P.; Zhang, X.Y.; Wang, S.Z.; Cao, Z.P.; Li, Y.M.; Li, H. Selection against abdominal fat percentage may increase intramuscular fat content in broilers. J. Anim. Breed Genet. 2016, 133, 422–428. [Google Scholar] [CrossRef]
- Chen, C.; Su, Z.; Li, Y.; Luan, P.; Wang, S.; Zhang, H.; Xiao, F.; Guo, H.; Cao, Z.; Li, H.; et al. Estimation of the genetic parameters of traits relevant to feed efficiency: Result from broiler lines divergent for high or low abdominal fat content. Poult. Sci. 2021, 100, 461–466. [Google Scholar] [CrossRef]
- Wang, J.; Zhu, B.; Wen, J.; Li, Q.; Zhao, G. Genome-Wide Association Study and Pathway Analysis for Heterophil/Lymphocyte (H/L) Ratio in Chicken. Genes 2020, 11, 1005. [Google Scholar] [CrossRef] [PubMed]
- Hu, G.; Wang, S.; Tian, J.; Chu, L.; Li, H. Epistatic effect between ACACA and FABP2 gene on abdominal fat traits in broilers. J Genet. Genom. 2010, 37, 505–512. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Li, H.; Wang, S.; Hu, X.; Gao, Y.; Wang, Q.; Li, N.; Wang, Y.; Zhang, H. Mapping quantitative trait loci affecting body weight and abdominal fat weight on chicken chromosome one. Poult Sci. 2007, 86, 1084–1089. [Google Scholar] [CrossRef] [PubMed]
- Zhao, G.P.; Chen, J.L.; Zheng, M.Q.; Wen, J.; Zhang, Y. Correlated responses to selection for increased intramuscular fat in a Chinese quality chicken line. Poult Sci. 2007, 86, 2309–2314. [Google Scholar] [CrossRef] [PubMed]
- Jiang, M.; Fan, W.L.; Xing, S.Y.; Wang, J.; Li, P.; Liu, R.R.; Li, Q.H.; Zheng, M.Q.; Cui, H.X.; Wen, J.; et al. Effects of balanced selection for intramuscular fat and abdominal fat percentage and estimates of genetic parameters. Poult Sci. 2017, 96, 282–287. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Liu, L.; Wang, J.; Cui, H.; Chu, H.; Bi, H.; Zhao, G.; Wen, J. Genome-wide association study of muscle glycogen in jingxing yellow chicken. Genes 2020, 11, 497. [Google Scholar] [CrossRef]
- Browning, S.R.; Browning, B.L. Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am. J. Hum. Genet. 2007, 81, 1084–1097. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Stephens, M. Genome-wide efficient mixed-model analysis for association studies. Nat Genet. 2012, 44, 821–824. [Google Scholar] [CrossRef] [PubMed]
- Nicodemus, K.K.; Liu, W.; Chase, G.A.; Tsai, Y.Y.; Fallin, M.D. Comparison of type I error for multiple test corrections in large single-nucleotide polymorphism studies using principal components versus haplotype blocking algorithms. BMC Genet. 2005, 6 (Suppl. S1), S78. [Google Scholar] [CrossRef]
- Danecek, P.; Auton, A.; Abecasis, G.; Albers, C.A.; Banks, E.; DePristo, M.A.; Handsaker, R.E.; Lunter, G.; Marth, G.T.; Sherry, S.T.; et al. The variant call format and VCFtools. Bioinformatics 2011, 27, 2156–2158. [Google Scholar] [CrossRef] [PubMed]
- Luo, N.; Shu, J.; Yuan, X.; Jin, Y.; Cui, H.; Zhao, G.; Wen, J. Differential regulation of intramuscular fat and abdominal fat deposition in chickens. BMC Genom. 2022, 23, 308. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Shim, H.; Chasman, D.I.; Smith, J.D.; Mora, S.; Ridker, P.M.; Nickerson, D.A.; Krauss, R.M.; Stephens, M. A multivariate genome-wide association analysis of 10 LDL subfractions, and their response to statin treatment, in 1868 Caucasians. PLoS ONE 2015, 10, e0120758. [Google Scholar] [CrossRef] [PubMed]
- Neilsen, P.M.; Noll, J.E.; Mattiske, S.; Bracken, C.P.; Gregory, P.A.; Schulz, R.B.; Lim, S.P.; Kumar, R.; Suetani, R.J.; Goodall, G.J.; et al. Mutant p53 drives invasion in breast tumors through up-regulation of miR-155. Oncogene 2013, 32, 2992–3000. [Google Scholar] [CrossRef]
- Chen, J.H.; Ren, X.Y.; Li, L.M.; Lu, S.Y.; Cheng, T.; Tan, L.T.; Liang, S.D.; He, D.L.; Luo, Q.B.; Nie, Q.H.; et al. The cell cycle pathway regulates chicken abdominal fat deposition as revealed by transcriptome sequencing. Yi Chuan 2019, 41, 962–973. [Google Scholar]
- Wang, W.; Li, Y.; Li, Z.; Wang, N.; Xiao, F.; Gao, H.; Guo, H.; Li, H.; Wang, S. Polymorphisms of KLF3 gene coding region and identification of their functionality for abdominal fat in chickens. Vet. Med. Sci. 2021, 7, 792–799. [Google Scholar] [CrossRef]
- Moreira, G.C.M.; Boschiero, C.; Cesar, A.S.M.; Reecy, J.M.; Godoy, T.F.; Pértille, F.; Ledur, M.C.; Moura, A.S.A.M.T.; Garrick, D.J.; Coutinho, L.L. Integration of genome wide association studies and whole genome sequencing provides novel insights into fat deposition in chicken. Sci. Rep. 2018, 8, 16222. [Google Scholar] [CrossRef]
- Park, H.B.; Jacobsson, L.; Wahlberg, P.; Siegel, P.B.; Andersson, L. QTL analysis of body composition and metabolic traits in an intercross between chicken lines divergently selected for growth. Physiol. Genom. 2006, 25, 216–223. [Google Scholar] [CrossRef]
- Zhou, J.; Yang, L.; Yu, J.; Zhang, K.; Xu, Z.; Cao, Z.; Luan, P.; Li, H.; Zhang, H. Association of PCSK1 gene polymorphisms with abdominal fat content in broilers. Anim. Sci. J. 2020, 91, e13371. [Google Scholar] [CrossRef]
- Huang, X.; Zhang, H.; Guo, X.; Zhu, Z.; Cai, H.; Kong, X. Insulin-like growth factor 2 mRNA-binding protein 1 (IGF2BP1) in cancer. J. Hematol. Oncol. 2018, 11, 88. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Luo, W.; Chi, X.; Zhang, L.; Ren, Q.; Wang, H.; Zhang, W. IGF2BP1 silencing inhibits proliferation and induces apoptosis of high glucose-induced non-small cell lung cancer cells by regulating Netrin-1. Arch. Biochem. Biophys. 2020, 693, 108581. [Google Scholar] [CrossRef]
- Li, C.; Xu, J.J.; Hu, H.T.; Shi, C.Y.; Yu, C.J.; Sheng, J.Z.; Wu, Y.T.; Huang, H.F. Amylin receptor insensitivity impairs hypothalamic POMC neuron differentiation in the male offspring of maternal high-fat diet-fed mice. Mol. Metab. 2021, 44, 101135. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Hu, H.; Tian, Y.; Li, J.; Scheben, A.; Zhang, C.; Li, Y.; Wu, J.; Yang, L.; Fan, X.; et al. The chicken pan-genome reveals gene content variation and a promoter region deletion in IGF2BP1 affecting body size. Mol. Biol. Evol. 2021, 38, 5066–5081. [Google Scholar] [CrossRef]
- Zhou, Z.; Li, M.; Cheng, H.; Fan, W.; Yuan, Z.; Gao, Q.; Xu, Y.; Guo, Z.; Zhang, Y.; Hu, J.; et al. An intercross population study reveals genes associated with body size and plumage color in ducks. Nat. Commun. 2018, 9, 2648. [Google Scholar] [CrossRef]
- Chen, J.; Ren, X.; Li, L.; Lu, S.; Chen, T.; Tan, L.; Liu, M.; Luo, Q.; Liang, S.; Nie, Q.; et al. Integrative analyses of mRNA expression profile reveal the involvement of IGF2BP1 in chicken adipogenesis. Int. J. Mol. Sci. 2019, 20, 2923. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Cheney, K.M.; Neilsen, P.M.; Schulz, R.B.; Callen, D.F. CBFA2T3-ZNF651, like CBFA2T3-ZNF652, functions as a transcriptional corepressor complex. FEBS Lett. 2010, 584, 859–864. [Google Scholar] [CrossRef]
- Coviello, A.D.; Haring, R.; Wellons, M.; Vaidya, D.; Lehtimäki, T.; Keildson, S.; Lunetta, K.L.; He, C.; Fornage, M.; Lagou, V.; et al. A genome-wide association meta-analysis of circulating sex hormone-binding globulin reveals multiple Loci implicated in sex steroid hormone regulation. PLoS Genet. 2012, 8, e1002805. [Google Scholar] [CrossRef]
- Wang, Y.; Bu, L.; Cao, X.; Qu, H.; Zhang, C.; Ren, J.; Huang, Z.; Zhao, Y.; Luo, C.; Hu, X.; et al. Genetic dissection of growth traits in a unique chicken advanced intercross line. Front Genet. 2020, 11, 894. [Google Scholar] [CrossRef]
- Li, Y.D.; Liu, X.; Li, Z.W.; Wang, W.J.; Li, Y.M.; Cao, Z.P.; Luan, P.; Xiao, F.; Gao, H.H.; Guo, H.S.; et al. A combination of genome-wide association study and selection signature analysis dissects the genetic architecture underlying bone traits in chickens. Animal 2021, 15, 100322. [Google Scholar] [CrossRef]
- Bhushan, B.; Michalopoulos, G.K. Role of epidermal growth factor receptor in liver injury and lipid metabolism: Emerging new roles for an old receptor. Chem. Biol. Interact. 2020, 324, 109090. [Google Scholar] [CrossRef]
- Choung, S.; Kim, J.M.; Joung, K.H.; Lee, E.S.; Kim, H.J.; Ku, B.J. Epidermal growth factor receptor inhibition attenuates non-alcoholic fatty liver disease in diet-induced obese mice. PLoS ONE 2019, 14, e0210828. [Google Scholar] [CrossRef] [PubMed]
- Uluçay, S.; Çam, F.S.; Batır, M.B.; Sütçü, R.; Bayturan, Ö.; Demircan, K. A novel association between TGFb1 and ADAMTS4 in coronary artery disease: A new potential mechanism in the progression of atherosclerosis and diabetes. Anatol. J. Cardiol. 2015, 15, 823–829. [Google Scholar] [CrossRef] [PubMed]
- Pan, K.; Lu, J.; Song, Y. Artesunate ameliorates cigarette smoke-induced airway remodelling via PPAR-γ/TGF-β1/Smad2/3 signalling pathway. Respir Res. 2021, 22, 91. [Google Scholar] [CrossRef] [PubMed]






| Trait | Group | N | Mean (SD) | CV (%) | Min | Max | p-Value |
|---|---|---|---|---|---|---|---|
| IMF(g) | control line | 252 | 1.84 ± 0.56 | 29.93 | 0.47 | 3.52 | <0.0001 |
| selection line | 268 | 2.15 ± 0.64 | 30.54 | 0.74 | 4.61 | ||
| TG(mg/g) | control line | 252 | 3.51 ± 0.85 | 28.49 | 1.65 | 6.08 | <0.0001 |
| selection line | 268 | 3.92 ± 1.12 | 24.24 | 1.73 | 8.60 | ||
| AFP (%) | control line | 252 | 5.21 ± 1.28 | 24.50 | 1.63 | 8.81 | 0.7453 |
| selection line | 268 | 5.30 ± 1.26 | 23.79 | 2.09 | 8.37 | ||
| AFW (g) | control line | 252 | 48.94 ± 14.26 | 29.14 | 8.60 | 92.70 | <0.0001 |
| selection line | 268 | 54.77 ± 14.96 | 27.31 | 18.20 | 96.30 | ||
| ECW (g) | control line | 252 | 882.08 ± 86.67 | 9.83 | 684.2 | 1091 | <0.0001 |
| selection line | 268 | 973.66 ± 84.94 | 8.72 | 740.4 | 1206.4 |
| SNP | CHR | Position | ALT/REF | MAF | β (SE) 1 | p-Value | PVE | Distance 2 | Gene |
|---|---|---|---|---|---|---|---|---|---|
| rs316720008 | 1 | 43,577,050 | C/T | 0.203 | −0.6383165(0.1291873) | 0.00000185 | 4.13% | intron2 | lncRNA |
| rs312351828 | 1 | 116,670,617 | T/C | 0.126 | −0.8267067(0.1729973) | 0.0000007 | 5.35% | intron29 | DMD |
| rs317324892 | 1 | 117,928,954 | A/T | 0.065 | 0.4439148(0.09234324) | 0.00000124 | 4.89% | intron9 | IL1RAPL1 |
| 1_125775571 | 1 | 125,775,571 | G/C | 0.117 | 0.5044212(0.1054399) | 0.00000196 | 3.77% | D51254 | ARHGAP6 |
| rs313755922 | 1 | 158,194,105 | C/G | 0.434 | 0.5432101(0.1122472) | 0.000000842 | 4.22% | intron1 | DACH1 |
| rs312621600 | 4 | 82,213,271 | A/G | 0.2 | −0.5663142(0.1179772) | 0.00000219 | 4.13% | U1957 | GRK4 |
| rs316613317 | 14 | 8,850,467 | G/C | 0.231 | −0.5989047(0.1222303) | 0.00000244 | 4.35% | intron7 | SYT17 |
| 27_5963734 | 27 | 5,963,734 | G/A | 0.279 | −0.5798911(0.1171215) | 0.0000023 | 3.73% | intron2 | ETV4 |
| rs13769190 | 27 | 5,971,903 | T/C | 0.19 | −0.5848534(0.1187516) | 0.00000258 | 5.41% | D582 | ETV4 |
| rs794259691 | 27 | 6,010,935 | C/T | 0.191 | −0.5642823(0.1175598) | 0.00000157 | 5.58% | intron1 | ZNF652 |
| rs312715211 | 27 | 6,017,027 | A/T | 0.285 | −0.5775431(0.1196338) | 0.00000192 | 4.29% | intron2 | ZNF652 |
| rs15242723 | 27 | 6,069,759 | A/G | 0.28 | −0.5986492(0.1190552) | 0.00000121 | 4.49% | intron12 | IGF2BP1 |
| rs314672842 | 27 | 6,091,289 | G/A | 0.216 | 0.4740275(0.09803836) | 0.00000203 | 4.06% | intron2 | GIP |
| rs737217409 | 27 | 6,106,019 | T/C | 0.24 | 0.507996(0.1029385) | 0.0000023 | 3.83% | intron2 | UBE2Z |
| rs317582031 | 27 | 6,118,076 | T/C | 0.227 | 0.5165271(0.1051145) | 0.00000157 | 3.34% | D4347 | UBE2Z |
| rs732151018 | 27 | 6,119,680 | A/G | 0.241 | 0.5115049(0.1063718) | 0.000000856 | 4.04% | D5951 | UBE2Z |
| SNP | CHR | Position | FST | 1 π (Group1) | 2 π (Group2) | Gene |
|---|---|---|---|---|---|---|
| rs312715211 | 27 | 6,017,027 | 0.108971 | 0.272997 | 0.471467 | ZNF652 |
| Pathway | Pathway Code | Corrected p-Value | Genes |
|---|---|---|---|
| MAPK signaling pathway | gga04010 | 0.000001450 | EGFR, TGFB1, PPP3CA, NGFR, etc. |
| Calcium signaling pathway | gga04020 | 0.000031000 | EGFR, MCU, PPP3CC, STIM1, etc. |
| Regulation of actin cytoskeleton | gga04810 | 0.000112941 | EGFR, CHRM5, MYLK2, MRAS, etc. |
| Focal adhesion | gga04510 | 0.000916584 | EGFR, FYN, MYLK2, COL9A3, etc. |
| Adherens junction | gga04520 | 0.001264711 | EGFR, BAIAP2, PTPRM, PTPRJ, etc. |
| Endocytosis | gga04144 | 0.001669978 | EGFR, RAB7A, GRK5, ZFYVE9, etc. |
| FoxO signaling pathway | gga04068 | 0.001849407 | TGFB1, EGFR, BCL6, CREBBP, etc. |
| Cytokine-cytokine receptor interaction | gga04060 | 0.016863281 | TGFB1, IL5, EDAR, BMP7, etc. |
| TGF-beta signaling pathway | gga04350 | 0.022547651 | TGFB1, SMAD9, SMAD5, DCN, etc. |
| AGE-RAGE signaling pathway in diabetic complications | gga04933 | 0.031877403 | TGFB1, COL4A1, AKT3, NOX1, etc. |
| ErbB signaling pathway | gga04012 | 0.031877403 | EGFR, GSK3B, PAK1, PAK3, etc. |
| Gap junction | gga04540 | 0.037408823 | EGFR, DRD2, ADCY9, CDK1, etc. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, Y.; Liu, X.; Wang, Y.; Liu, L.; Wang, Y.; Zhao, G.; Wen, J.; Cui, H. Genome-Wide Association Study Revealed the Effect of rs312715211 in ZNF652 Gene on Abdominal Fat Percentage of Chickens. Biology 2022, 11, 1849. https://doi.org/10.3390/biology11121849
Zhu Y, Liu X, Wang Y, Liu L, Wang Y, Zhao G, Wen J, Cui H. Genome-Wide Association Study Revealed the Effect of rs312715211 in ZNF652 Gene on Abdominal Fat Percentage of Chickens. Biology. 2022; 11(12):1849. https://doi.org/10.3390/biology11121849
Chicago/Turabian StyleZhu, Yuting, Xiaojing Liu, Yongli Wang, Lu Liu, Yidong Wang, Guiping Zhao, Jie Wen, and Huanxian Cui. 2022. "Genome-Wide Association Study Revealed the Effect of rs312715211 in ZNF652 Gene on Abdominal Fat Percentage of Chickens" Biology 11, no. 12: 1849. https://doi.org/10.3390/biology11121849
APA StyleZhu, Y., Liu, X., Wang, Y., Liu, L., Wang, Y., Zhao, G., Wen, J., & Cui, H. (2022). Genome-Wide Association Study Revealed the Effect of rs312715211 in ZNF652 Gene on Abdominal Fat Percentage of Chickens. Biology, 11(12), 1849. https://doi.org/10.3390/biology11121849




