Sexually Dimorphic Gene Expression in X and Y Sperms Instructs Sexual Dimorphism of Embryonic Genome Activation in Yellow Catfish (Pelteobagrus fulvidraco)
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Fish and Breeding
2.2. Preparation of Percoll Gradient-Centrifuged Sperm
2.3. RNA-seq of X/Y Sperm and Data Analysis
2.4. WGBS of X/Y Sperm and Data Analysis
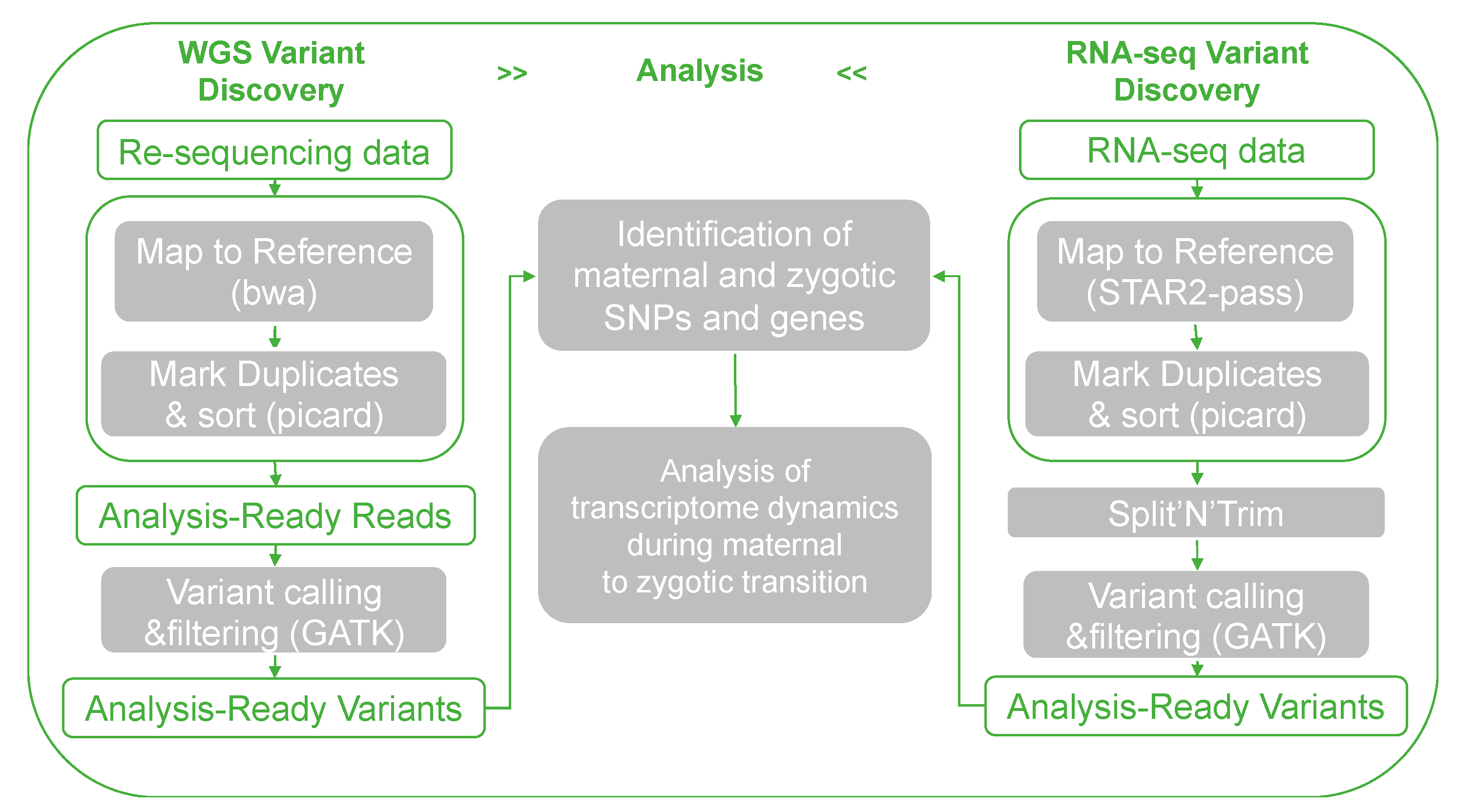
2.5. Identification of Maternal, Zygote, and Paternal SNPs and Genes in Early Embryonic Development through WGRS and RNA-seq
2.6. Statistics
3. Results
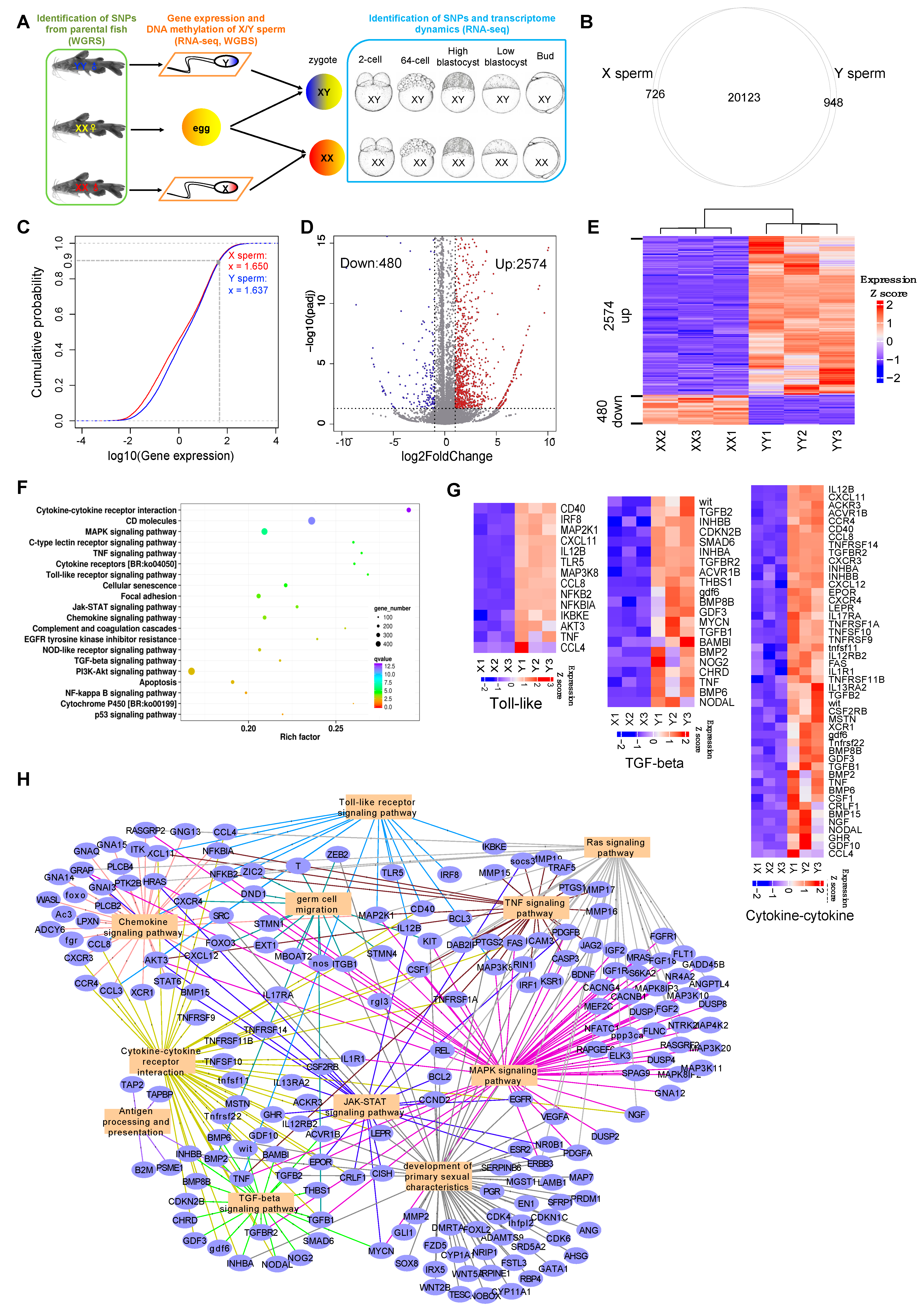
3.1. Gene Expression Differences between X and Y Sperm of Yellow Catfish
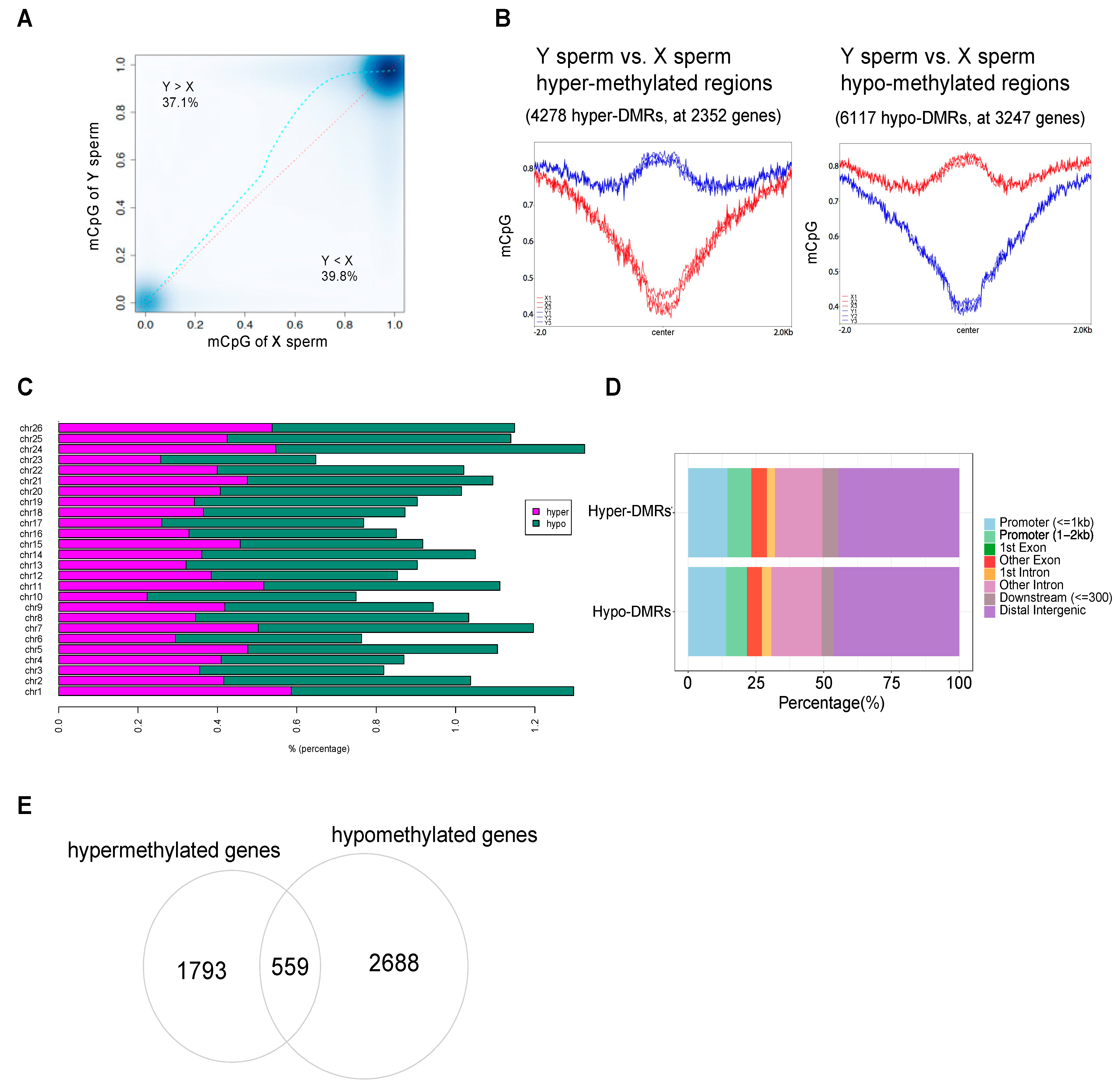
3.2. Genome-Wide DNA Methylation Differences between X and Y Sperm of Yellow Catfish
3.3. Dissection of DEGs Regulated by Methylation Modification in X/Y Sperm
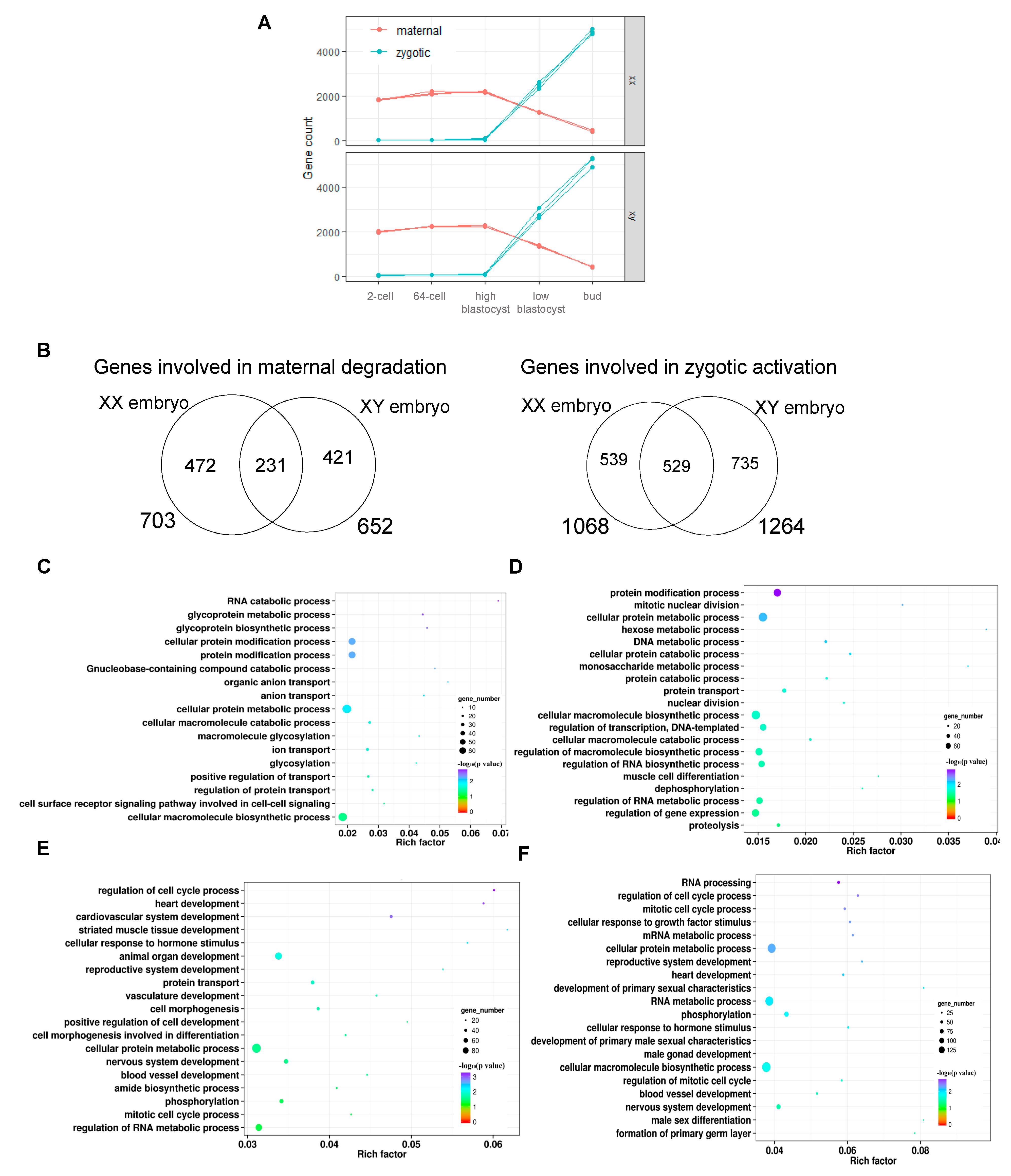
3.4. Identification and Profiling of Maternal-to-Zygotic Transition (MZT) of XX and XY Yellow Catfish during Early Embryonic Development
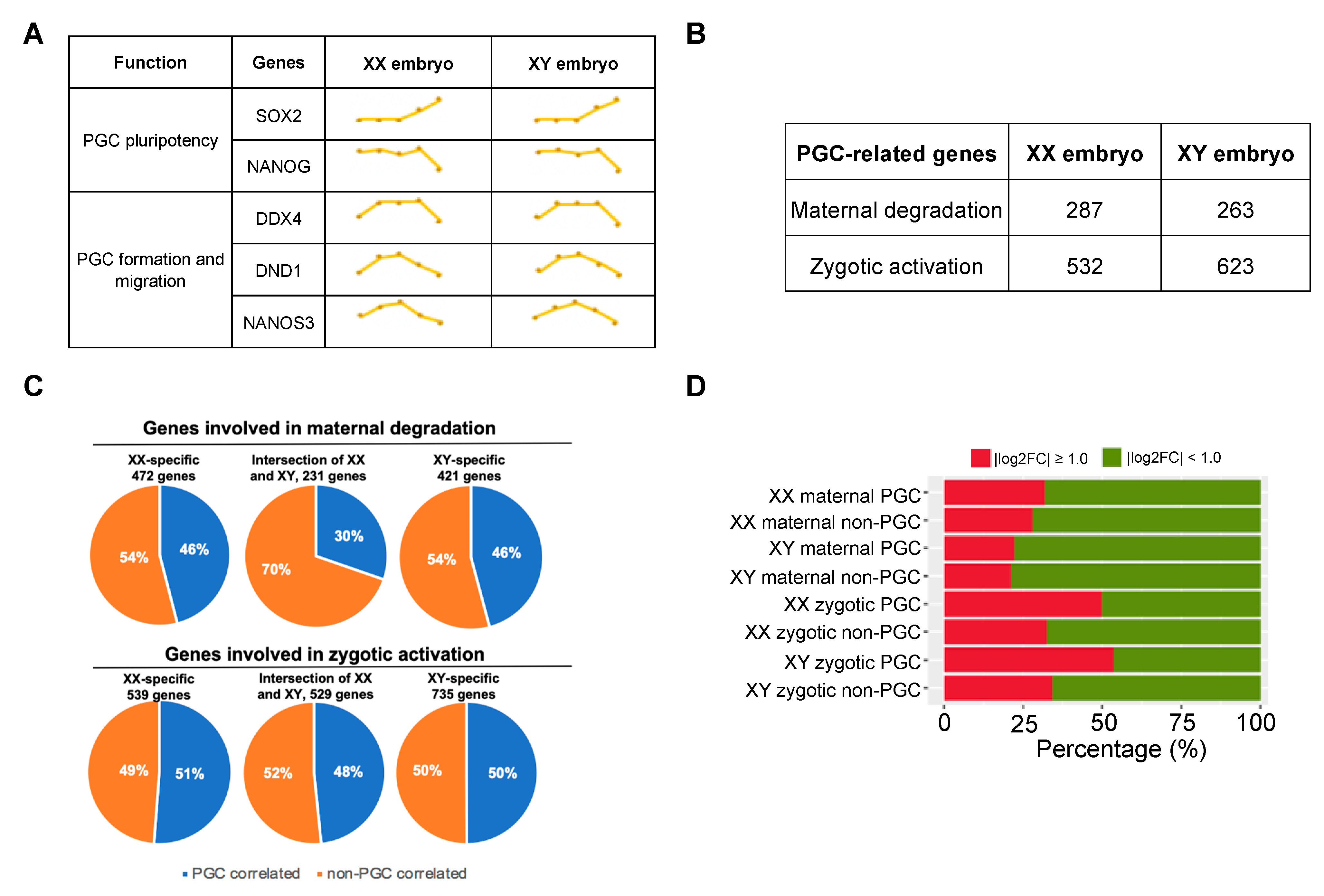
3.5. Comparative Analysis of Sex Determination Genes and PGC-Related Genes of XX and XY Embryos during the Maternal-to-Zygotic Transition
3.6. Detecting Influence of Paternal Inheritance on Sexual Dimorphism Genome Activation of Yellow Catfish during Embryonic Development
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Shettles, L.B. Nuclear morphology of human spermatozoa. Nature 1960, 186, 648–649. [Google Scholar] [CrossRef] [PubMed]
- Cui, K.H.; Matthews, C.D. X Larger Than Y. Nature 1993, 366, 117–118. [Google Scholar] [CrossRef] [PubMed]
- Cui, K.H. Size differences between human X and Y spermatozoa and prefertilization diagnosis. Mol. Hum. Reprod 1997, 3, 61–67. [Google Scholar] [CrossRef] [PubMed]
- Hossain, A.M.; Barik, S.; Kulkarni, P.M. Lack of significant morphological differences between human X and Y spermatozoa and their precursor cells (spermatids) exposed to different prehybridization treatments. J. Androl. 2001, 22, 119–123. [Google Scholar] [CrossRef]
- You, Y.A.; Kwon, W.S.; Rahman, M.S.; Park, Y.J.; Kim, Y.J.; Pang, M.G. Sex chromosome-dependent differential viability of human spermatozoa during prolonged incubation. Hum. Reprod 2017, 32, 1183–1191. [Google Scholar] [CrossRef]
- Carvalho, J.O.; Silva, L.P.; Sartori, R.; Dode, M.A.N. Nanoscale Differences in the Shape and Size of X and Y Chromosome-Bearing Bovine Sperm Heads Assessed by Atomic Force Microscopy. PLoS ONE 2013, 8, e59387. [Google Scholar] [CrossRef]
- Rahman, M.S.; Pang, M.G. New Biological Insights on X and Y Chromosome-Bearing Spermatozoa. Front. Cell Dev. Biol. 2019, 7, 388. [Google Scholar] [CrossRef]
- Alminana, C.; Caballero, I.; Heath, P.R.; Maleki-Dizaji, S.; Parrilla, I.; Cuello, C.; Gil, M.A.; Vazquez, J.L.; Vazquez, J.M.; Roca, J.; et al. The battle of the sexes starts in the oviduct: Modulation of oviductal transcriptome by X and Y-bearing spermatozoa. BMC Genom. 2014, 15, 293. [Google Scholar] [CrossRef]
- Chen, X.; Yue, Y.; He, Y.; Zhu, H.; Hao, H.; Zhao, X.; Qin, T.; Wang, D. Identification and characterization of genes differentially expressed in X and Y sperm using suppression subtractive hybridization and cDNA microarray. Mol. Reprod Dev. 2014, 81, 908–917. [Google Scholar] [CrossRef]
- De Canio, M.; Soggiu, A.; Piras, C.; Bonizzi, L.; Galli, A.; Urbani, A.; Roncada, P. Differential protein profile in sexed bovine semen: Shotgun proteomics investigation. Mol. Biosyst. 2014, 10, 1264–1271. [Google Scholar] [CrossRef]
- Scott, C.; de Souza, F.F.; Aristizabal, V.H.V.; Hethrington, L.; Krisp, C.; Molloy, M.; Baker, M.A.; Dell’Aqua, J.A.J. Proteomic profile of sex-sorted bull sperm evaluated by SWATH-MS analysis. Anim. Reprod Sci. 2018, 198, 121–128. [Google Scholar] [CrossRef]
- Nagahama, Y.; Chakraborty, T.; Paul-Prasanth, B.; Ohta, K.; Nakamura, M. Sex determination, gonadal sex differentiation, and plasticity in vertebrate species. Physiol. Rev. 2021, 101, 1237–1308. [Google Scholar] [CrossRef] [PubMed]
- Vandenbon, A.; Kumagai, Y.; Lin, M.; Suzuki, Y.; Nakai, K. Waves of chromatin modifications in mouse dendritic cells in response to LPS stimulation. Genome Biol. 2018, 19, 138. [Google Scholar] [CrossRef] [PubMed]
- Xiong, S.; Jing, J.; Wu, J.; Ma, W.; Dawar, F.U.; Mei, J.; Gui, J.F. Characterization and sexual dimorphic expression of Cytochrome P450 genes in the hypothalamic-pituitary-gonad axis of yellow catfish. Gen. Comp. Endocrinol. 2015, 216, 90–97. [Google Scholar] [CrossRef]
- Capel, B. Vertebrate sex determination: Evolutionary plasticity of a fundamental switch. Nat. Rev. Genet. 2017, 18, 675–689. [Google Scholar] [CrossRef] [PubMed]
- Jukam, D.; Shariati, S.A.M.; Skotheim, J.M. Zygotic Genome Activation in Vertebrates. Dev. Cell 2017, 42, 316–332. [Google Scholar] [CrossRef]
- Gert, K.R.; Quio, L.E.C.; Novatchkova, M.; Guo, Y.; Cairns, B.R.; Pauli, A. Reciprocal zebrafish-medaka hybrids reveal maternal control of zygotic genome activation timing. bioRxiv 2021, 11, 467109. [Google Scholar]
- Ge, S.; Dan, C.; Xiong, Y.; Gong, G.; Li, X. Identifying difference in primordial germ cells between XX female and XY male yellow catfish embryos. Gene 2020, 761, 145037. [Google Scholar] [CrossRef]
- Kocer, A.; Reichmann, J.; Best, D.; Adams, I.R. Germ cell sex determination in mammals. Mol. Hum. Reprod 2009, 15, 205–213. [Google Scholar] [CrossRef]
- Slanchev, K.; Stebler, J.; de la Cueva-Mendez, G.; Raz, E. Development without germ cells: The role of the germ line in zebrafish sex differentiation. Proc. Natl. Acad. Sci. USA 2005, 102, 4074–4079. [Google Scholar] [CrossRef]
- Lesch, B.J.; Page, D.C. Genetics of germ cell development. Nat. Rev. Genet. 2012, 13, 781–794. [Google Scholar] [CrossRef] [PubMed]
- Fan, R.A.; Ran, M.A.; Rui, X.; Jie, M.A. m6A reader Igf2bp3 enables germ plasm assembly by m 6 A-dependent regulation of gene expression in zebrafish. Sci. Bull. 2021, 66, 1119–1128. [Google Scholar]
- Zou, Y.; Geuverink, E.; Beukeboom, L.W.; Verhulst, E.C.; van de Zande, L. A chimeric gene paternally instructs female sex determination in the haplodiploid wasp Nasonia. Science 2020, 370, 1115–1118. [Google Scholar] [CrossRef] [PubMed]
- Li, X.Y.; Mei, J.; Ge, C.T.; Liu, X.L.; Gui, J.F. Sex determination mechanisms and sex control approaches in aquaculture animals. Sci. China Life Sci. 2022, 65, 1091–1122. [Google Scholar] [CrossRef] [PubMed]
- Lowe, R.; Gemma, C.; Rakyan, V.K.; Holland, M.L. Sexually dimorphic gene expression emerges with embryonic genome activation and is dynamic throughout development. BMC Genom. 2015, 16, 295. [Google Scholar] [CrossRef]
- Gong, G.; Xiong, Y.; Xiao, S.; Li, X.-Y.; Huang, P.; Liao, Q.; Han, Q.; Lin, Q.; Dan, C.; Zhou, L. Origin and chromatin remodelling of young X/Y sex chromosomes in catfish with sexual plasticity. Natl. Sci. Rev. 2022, nwac239. [Google Scholar] [CrossRef]
- Sardina, J.L.; Collombet, S.; Tian, T.V.; Gomez, A.; Di Stefano, B.; Berenguer, C.; Brumbaugh, J.; Stadhouders, R.; Segura-Morales, C.; Gut, M.; et al. Transcription Factors Drive Tet2-Mediated Enhancer Demethylation to Reprogram Cell Fate. Cell Stem. Cell 2018, 23, 727–741.e9. [Google Scholar] [CrossRef]
- Liu, H.Q.; Guan, B.; Xu, J.; Hou, C.C.; Tian, H.; Chen, H.X. Genetic manipulation of sex ratio for the large-scale breeding of YY super-male and XY all-male yellow catfish (Pelteobagrus fulvidraco (Richardson)). Mar. Biotechnol. 2013, 15, 321–328. [Google Scholar] [CrossRef]
- Dan, C.; Mei, J.; Wang, D.; Gui, J.F. Genetic differentiation and efficient sex-specific marker development of a pair of Y- and X-linked markers in yellow catfish. Int. J. Biol. Sci. 2013, 9, 1043–1049. [Google Scholar] [CrossRef]
- Xiong, Y.; Wang, S.; Gui, J.F.; Mei, J. Artificially induced sex-reversal leads to transition from genetic to temperature-dependent sex determination in fish species. Sci. China Life Sci. 2020, 63, 157–159. [Google Scholar] [CrossRef]
- Xiong, Y.; Han, Q.Q.; Liu, Y.; Wang, S.; Yang, J.H.; Jiang, W.; Hu, J.Q.; Chen, J.; Li, P.; Mei, J. Biotechnological manipulation of the transition from genetic to temperature-dependent sex determination to obtain high quality neomale in aquaculture. Aquaculture 2022, 560, 738471. [Google Scholar] [CrossRef]
- Mei, J.; Gui, J.F. Genetic basis and biotechnological manipulation of sexual dimorphism and sex determination in fish. Sci. China Life Sci. 2015, 58, 124–136. [Google Scholar] [CrossRef] [PubMed]
- Chu, Z.; Guo, W.; Hu, W.; Mei, J. Delayed elimination of paternal mtDNA in the interspecific hybrid of Pelteobagrus fulvidraco and Pelteobagrus vachelli during early embryogenesis. Gene 2019, 704, 1–7. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. ClusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Krueger, F.; Andrews, S.R. Bismark: A flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 2011, 27, 1571–1572. [Google Scholar] [CrossRef]
- Schultz, M.D.; He, Y.; Whitaker, J.W.; Hariharan, M.; Mukamel, E.A.; Leung, D.; Rajagopal, N.; Nery, J.R.; Urich, M.A.; Chen, H.; et al. Human body epigenome maps reveal noncanonical DNA methylation variation. Nature 2015, 523, 212–216. [Google Scholar] [CrossRef]
- Akalin, A.; Kormaksson, M.; Li, S.; Garrett-Bakelman, F.E.; Figueroa, M.E.; Melnick, A.; Mason, C.E. MethylKit: A comprehensive R package for the analysis of genome-wide DNA methylation profiles. Genome Biol. 2012, 13, R87. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.G.; He, Q.Y. ChIPseeker: An R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics 2015, 31, 2382–2383. [Google Scholar] [CrossRef] [PubMed]
- Ramirez, F.; Dundar, F.; Diehl, S.; Gruning, B.A.; Manke, T. DeepTools: A flexible platform for exploring deep-sequencing data. Nucleic Acids Res. 2014, 42, W187–W191. [Google Scholar] [CrossRef] [PubMed]
- Harvey, S.A.; Sealy, I.; Kettleborough, R.; Fenyes, F.; White, R.; Stemple, D.; Smith, J.C. Identification of the zebrafish maternal and paternal transcriptomes. Development 2013, 140, 2703–2710. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- Wingett, S.W.; Andrews, S. FastQ Screen: A tool for multi-genome mapping and quality control. F1000Research 2018, 7, 1338. [Google Scholar] [CrossRef] [PubMed]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; Genome Project Data Processing, S. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Van der Auwera, G.A.; Carneiro, M.O.; Hartl, C.; Poplin, R.; Del Angel, G.; Levy-Moonshine, A.; Jordan, T.; Shakir, K.; Roazen, D.; Thibault, J.; et al. From FastQ data to high confidence variant calls: The Genome Analysis Toolkit best practices pipeline. Curr. Protoc. Bioinform. 2013, 43, 11.0.1–11.0.33. [Google Scholar]
- Sereshki, N.; Andalib, A.; Ghahiri, A.; Mehrabian, F.; Sherkat, R.; Rezaei, A. Decreased Toll-like Receptor (TLR) 2 and 4 Expression in Spermatozoa in Couples with Unexplained Recurrent Spontaneous Abortion (URSA). Iran J. Allergy Asthma. Immunol. 2019, 18, 701–706. [Google Scholar] [CrossRef]
- Umehara, T.; Tsujita, N.; Shimada, M. Activation of Toll-like receptor 7/8 encoded by the X chromosome alters sperm motility and provides a novel simple technology for sexing sperm. PLoS Biol. 2019, 17, e3000398. [Google Scholar] [CrossRef]
- Vastenhouw, N.L.; Cao, W.X.; Lipshitz, H.D. The maternal-to-zygotic transition revisited. Development 2019, 146, dev161471. [Google Scholar] [CrossRef] [PubMed]
- Saitou, M.; Yamaji, M. Primordial germ cells in mice. Cold Spring Harb. Perspect Biol. 2012, 4, a008375. [Google Scholar] [CrossRef] [PubMed]
- Pereda, J.; Zorn, T.; Soto-Suazo, M. Migration of human and mouse primordial germ cells and colonization of the developing ovary: An ultrastructural and cytochemical study. Microsc. Res. Tech. 2006, 69, 386–395. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.B.; Wang, F.C.; Xu, Q.H.; Shi, J.C.; Zhang, X.X.; Lu, X.K.; Zhao, Z.A.; Gao, Z.; Ma, H.X.; Duan, E.K.; et al. BCAS2 is involved in alternative mRNA splicing in spermatogonia and the transition to meiosis. Nat. Commun. 2017, 8, 14182. [Google Scholar] [CrossRef]
- Legrand, J.M.D.; Chan, A.L.; La, H.M.; Rossello, F.J.; Anko, M.L.; Fuller-Pace, F.V.; Hobbs, R.M. DDX5 plays essential transcriptional and post-transcriptional roles in the maintenance and function of spermatogonia. Nat. Commun. 2019, 10, 2278. [Google Scholar] [CrossRef] [PubMed]
- Imarazene, B.; Beille, S.; Jouanno, E.; Branthonne, A.; Thermes, V.; Thomas, M.; Herpin, A.; Retaux, S.; Guiguen, Y. Primordial Germ Cell Migration and Histological and Molecular Characterization of Gonadal Differentiation in Pachon Cavefish Astyanax mexicanus. Sex Dev. 2020, 14, 80–98. [Google Scholar] [CrossRef]
- Whiteley, S.L.; Holleley, C.E.; Wagner, S.; Blackburn, J.; Deveson, I.W.; Marshall Graves, J.A.; Georges, A. Two transcriptionally distinct pathways drive female development in a reptile with both genetic and temperature dependent sex determination. PLoS Genet. 2021, 17, e1009465. [Google Scholar] [CrossRef]
- Liu, B.; Hu, F.F.; Zhang, Q.; Hu, H.; Ye, Z.; Tang, Q.; Guo, A.Y. Genomic landscape and mutational impacts of recurrently mutated genes in cancers. Mol. Genet. Genom. Med. 2018, 6, 910–923. [Google Scholar] [CrossRef]
- Moon, J.Y.; Nam, B.H.; Kong, H.J.; Kim, Y.O.; Kim, W.J.; Kim, B.S.; Kim, K.K.; Lee, S.J. Maximal transcriptional activation of piscine soluble Toll-like receptor 5 by the NF-kappaB subunit p65 and flagellin. Fish Shellfish Immunol. 2011, 31, 881–886. [Google Scholar] [CrossRef]
- Young, J.C.; Wakitani, S.; Loveland, K.L. TGF-beta superfamily signaling in testis formation and early male germline development. Semin. Cell Dev. Biol. 2015, 45, 94–103. [Google Scholar] [CrossRef]
- Jorgensen, A.; Macdonald, J.; Nielsen, J.E.; Kilcoyne, K.R.; Perlman, S.; Lundvall, L.; Langhoff Thuesen, L.; Juul Hare, K.; Frederiksen, H.; Andersson, A.M.; et al. Nodal Signaling Regulates Germ Cell Development and Establishment of Seminiferous Cords in the Human Fetal Testis. Cell Rep. 2018, 25, 1924–1937.e4. [Google Scholar] [CrossRef] [PubMed]
- Smith, Z.D.; Meissner, A. DNA methylation: Roles in mammalian development. Nat. Rev. Genet. 2013, 14, 204–220. [Google Scholar] [CrossRef] [PubMed]
- Tessarz, P.; Kouzarides, T. Histone core modifications regulating nucleosome structure and dynamics. Nat. Rev. Mol. Cell Biol. 2014, 15, 703. [Google Scholar] [CrossRef] [PubMed]
- Stephanie, C.; Tang, Q.; Joun, L.; Taylor, S.J.; Zhao, Y.; Ulrich, S.; Zheng, D.; Meeladm, D. The DNA dioxygenase Tet1 regulates H3K27 modification and embryonic stem cell biology independent of its catalytic activity. Nucleic Acids Res. 2022, 6, 3169–3189. [Google Scholar]
- Domingos, J.A.; Budd, A.M.; Banh, Q.Q.; Goldsbury, J.A.; Zenger, K.R.; Jerry, D.R. Sex-specific dmrt1 and cyp19a1 methylation and alternative splicing in gonads of the protandrous hermaphrodite barramundi. PLoS ONE 2018, 13, e0204182. [Google Scholar] [CrossRef]
- Wu, J.; Xiong, S.; Jing, J.; Chen, X.; Wang, W.; Gui, J.F.; Mei, J. Comparative Transcriptome Analysis of Differentially Expressed Genes and Signaling Pathways between XY and YY Testis in Yellow Catfish. PLoS ONE 2015, 10, e0134626. [Google Scholar] [CrossRef]
- Liu, J.; Zhang, W.; Sun, Y.; Wang, Z.; Zhang, Q.; Wang, X. Molecular characterization and expression profiles of GATA6 in tongue sole (Cynoglossus semilaevis). Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2016, 198, 19–26. [Google Scholar] [CrossRef]
- Bennett-Toomey, J.; Stocco, C. GATA Regulation and Function during the Ovarian Life Cycle. Vitam. Horm. 2018, 107, 193–225. [Google Scholar]
- Sone, R.; Taimatsu, K.; Ohga, R.; Nishimura, T.; Tanaka, M.; Kawahara, A. Critical roles of the ddx5 gene in zebrafish sex differentiation and oocyte maturation. Sci. Rep. 2020, 10, 14157. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xiong, Y.; Wang, D.-Y.; Guo, W.; Gong, G.; Chen, Z.-X.; Tang, Q.; Mei, J. Sexually Dimorphic Gene Expression in X and Y Sperms Instructs Sexual Dimorphism of Embryonic Genome Activation in Yellow Catfish (Pelteobagrus fulvidraco). Biology 2022, 11, 1818. https://doi.org/10.3390/biology11121818
Xiong Y, Wang D-Y, Guo W, Gong G, Chen Z-X, Tang Q, Mei J. Sexually Dimorphic Gene Expression in X and Y Sperms Instructs Sexual Dimorphism of Embryonic Genome Activation in Yellow Catfish (Pelteobagrus fulvidraco). Biology. 2022; 11(12):1818. https://doi.org/10.3390/biology11121818
Chicago/Turabian StyleXiong, Yang, Dan-Yang Wang, Wenjie Guo, Gaorui Gong, Zhen-Xia Chen, Qin Tang, and Jie Mei. 2022. "Sexually Dimorphic Gene Expression in X and Y Sperms Instructs Sexual Dimorphism of Embryonic Genome Activation in Yellow Catfish (Pelteobagrus fulvidraco)" Biology 11, no. 12: 1818. https://doi.org/10.3390/biology11121818
APA StyleXiong, Y., Wang, D.-Y., Guo, W., Gong, G., Chen, Z.-X., Tang, Q., & Mei, J. (2022). Sexually Dimorphic Gene Expression in X and Y Sperms Instructs Sexual Dimorphism of Embryonic Genome Activation in Yellow Catfish (Pelteobagrus fulvidraco). Biology, 11(12), 1818. https://doi.org/10.3390/biology11121818





