Comparison of Reverse Transcriptase (RT) Activities of Various M-MuLV RTs for RT-LAMP Assays
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Standard Plasmids with Genome Fragments of SARS-CoV-2 and MS2 Phage
2.2. In Vitro RNA Synthesis
2.3. Isolation of MS2 Phage RNA
2.4. Expression and Purification of Reverse Transcriptases
2.5. RNA-Dependent DNA-Polymerase Activity Measurement
2.6. Droplet Digital PCR
2.7. Real-Time Reverse-Transcription Loop-Mediated Isothermal Amplification (RT-LAMP)
3. Results
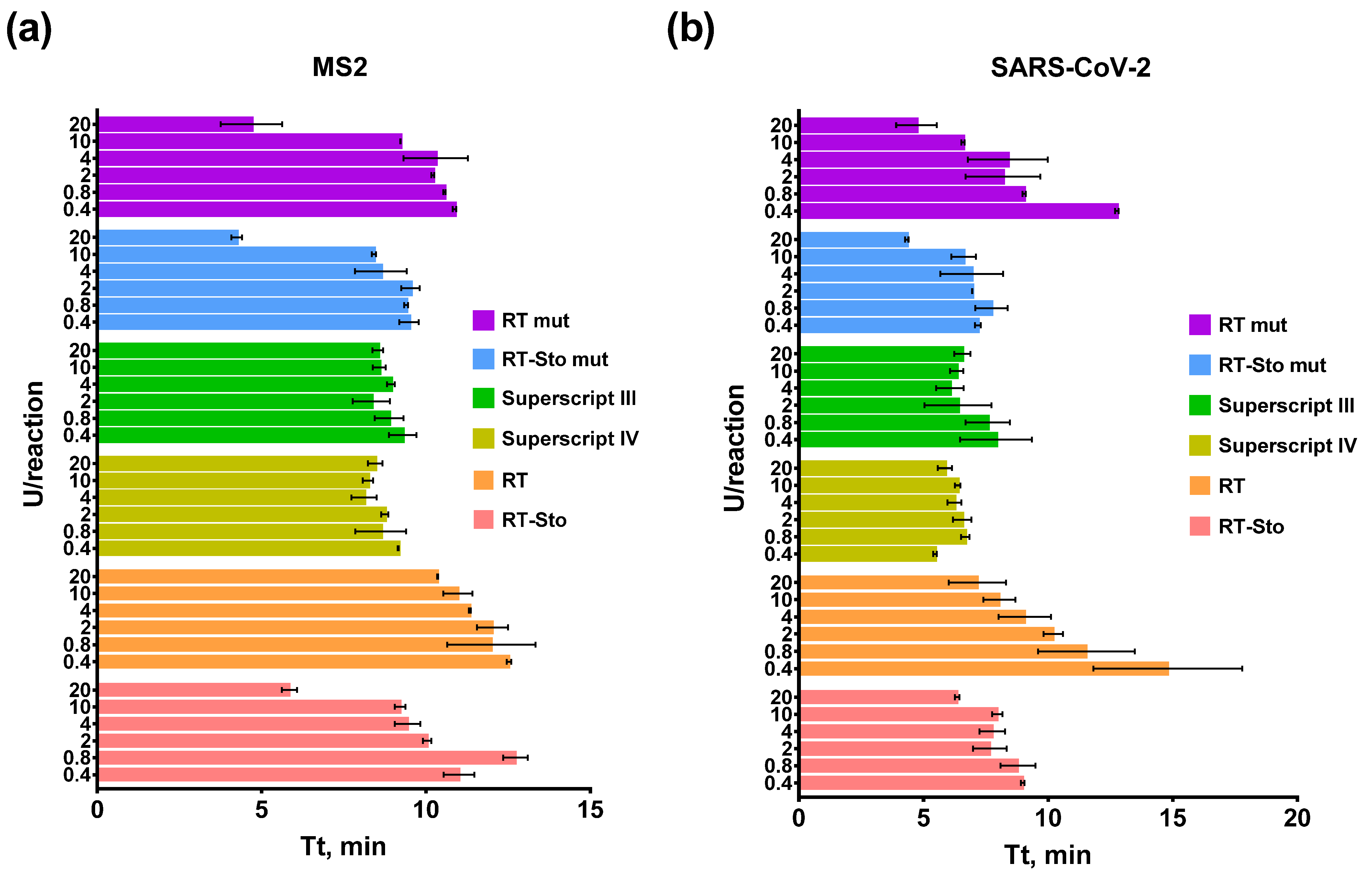
3.1. Titration of RTs in RT-LAMP
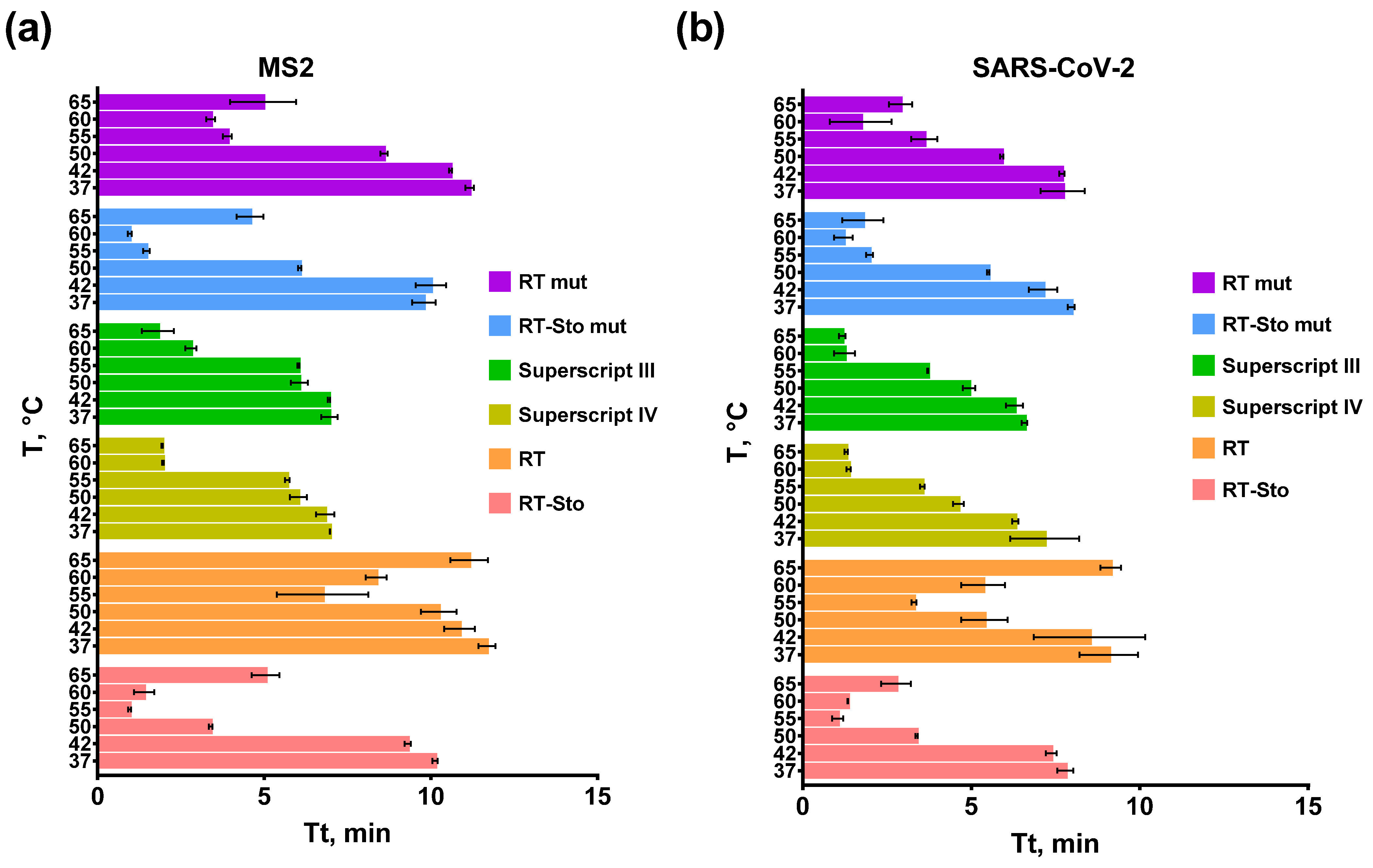
3.2. Optimal Temperature of the Reverse Transcription Step in RT-LAMP
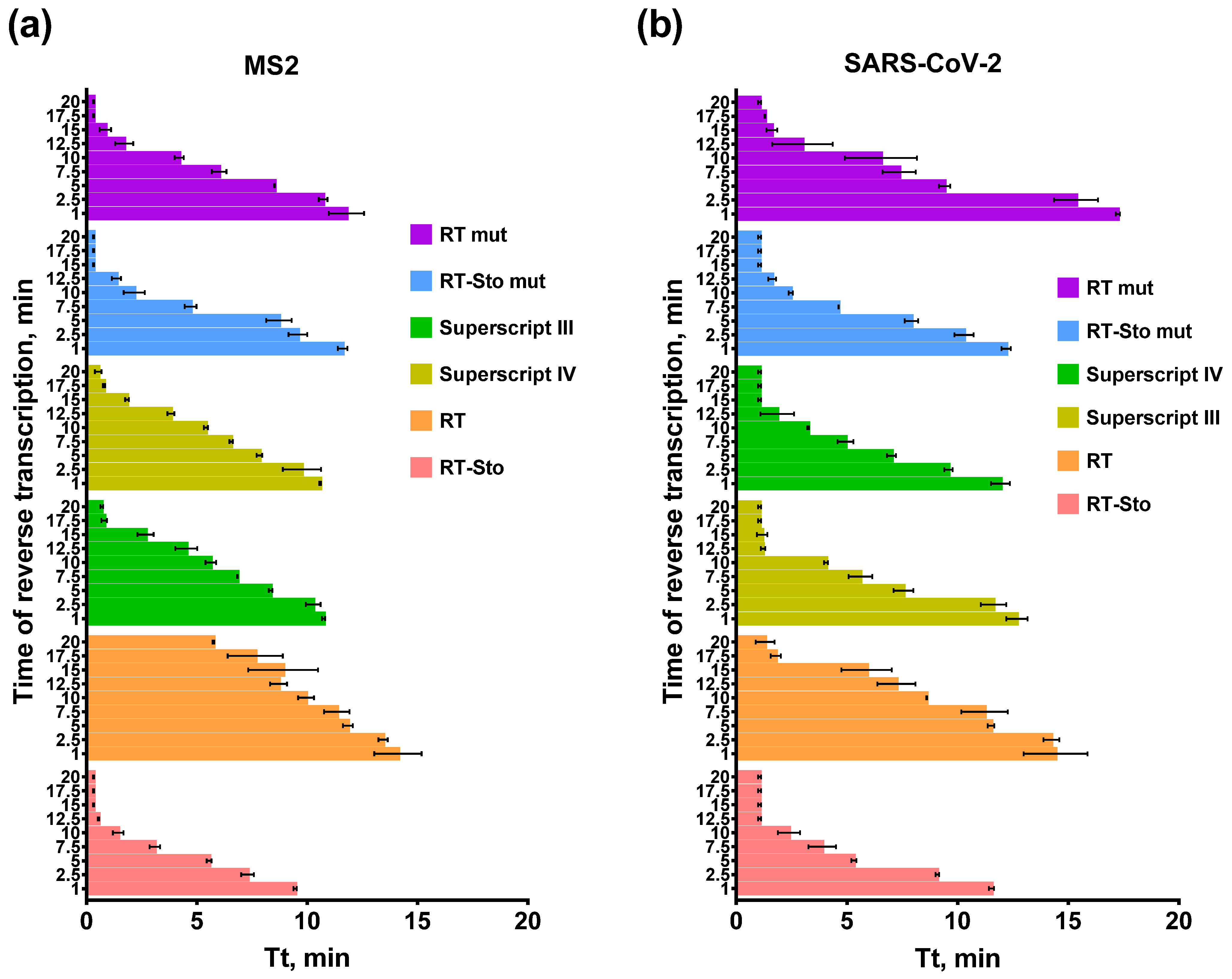
3.3. Optimal Time of the Reverse Transcription Step in RT-LAMP
3.4. RNA Titration in RT-LAMP
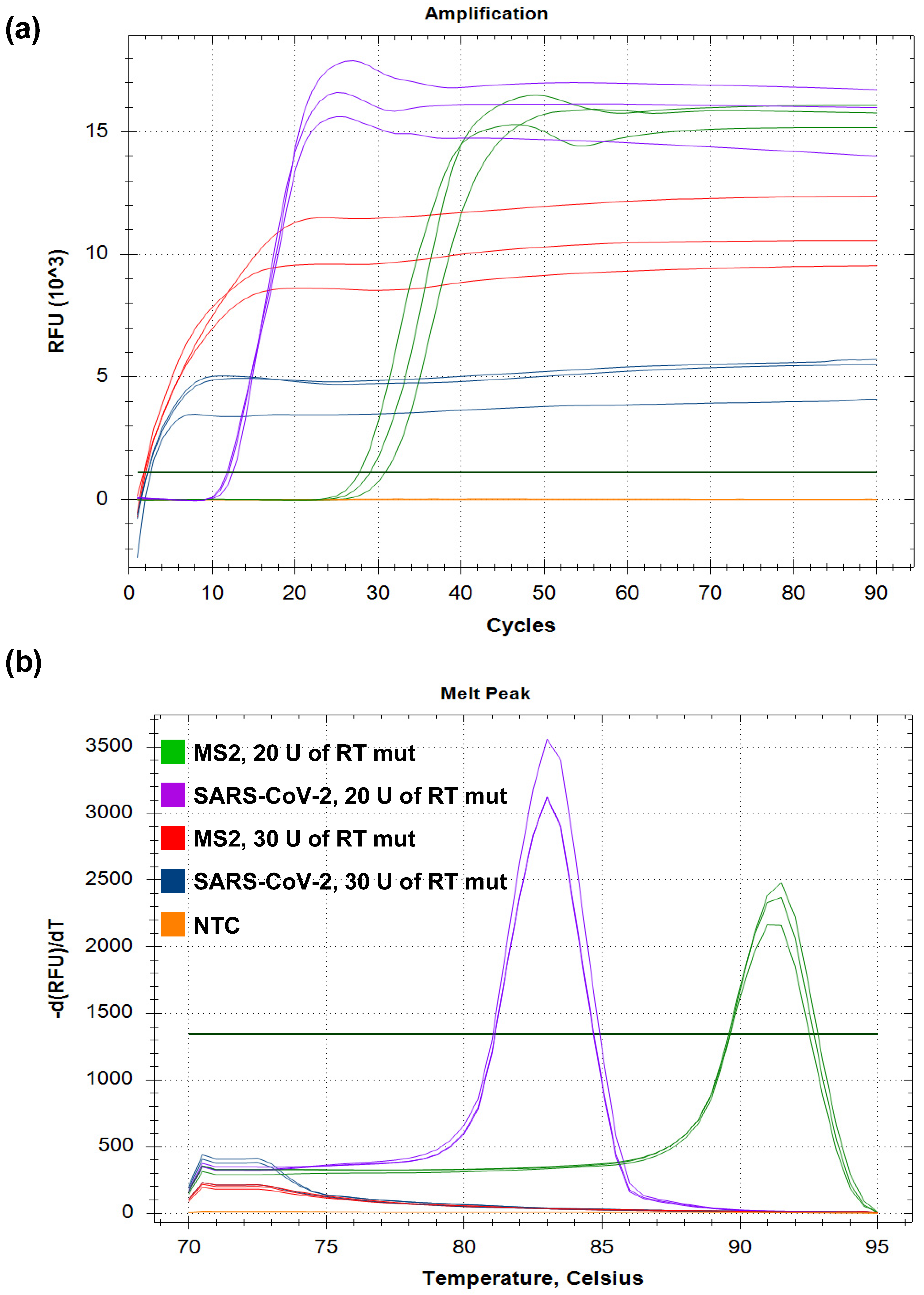
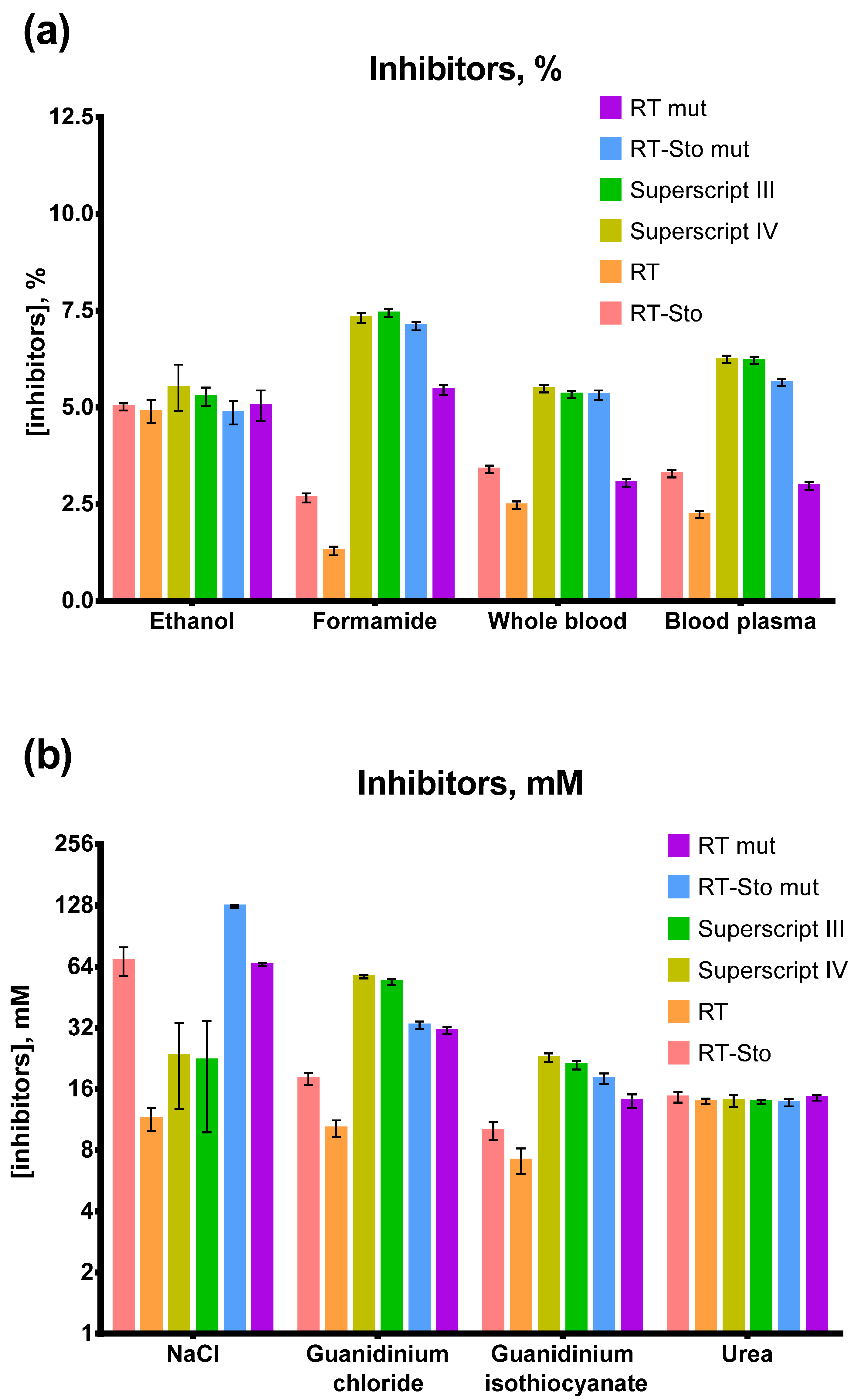
3.5. Inhibitors in RT-LAMP
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lingner, J. Reverse Transcriptase Motifs in the Catalytic Subunit of Telomerase. Science 1997, 276, 561–567. [Google Scholar] [CrossRef] [PubMed]
- Menéndez-Arias, L.; Sebastián-Martín, A.; Álvarez, M. Viral Reverse Transcriptases. Virus Res. 2016, 234, 153–176. [Google Scholar] [CrossRef] [PubMed]
- Baltimore, D. RNA-Dependent DNA Polymerase in Virions of RNA Tumour Viruses. Nature 1970, 226, 1209–1211. [Google Scholar] [CrossRef] [PubMed]
- Temin, H.M.; Mizutani, S. RNA-Dependent DNA Polymerase in Virions of Rous Sarcoma Virus. Nature 1970, 226, 1211–1213. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Liu, F.; Pyle, A.M. An Ultraprocessive, Accurate Reverse Transcriptase Encoded by a Metazoan Group II Intron. RNA 2018, 24, 183–195. [Google Scholar] [CrossRef]
- Mohr, S.; Ghanem, E.; Smith, W.; Sheeter, D.; Qin, Y.; King, O.; Polioudakis, D.; Iyer, V.R.; Hunicke-Smith, S.; Swamy, S.; et al. Thermostable Group II Intron Reverse Transcriptase Fusion Proteins and Their Use in CDNA Synthesis and Next-Generation RNA Sequencing. RNA 2013, 19, 958–970. [Google Scholar] [CrossRef]
- Di Marzo Veronese, F.; Copeland, T.; DeVico, A.; Rahman, R.; Oroszlan, S.; Gallo, R.; Sarngadharan, M. Characterization of Highly Immunogenic P66/P51 as the Reverse Transcriptase of HTLV-III/LAV. Science 1986, 231, 1289–1291. [Google Scholar] [CrossRef]
- Houts, G.E.; Miyagi, M.; Ellis, C.; Beard, D.; Beard, J.W. Reverse Transcriptase from Avian Myeloblastosis Virus. J. Virol. 1979, 29, 517–522. [Google Scholar] [CrossRef]
- Roth, M.J.; Tanese, N.; Goff, S.P. Purification and Characterization of Murine Retroviral Reverse Transcriptase Expressed in Escherichia Coli. J. Biol. Chem. 1985, 260, 9326–9335. [Google Scholar] [CrossRef]
- Kaushik, N.; Singh, K.; Alluru, I.; Modak, M.J. Tyrosine 222, a Member of the YXDD Motif of MuLV RT, Is Catalytically Essential and Is a Major Component of the Fidelity Center. Biochemistry 1999, 38, 2617–2627. [Google Scholar] [CrossRef]
- Gerard, G.F.; Potter, R.J.; Smith, M.D.; Rosenthal, K.; Dhariwal, G.; Lee, J.; Chatterjee, D.K. The Role of Template-Primer in Protection of Reverse Transcriptase from Thermal Inactivation. Nucleic Acids Res. 2002, 30, 3118–3129. [Google Scholar] [CrossRef] [PubMed]
- Skasko, M.; Weiss, K.K.; Reynolds, H.M.; Jamburuthugoda, V.; Lee, K.; Kim, B. Mechanistic Differences in RNA-Dependent DNA Polymerization and Fidelity between Murine Leukemia Virus and HIV-1 Reverse Transcriptases. J. Biol. Chem. 2005, 280, 12190–12200. [Google Scholar] [CrossRef] [PubMed]
- Oz-Gleenberg, I.; Herzig, E.; Voronin, N.; Hizi, A. Substrate Variations That Affect the Nucleic Acid Clamp Activity of Reverse Transcriptases. FEBS J. 2012, 279, 1894–1903. [Google Scholar] [CrossRef] [PubMed]
- Whiting, S.H.; Champoux, J.J. Strand Displacement Synthesis Capability of Moloney Murine Leukemia Virus Reverse Transcriptase. J. Virol. 1994, 68, 4747–4758. [Google Scholar] [CrossRef]
- Chen, D.; Patton, J.T. Reverse Transcriptase Adds Nontemplated Nucleotides to CDNAs during 5′-RACE and Primer Extension. Biotechniques 2001, 30, 574–580, 582. [Google Scholar] [CrossRef]
- Arezi, B.; Hogrefe, H. Novel Mutations in Moloney Murine Leukemia Virus Reverse Transcriptase Increase Thermostability through Tighter Binding to Template-Primer. Nucleic Acids Res. 2009, 37, 473–481. [Google Scholar] [CrossRef]
- Baranauskas, A.; Paliksa, S.; Alzbutas, G.; Vaitkevicius, M.; Lubiene, J.; Letukiene, V.; Burinskas, S.; Sasnauskas, G.; Skirgaila, R. Generation and Characterization of New Highly Thermostable and Processive M-MuLV Reverse Transcriptase Variants. Protein Eng. Des. Sel. 2012, 25, 657–668. [Google Scholar] [CrossRef]
- Yasukawa, K.; Mizuno, M.; Konishi, A.; Inouye, K. Increase in Thermal Stability of Moloney Murine Leukaemia Virus Reverse Transcriptase by Site-Directed Mutagenesis. J. Biotechnol. 2010, 150, 299–306. [Google Scholar] [CrossRef]
- Yasukawa, K.; Nemoto, D.; Inouye, K. Comparison of the Thermal Stabilities of Reverse Transcriptases from Avian Myeloblastosis Virus and Moloney Murine Leukaemia Virus. J. Biochem. 2008, 143, 261–268. [Google Scholar] [CrossRef]
- Oscorbin, I.P.; Wong, P.F.; Boyarskikh, U.A.; Khrapov, E.A.; Filipenko, M.L. The Attachment of a DNA-Binding Sso7d-like Protein Improves Processivity and Resistance to Inhibitors of M-MuLV Reverse Transcriptase. FEBS Lett. 2020, 594, 4338–4356. [Google Scholar] [CrossRef]
- Baba, M.; Kakue, R.; Leucht, C.; Rasor, P.; Walch, H.; Ladiges, D.; Bell, C.; Kojima, K.; Takita, T.; Yasukawa, K. Further Increase in Thermostability of Moloney Murine Leukemia Virus Reverse Transcriptase by Mutational Combination. Protein Eng. Des. Sel. 2017, 30, 551–557. [Google Scholar] [CrossRef] [PubMed]
- Arezi, B.; McCarthy, M.; Hogrefe, H. Mutant of Moloney Murine Leukemia Virus Reverse Transcriptase Exhibits Higher Resistance to Common RT-QPCR Inhibitors. Anal. Biochem. 2010, 400, 301–303. [Google Scholar] [CrossRef] [PubMed]
- Notomi, T.; Okayama, H.; Masubuchi, H.; Yonekawa, T.; Watanabe, K.; Amino, N.; Hase, T. Loop-Mediated Isothermal Amplification of DNA. Nucleic Acids Res. 2000, 28, E63. [Google Scholar] [CrossRef] [PubMed]
- Francois, P.; Tangomo, M.; Hibbs, J.; Bonetti, E.-J.; Boehme, C.C.; Notomi, T.; Perkins, M.D.; Schrenzel, J. Robustness of a Loop-Mediated Isothermal Amplification Reaction for Diagnostic Applications. FEMS Immunol. Med. Microbiol. 2011, 62, 41–48. [Google Scholar] [CrossRef]
- Moore, K.J.M.; Cahill, J.; Aidelberg, G.; Aronoff, R.; Bektaş, A.; Bezdan, D.; Butler, D.J.; Chittur, S.V.; Codyre, M.; Federici, F.; et al. Loop-Mediated Isothermal Amplification Detection of SARS-CoV-2 and Myriad Other Applications. J. Biomol. Tech. 2021, 32, 228–275. [Google Scholar] [CrossRef]
- Poon, L.L.M.; Leung, C.S.W.; Chan, K.H.; Lee, J.H.C.; Yuen, K.Y.; Guan, Y.; Peiris, J.S.M. Detection of Human Influenza A Viruses by Loop-Mediated Isothermal Amplification. J. Clin. Microbiol. 2005, 43, 427–430. [Google Scholar] [CrossRef]
- Modak, S.S.; Barber, C.A.; Geva, E.; Abrams, W.R.; Malamud, D.; Ongagna, Y.S.Y. Rapid Point-of-Care Isothermal Amplification Assay for the Detection of Malaria without Nucleic Acid Purification. Infect. Dis. (Auckl.) 2016, 9, 1–9. [Google Scholar] [CrossRef]
- The Use of Loop-Mediated Isothermal Amplification (TB-LAMP) for the Diagnosis of Pulmonary Tuberculosis: Policy Guidance; World Health Organization: Geneva, Switzerland, 2016; ISBN 9789241511186.
- Guo, X.G.; Zhou, Y.Z.; Li, Q.; Wang, W.; Wen, J.Z.; Zheng, L.; Wang, Q. Rapid and Reliable Diagnostic Method to Detect Zika Virus by Real-Time Fluorescence Reverse Transcription Loop-Mediated Isothermal Amplification. AMB Express 2018, 8, 60. [Google Scholar] [CrossRef]
- Siriyasatien, P.; Wacharapluesadee, S.; Kraivichian, K.; Suwanbamrung, C.; Sutthanont, N.; Cantos-Barreda, A.; Phumee, A. Development and Evaluation of a Visible Reverse Transcription-Loop-Mediated Isothermal Amplification (RT-LAMP) for the Detection of Asian Lineage ZIKV in Field-Caught Mosquitoes. Acta Trop. 2022, 236, 106691. [Google Scholar] [CrossRef]
- Lalli, M.A.; Langmade, J.S.; Chen, X.; Fronick, C.C.; Sawyer, C.S.; Burcea, L.C.; Wilkinson, M.N.; Fulton, R.S.; Heinz, M.; Buchser, W.J.; et al. Rapid and Extraction-Free Detection of SARS-CoV-2 from Saliva by Colorimetric Reverse-Transcription Loop-Mediated Isothermal Amplification. Clin. Chem. 2021, 67, 415–424. [Google Scholar] [CrossRef]
- Dao Thi, V.L.; Herbst, K.; Boerner, K.; Meurer, M.; Kremer, L.P.; Kirrmaier, D.; Freistaedter, A.; Papagiannidis, D.; Galmozzi, C.; Stanifer, M.L.; et al. A Colorimetric RT-LAMP Assay and LAMP-Sequencing for Detecting SARS-CoV-2 RNA in Clinical Samples. Sci. Transl. Med. 2020, 12, eabc7075. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Tanner, N.A. Development of Multiplexed Reverse-Transcription Loop-Mediated Isothermal Amplification for Detection of SARS-CoV-2 and Influenza Viral RNA. Biotechniques 2021, 70, 167–174. [Google Scholar] [CrossRef]
- Jang, W.S.; Lim, D.H.; Yoon, J.; Kim, A.; Lim, M.; Nam, J.; Yanagihara, R.; Ryu, S.-W.; Jung, B.K.; Ryoo, N.-H.; et al. Development of a Multiplex Loop-Mediated Isothermal Amplification (LAMP) Assay for on-Site Diagnosis of SARS-CoV-2. PLoS ONE 2021, 16, e0248042. [Google Scholar] [CrossRef] [PubMed]
- Yan, C.; Cui, J.; Huang, L.; Du, B.; Chen, L.; Xue, G.; Li, S.; Zhang, W.; Zhao, L.; Sun, Y.; et al. Rapid and Visual Detection of 2019 Novel Coronavirus (SARS-CoV-2) by a Reverse Transcription Loop-Mediated Isothermal Amplification Assay. Clin. Microbiol. Infect. 2020, 26, 773–779. [Google Scholar] [CrossRef] [PubMed]
- Broughton, J.; Deng, X.; Yu, G.; Fasching, C.; Singh, J.; Streithorst, J.; Granados, A.; Sotomayor-Gonzalez, A.; Zorn, K.; Gopez, A.; et al. Rapid Detection of 2019 Novel Coronavirus SARS-CoV-2 Using a CRISPR-Based DETECTR Lateral Flow Assay. medRxiv 2020. [Google Scholar] [CrossRef]
- Park, G.S.; Ku, K.; Baek, S.H.; Kim, S.J.; Kim, S.I.; Kim, B.T.; Maeng, J.S. Development of Reverse Transcription Loop-Mediated Isothermal Amplification Assays Targeting Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2). J. Mol. Diagn. 2020, 22, 729–735. [Google Scholar] [CrossRef]
- Kellner, M.J.; Ross, J.J.; Schnabl, J.; Dekens, M.P.S.; Matl, M.; Heinen, R.; Grishkovskaya, I.; Bauer, B.; Stadlmann, J.; Menéndez-Arias, L.; et al. A Rapid, Highly Sensitive and Open-Access SARS-CoV-2 Detection Assay for Laboratory and Home Testing. Front. Mol. Biosci. 2022, 9, 801309. [Google Scholar] [CrossRef]
- Nie, X. Reverse Transcription Loop-Mediated Isothermal Amplification of DNA for Detection of Potato Virus Y. Plant Dis. 2005, 89, 605–610. [Google Scholar] [CrossRef]
- Jiang, M.; Pan, W.; Arasthfer, A.; Fang, W.; Ling, L.; Fang, H.; Daneshnia, F.; Yu, J.; Liao, W.; Pei, H.; et al. Development and Validation of a Rapid, Single-Step Reverse Transcriptase Loop-Mediated Isothermal Amplification (RT-LAMP) System Potentially to Be Used for Reliable and High-Throughput Screening of COVID-19. Front. Cell. Infect. Microbiol. 2020, 10, 331. [Google Scholar] [CrossRef]
- Stemmer, W.P.C.; Crameri, A.; Ha, K.D.; Brennan, T.M.; Heyneker, H.L. Single-Step Assembly of a Gene and Entire Plasmid from Large Numbers of Oligodeoxyribonucleotides. Gene 1995, 164, 49–53. [Google Scholar] [CrossRef]
- Sambrook, J.; Russell, D.W. Purification of Nucleic Acids by Extraction with Phenol:Chloroform. CSH Protoc. 2006, 2006, pdb.prot4455. [Google Scholar] [CrossRef] [PubMed]
- Evans, G.A. Molecular Cloning: A Laboratory Manual (Second Edition): Volumes 1, 2, and 3. Current Protocols in Molecular Biology: Volumes 1 and 2; Cold Spring Harbor Laboratory Press: New York, NY, USA, 1990; Volume 61, ISBN 978-0879693091. [Google Scholar]
- Corman, V.M.; Landt, O.; Kaiser, M.; Molenkamp, R.; Meijer, A.; Chu, D.K.; Bleicker, T.; Brünink, S.; Schneider, J.; Schmidt, M.L.; et al. Detection of 2019 Novel Coronavirus (2019-NCoV) by Real-Time RT-PCR. Eurosurveillance 2020, 25, 2000045. [Google Scholar] [CrossRef] [PubMed]
- Oscorbin, I.P.; Shevelev, G.Y.; Pronyaeva, K.A.; Stepanov, A.A.; Shamovskaya, D.V.; Mishukova, O.V.; Pyshnyi, D.V.; Filipenko, M.L. Detection of SARS-CoV-2 RNA by a Multiplex Reverse-Transcription Loop-Mediated Isothermal Amplification Coupled with Melting Curves Analysis. Int. J. Mol. Sci. 2021, 22, 5743. [Google Scholar] [CrossRef] [PubMed]
- Oscorbin, I.P.; Boyarskikh, U.A.; Filipenko, M.L. Large Fragment of DNA Polymerase I from Geobacillus Sp. 777: Cloning and Comparison with DNA Polymerases I in Practical Applications. Mol. Biotechnol. 2015, 57, 947–959. [Google Scholar] [CrossRef] [PubMed]
- Bektaş, A.; Covington, M.F.; Aidelberg, G.; Arce, A.; Matute, T.; Núñez, I.; Walsh, J.; Boutboul, D.; Delaugerre, C.; Lindner, A.B.; et al. Accessible LAMP-Enabled Rapid Test (ALERT) for Detecting SARS-CoV-2. Viruses 2021, 13, 742. [Google Scholar] [CrossRef]
- Kotewicz, M.; Gerard, G. Cloned Genes Encoding Reverse Transcriptase Lacking RNase H Activity 2. U.S. Patent 5244797B1, 14 September 1993. [Google Scholar]
- Potter, R.J.; Rosenthal, K. High Fidelity Reverse Transcriptases and Uses Thereof 3. U.S. Patent 7056716B2, 6 June 2006. [Google Scholar]
- Rogers, J.; Potter, J. Reverse Transcriptases for Use in High Temperature Nucleic Acid Synthesis 5. U.S. Patent 9663770B2, 30 May 2017. [Google Scholar]
- Katano, Y.; Li, T.; Baba, M.; Nakamura, M.; Ito, M.; Kojima, K.; Takita, T.; Yasukawa, K. Generation of Thermostable Moloney Murine Leukemia Virus Reverse Transcriptase Variants Using Site Saturation Mutagenesis Library and Cell-Free Protein Expression System. Biosci. Biotechnol. Biochem. 2017, 81, 2339–2345. [Google Scholar] [CrossRef]
- Skirgaila, R.; Pudzaitis, V.; Paliksa, S.; Vaitkevicius, M.; Janulaitis, A. Compartmentalization of Destabilized Enzyme-MRNA-Ribosome Complexes Generated by Ribosome Display: A Novel Tool for the Directed Evolution of Enzymes. Protein Eng. Des. Sel. 2013, 26, 453–461. [Google Scholar] [CrossRef][Green Version]
- Janulaitis, A.; Skirgaila, R.; Siksniene, D. Production of Nucleic Acid 102. U.S. Patent 8835148B2, 16 September 2014. [Google Scholar]
- Stahlberg, A. Comparison of Reverse Transcriptases in Gene Expression Analysis. Clin. Chem. 2004, 50, 1678–1680. [Google Scholar] [CrossRef]
- Sieber, M.W.; Recknagel, P.; Glaser, F.; Witte, O.W.; Bauer, M.; Claus, R.A.; Frahm, C. Substantial Performance Discrepancies among Commercially Available Kits for Reverse Transcription Quantitative Polymerase Chain Reaction: A Systematic Comparative Investigator-Driven Approach. Anal. Biochem. 2010, 401, 303–311. [Google Scholar] [CrossRef]
- Moison, C.; Arimondo, P.B.; Guieysse-Peugeot, A.-L. Commercial Reverse Transcriptase as Source of False-Positive Strand-Specific RNA Detection in Human Cells. Biochimie 2011, 93, 1731–1737. [Google Scholar] [CrossRef]
- Borst, A.; Box, A.T.A.; Fluit, A.C. False-Positive Results and Contamination in Nucleic Acid Amplification Assays: Suggestions for a Prevent and Destroy Strategy. Eur. J. Clin. Microbiol. Infect. Dis. 2004, 23, 289–299. [Google Scholar] [CrossRef] [PubMed]
- Kotewicz, M.L.; Sampson, C.M.; D’Alessio, J.M.; Gerard, G.F. Isolation of Cloned Moloney Murine Leukemia Virus Reverse Transcriptase Lacking Ribonuclease H Activity. Nucleic Acids Res. 1988, 16, 265–277. [Google Scholar] [CrossRef] [PubMed]
- Misra, V.K.; Hecht, J.L.; Sharp, K.A.; Friedman, R.A.; Honig, B. Salt Effects on Protein-DNA Interactions. The Lambda CI Repressor and EcoRI Endonuclease. J. Mol. Biol. 1994, 238, 264–280. [Google Scholar] [CrossRef] [PubMed]
- Kutnowski, N.; Shmulevich, F.; Davidov, G.; Shahar, A.; Bar-Zvi, D.; Eichler, J.; Zarivach, R.; Shaanan, B. Specificity of Protein–DNA Interactions in Hypersaline Environment: Structural Studies on Complexes of Halobacterium Salinarum Oxidative Stress-Dependent Protein HsRosR. Nucleic Acids Res. 2019, 47, 8860–8873. [Google Scholar] [CrossRef]
- Minshall, N.; Git, A. Enzyme- and Gene-Specific Biases in Reverse Transcription of RNA Raise Concerns for Evaluating Gene Expression. Sci. Rep. 2020, 10, 8151. [Google Scholar] [CrossRef]
- Nardon, E.; Donada, M.; Bonin, S.; Dotti, I.; Stanta, G. Higher Random Oligo Concentration Improves Reverse Transcription Yield of CDNA from Bioptic Tissues and Quantitative RT-PCR Reliability. Exp. Mol. Pathol. 2009, 87, 146–151. [Google Scholar] [CrossRef]
- Stangegaard, M.; Dufva, I.H.; Dufva, M. Reverse Transcription Using Random Pentadecamer Primers Increases Yield and Quality of Resulting CDNA. Biotechniques 2006, 40, 649–657. [Google Scholar] [CrossRef]
- Spiess, A.-N.; Ivell, R. A Highly Efficient Method for Long-Chain CDNA Synthesis Using Trehalose and Betaine. Anal. Biochem. 2002, 301, 168–174. [Google Scholar] [CrossRef]
- Jucá, M.B.; Aoyama, H. Effect of Dimethyl Sulfoxide on Reverse Transcriptase Activity. Braz. J. Med. Biol. Res. 1995, 28, 285–290. [Google Scholar]

| Name | Alteration |
|---|---|
| RT | Truncated RNAse H domain |
| RT-Sto | Truncated RNAse H domain, fusion with Sto7d protein |
| RT mut | Truncated RNAse H domain, mutations L139P, D200N, T330P |
| RT-Sto-mut | Truncated RNAse H domain, mutations L139P, D200N, T330P, fusion with Sto7d protein |
| Superscript III | Mutations H204R, T306K, F309N, V223H, D524G, E562Q, and D583N |
| Superscript IV | Mutations P51L, S67R, E69K, T197A, H204R, E302K, F309N, W313F, T330P, L435G, N454K, D524G, D583N, H594Q, D653N, and L671P |
| Name | 5′-Sequence-3′ |
|---|---|
| CoR2-F2 | TGCAACTGAGGGAGCCTTG |
| CoR2-B2 | TGGAGTTGAATTTCTTGAACTG |
| CoR2-LF | CGGCAGTCAAGCCTCTTCTC |
| CoR2-LB | ATTGTTAGCAGGATTGCGGGT |
| CoR2-FIP | GGAAGTTGTAGCACGATTGCAGATACACCAAAAGATCACATTGG |
| CoR2-BIP | GCTTCTACGCAGAAGGGAGCATGCGACTACGTGATGAGGAA |
| MS2-2-F3 | TGCCTGTAAGGAGCCTGAT |
| MS2-2-B3 | TGAGCGGATACGATCGAGAT |
| MS2-2-LB | GTCTATACCAACGGATTTGAGCC |
| MS2-2-LF | GCATCCGATTCCATCTCCGAT |
| MS2-1-FIP | CTCCTGAGGGAATGTGGGAACCCCGGCGTGCGCGTTAT |
| MS2-1-BIP | GCCAGCGAGCTCTCCTCGGGCACCCGTGCTCTTTCGA |
| E_Sarb_F | ACAGGTACGTTAATAGTTAATAGCGT |
| E_Sarb_R | ATATTGCAGCAGTACGCACACA |
| E_Sarb_P | HEX-ACACTAGCCATCCTTACTGCGCTTCG-BHQ2 |
| MS2-5-F | GTACGAGGAGAAAGCCGGTTTC |
| MS2-5-R | GTTCTGCGGCACTTCGATG |
| MS2-5-P | FAM-TCCCTCGACGCACGCTCCTGCT-BHQ1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Oscorbin, I.P.; Novikova, L.M.; Filipenko, M.L. Comparison of Reverse Transcriptase (RT) Activities of Various M-MuLV RTs for RT-LAMP Assays. Biology 2022, 11, 1809. https://doi.org/10.3390/biology11121809
Oscorbin IP, Novikova LM, Filipenko ML. Comparison of Reverse Transcriptase (RT) Activities of Various M-MuLV RTs for RT-LAMP Assays. Biology. 2022; 11(12):1809. https://doi.org/10.3390/biology11121809
Chicago/Turabian StyleOscorbin, Igor P., Lidiya M. Novikova, and Maxim L. Filipenko. 2022. "Comparison of Reverse Transcriptase (RT) Activities of Various M-MuLV RTs for RT-LAMP Assays" Biology 11, no. 12: 1809. https://doi.org/10.3390/biology11121809
APA StyleOscorbin, I. P., Novikova, L. M., & Filipenko, M. L. (2022). Comparison of Reverse Transcriptase (RT) Activities of Various M-MuLV RTs for RT-LAMP Assays. Biology, 11(12), 1809. https://doi.org/10.3390/biology11121809








