Processing of Fluorescent Proteins May Prevent Detection of Prion Particles in [PSI+] Cells
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Plasmids
2.2. Yeast Strains
2.3. Media and Cultivation
2.4. Fluorescence Microscopy
2.5. Protein Analysis
3. Results
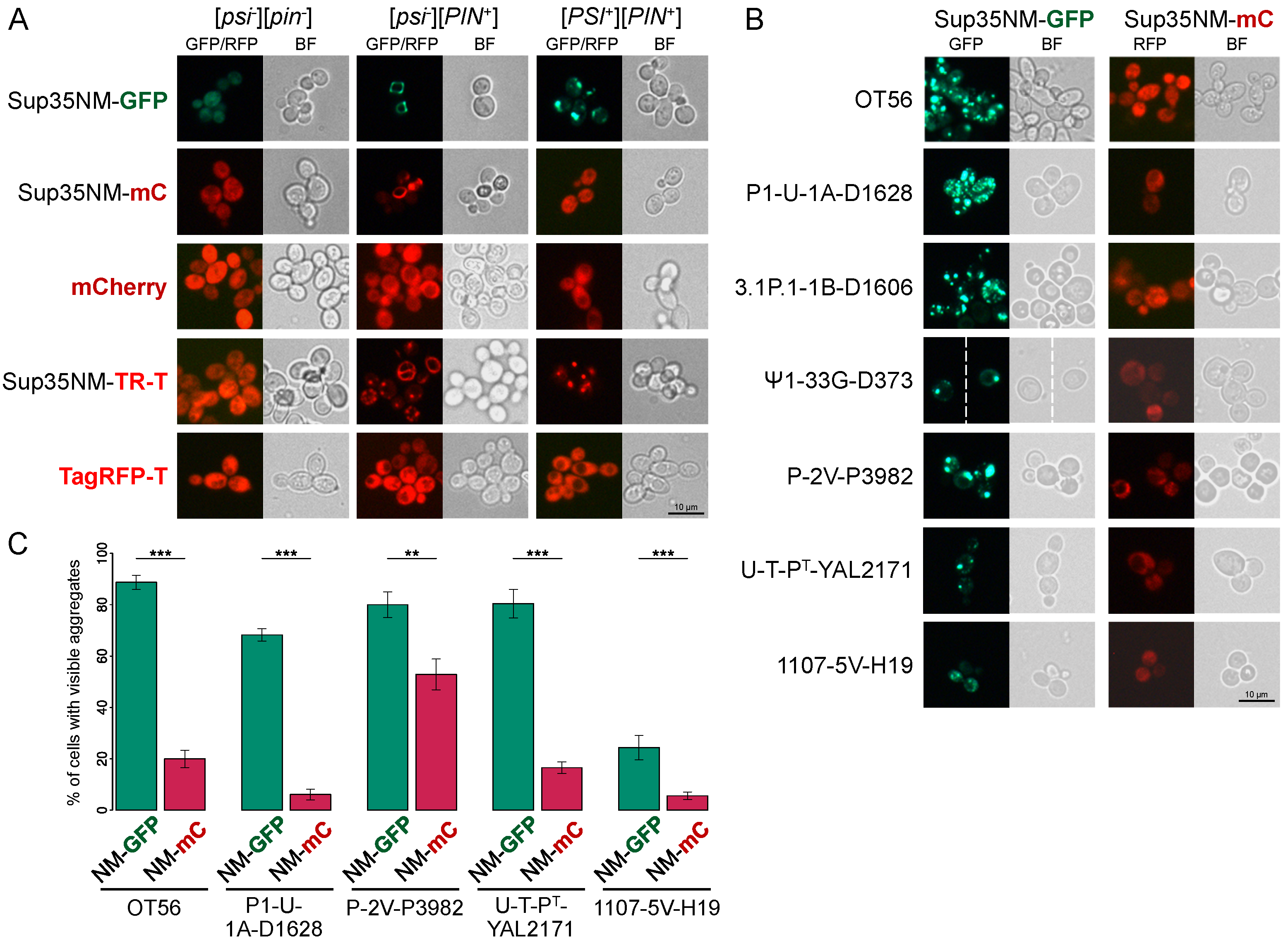
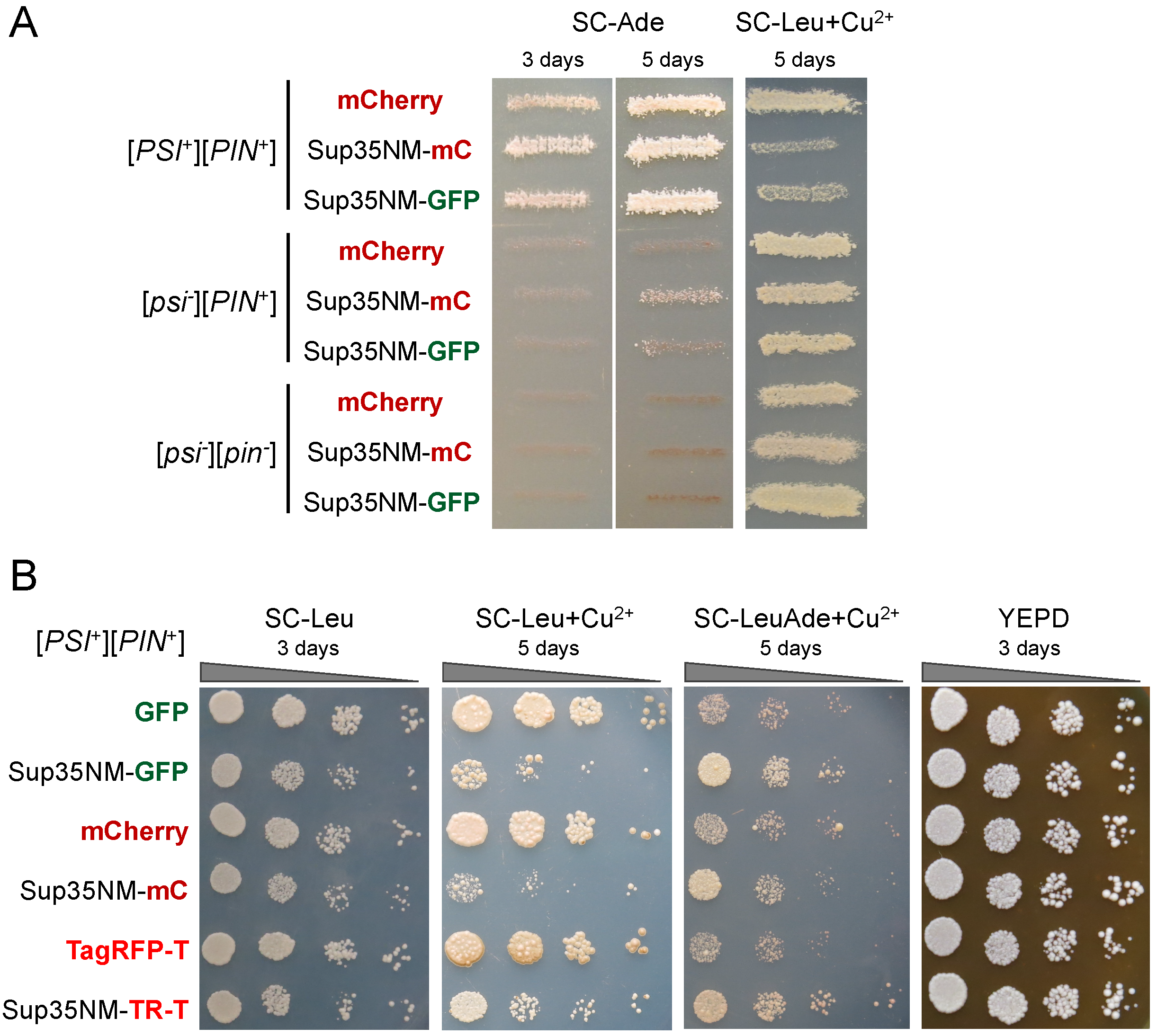
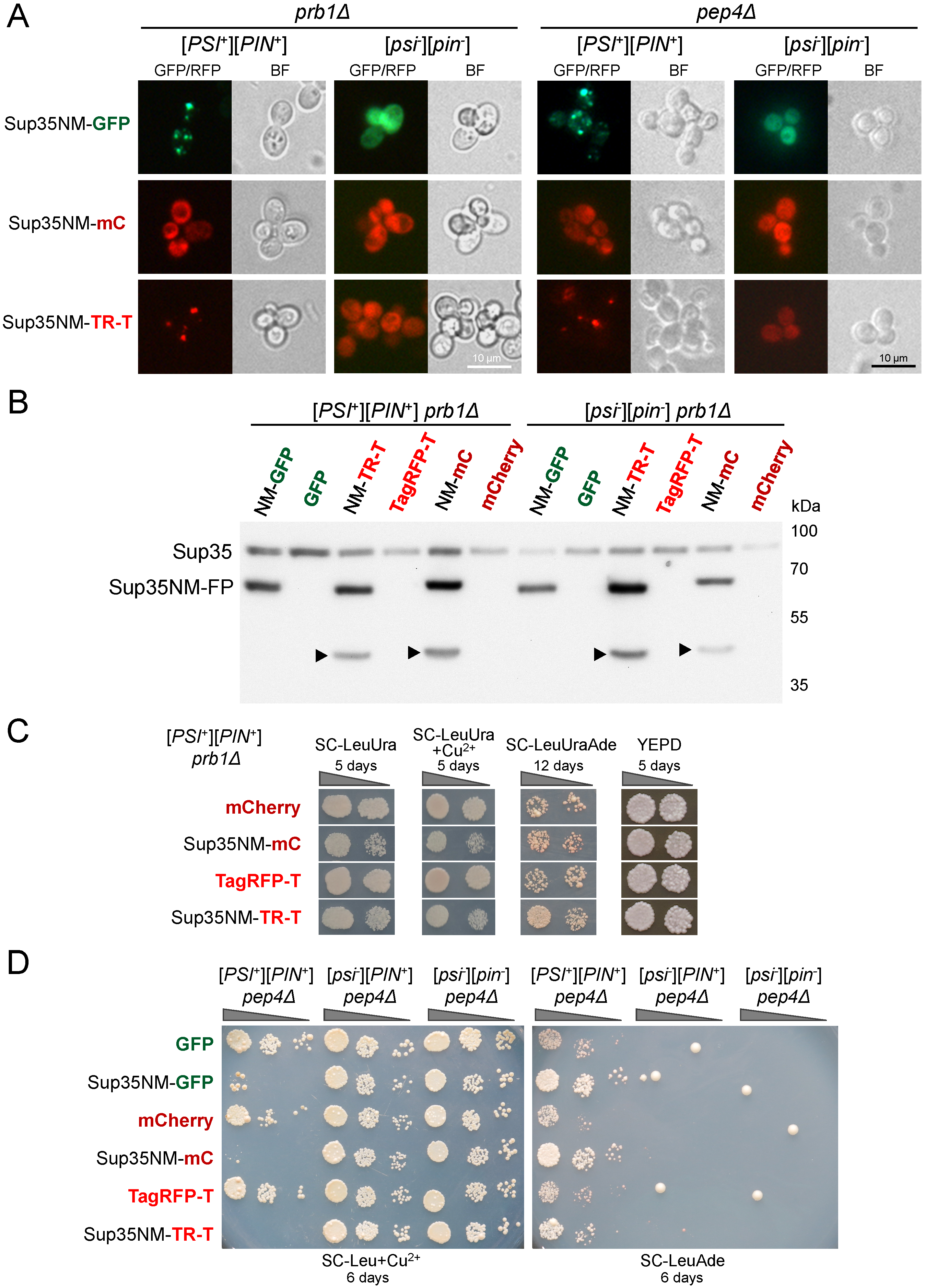
3.1. Sup35NM-mCherry Does Not Allow Detection of the [PSI+] Aggregates
3.2. Fusion to mCherry Does Not Affect Sup35NM Prion Properties
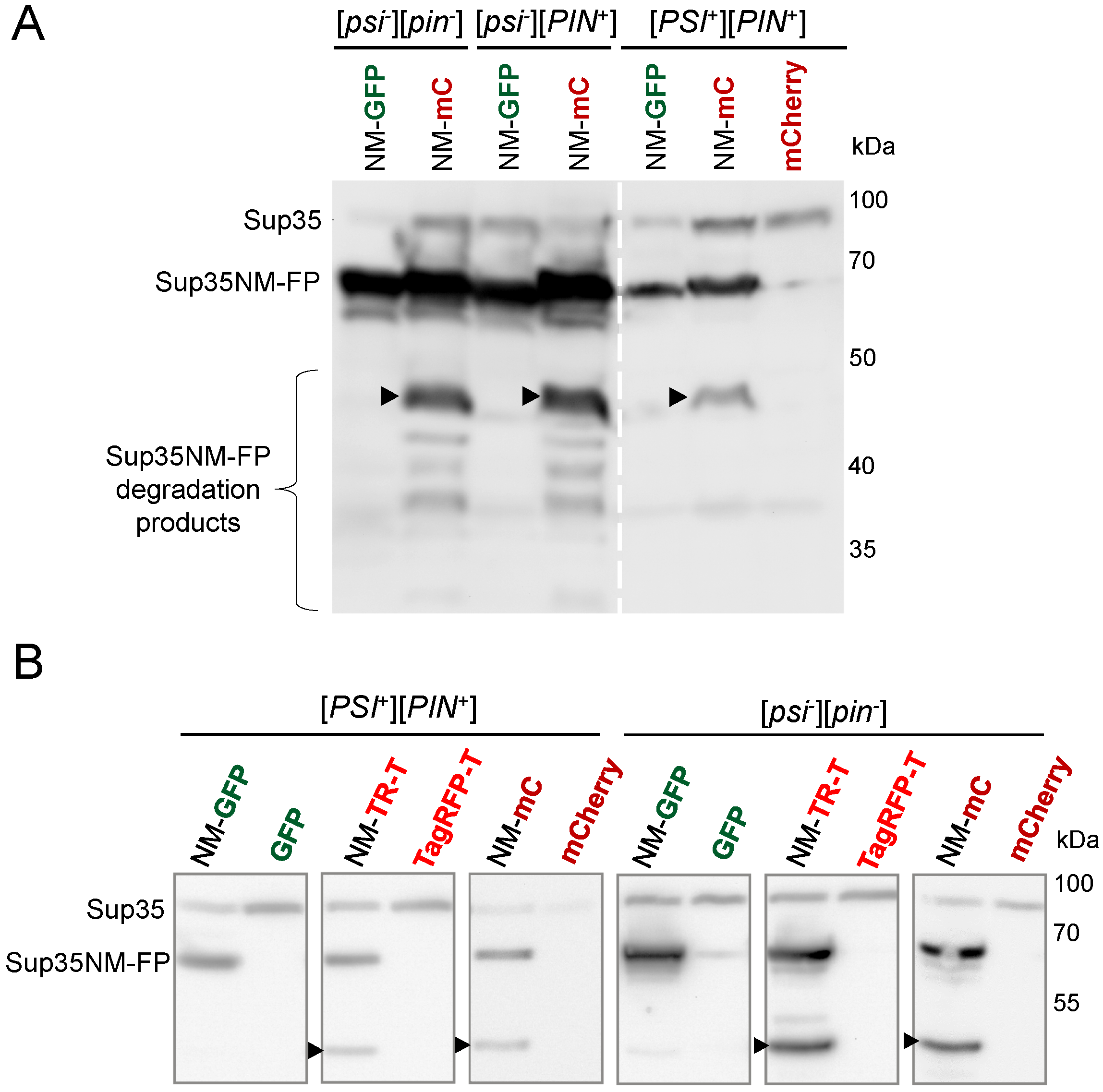
3.3. Sup35NM-mCherry and Sup35NM-TagRFP-T Are Partially Degraded in Yeast Cells
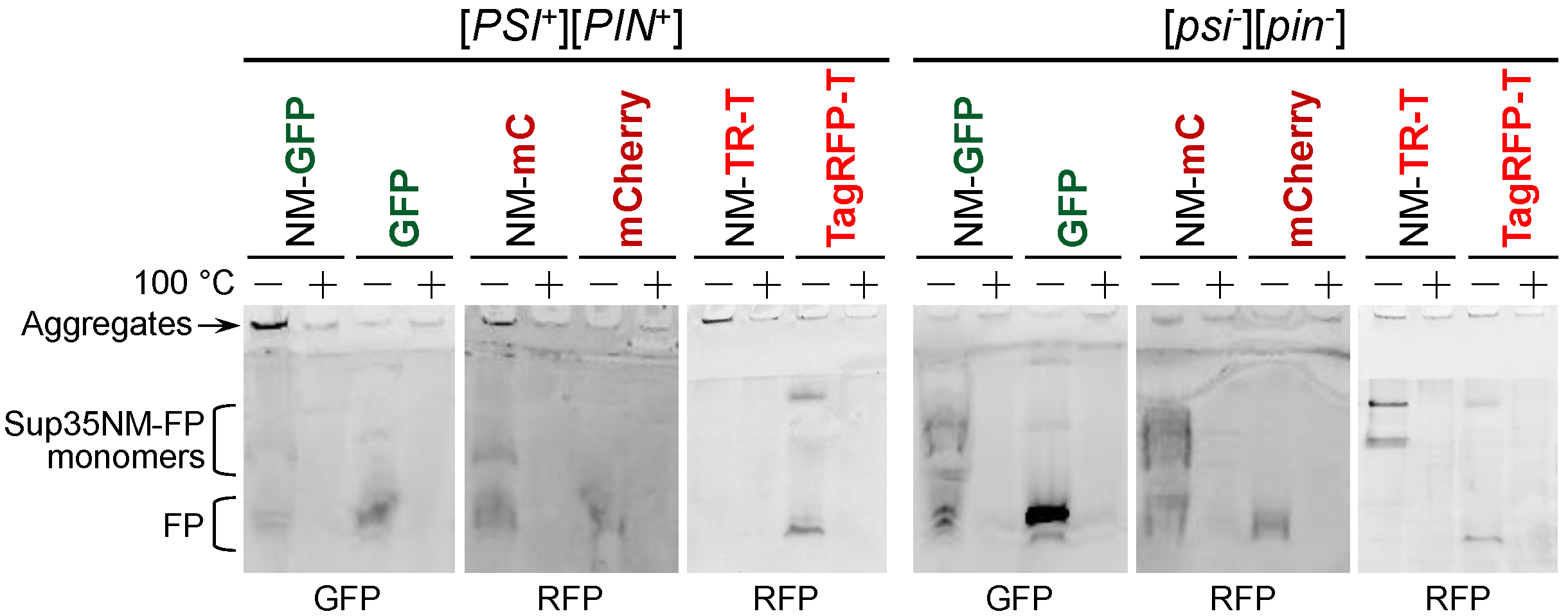
3.4. Products of Sup35NM-mCherry Degradation Retain Fluorescent Properties thus Interfering with Detection of the [PSI+] Aggregates
3.5. Degradation of Sup35NM-mCherry and Sup35NM-TagRFP-T Is Not Caused by Vacuolar Proteases
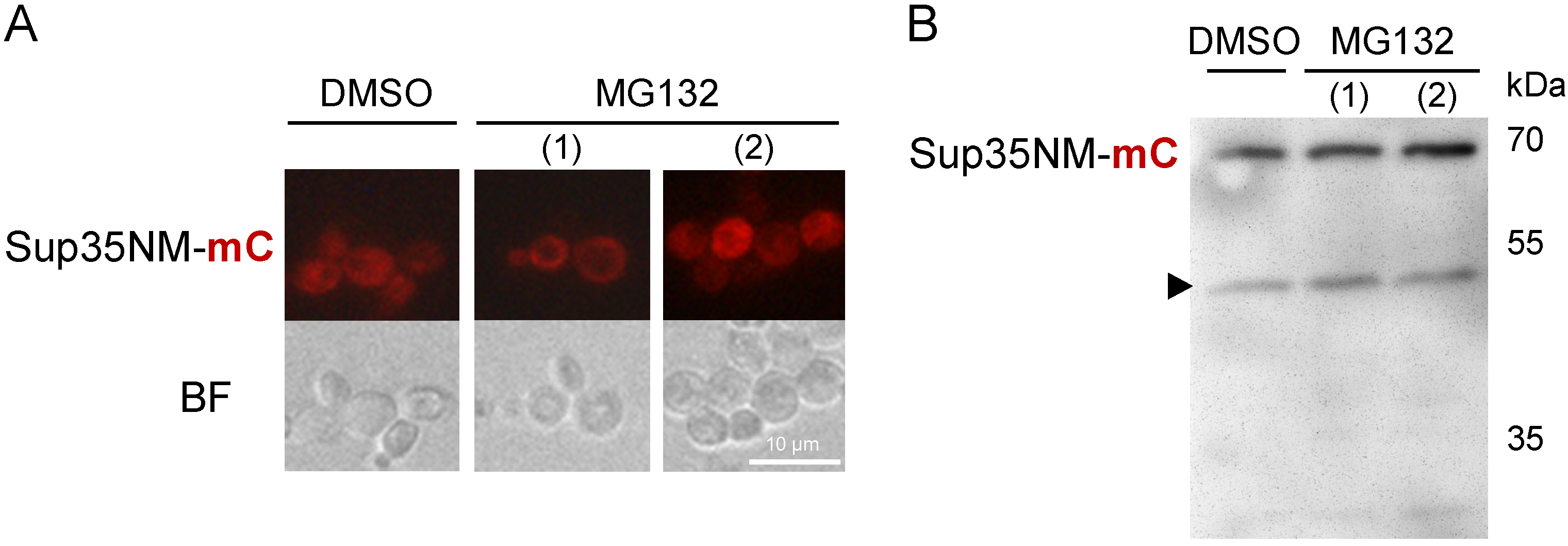
3.6. Sup35NM-mCherry Degradation Is Not Affected by the Ubiquitin-Proteasomal Pathway
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
Abbreviations
| aa | Amino acid |
| GFP | Green fluorescent protein |
| GuHCl | Guanidine hydrochloride |
| RFP | Red fluorescent protein |
References
- Wickner, R.B.; Kryndushkin, D.; Shewmaker, F.; McGlinchey, R.; Edskes, H.K. Study of amyloids using yeast. Methods Mol. Biol. 2018, 1779, 313–339. [Google Scholar] [CrossRef] [PubMed]
- Liebman, S.W.; Chernoff, Y.O. Prions in yeast. Genetics 2012, 191, 1041–1072. [Google Scholar] [CrossRef] [PubMed]
- Wickner, R.B.; Edskes, H.K.; Gorkovskiy, A.; Bezsonov, E.E.; Stroobant, E.E. Yeast and fungal prions: Amyloid-handling systems, amyloid structure, and prion biology. Adv. Genet. 2016, 93, 191–236. [Google Scholar] [CrossRef] [PubMed]
- Naeimi, W.R.; Serio, T.R. Beyond amyloid fibers: Accumulation, biological relevance, and regulation of higher-order prion architectures. Viruses 2022, 14, 1635. [Google Scholar] [CrossRef]
- Matiiv, A.B.; Trubitsina, N.P.; Matveenko, A.G.; Barbitoff, Y.A.; Zhouravleva, G.A.; Bondarev, S.A. Structure and polymorphism of amyloid and amyloid-like aggregates. Biochemistry 2022, 87, 450–463. [Google Scholar] [CrossRef]
- Cox, B.; Tuite, M. The life of [PSI]. Curr. Genet. 2018, 64, 1–8. [Google Scholar] [CrossRef]
- Paushkin, S.V.; Kushnirov, V.V.; Smirnov, V.N.; Ter-Avanesyan, M.D. Propagation of the yeast prion-like [psi+] determinant is mediated by oligomerization of the SUP35-encoded polypeptide chain release factor. EMBO J. 1996, 15, 3127–3134. [Google Scholar] [CrossRef]
- Ter-Avanesyan, M.D.; Kushnirov, V.V.; Dagkesamanskaya, A.R.; Didichenko, S.A.; Chernoff, Y.O.; Inge-Vechtomov, S.G.; Smirnov, V.N. Deletion analysis of the SUP35 gene of the yeast Saccharomyces cerevisiae reveals two non-overlapping functional regions in the encoded protein. Mol. Microbiol. 1993, 7, 683–692. [Google Scholar] [CrossRef]
- Zhouravleva, G.; Frolova, L.; Le Goff, X.; Le Guellec, R.; Inge-Vechtomov, S.; Kisselev, L.; Philippe, M. Termination of translation in eukaryotes is governed by two interacting polypeptide chain release factors, eRF1 and eRF3. EMBO J. 1995, 14, 4065–4072. [Google Scholar] [CrossRef]
- Stansfield, I.; Jones, K.M.; Kushnirov, V.V.; Dagkesamanskaya, A.R.; Poznyakovski, A.I.; Paushkin, S.V.; Nierras, C.R.; Cox, B.S.; Ter-Avanesyan, M.D.; Tuite, M.F. The products of the SUP45 (eRF1) and SUP35 genes interact to mediate translation termination in Saccharomyces cerevisiae. EMBO J. 1995, 14, 4365–4373. [Google Scholar] [CrossRef]
- Franzmann, T.M.; Jahnel, M.; Pozniakovsky, A.; Mahamid, J.; Holehouse, A.S.; Nüske, E.; Richter, D.; Baumeister, W.; Grill, S.W.; Pappu, R.V.; et al. Phase separation of a yeast prion protein promotes cellular fitness. Science 2018, 359, eaao5654. [Google Scholar] [CrossRef]
- Barbitoff, Y.A.; Matveenko, A.G.; Zhouravleva, G.A. Differential interactions of molecular chaperones and yeast prions. J. Fungi 2022, 8, 122. [Google Scholar] [CrossRef] [PubMed]
- Derkatch, I.L.; Chernoff, Y.O.; Kushnirov, V.V.; Inge-Vechtomov, S.G.; Liebman, S.W. Genesis and variability of [PSI] prion factors in Saccharomyces cerevisiae. Genetics 1996, 144, 1375–1386. [Google Scholar] [CrossRef] [PubMed]
- Derkatch, I.L.; Bradley, M.E.; Zhou, P.; Chernoff, Y.O.; Liebman, S.W. Genetic and environmental factors affecting the de novo appearance of the [PSI+] prion in Saccharomyces cerevisiae. Genetics 1997, 147, 507–519. [Google Scholar] [CrossRef] [PubMed]
- Derkatch, I.L.; Bradley, M.E.; Hong, J.Y.; Liebman, S.W. Prions affect the appearance of other prions: The story of [PIN+]. Cell 2001, 106, 171–182. [Google Scholar] [CrossRef]
- Tanaka, M.; Chien, P.; Naber, N.; Cooke, R.; Weissman, J.S. Conformational variations in an infectious protein determine prion strain differences. Nature 2004, 428, 323–328. [Google Scholar] [CrossRef]
- Chernoff, Y.O.; Inge-Vechtomov, S.G.; Derkach, I.L.; Ptyushkina, M.V.; Tarunina, O.V.; Dagkesamanskaya, A.R.; Ter-Avanesyan, M.D. Dosage-dependent translational suppression in yeast Saccharomyces cerevisiae. Yeast 1992, 8, 489–499. [Google Scholar] [CrossRef]
- Kryndushkin, D.S.; Alexandrov, I.M.; Ter-Avanesyan, M.D.; Kushnirov, V.V. Yeast [PSI+] Prion Aggregates Are Formed by Small Sup35 Polymers Fragmented by Hsp104. J. Biol. Chem. 2003, 278, 49636–49643. [Google Scholar] [CrossRef]
- Drozdova, P.B.; Barbitoff, Y.A.; Belousov, M.V.; Skitchenko, R.K.; Rogoza, T.M.; Leclercq, J.Y.; Kajava, A.V.; Matveenko, A.G.; Zhouravleva, G.A.; Bondarev, S.A. Estimation of amyloid aggregate sizes with semi-denaturing detergent agarose gel electrophoresis and its limitations. Prion 2020, 14, 118–128. [Google Scholar] [CrossRef]
- Greene, L.E.; Park, Y.N.; Masison, D.C.; Eisenberg, E. Application of GFP-labeling to study prions in yeast. Protein Pept. Lett. 2009, 16, 635–641. [Google Scholar] [CrossRef][Green Version]
- Liu, J.J.; Lindquist, S. Oligopeptide-repeat expansions modulate ’protein-only’ inheritance in yeast. Nature 1999, 400, 573–576. [Google Scholar] [CrossRef] [PubMed]
- Zhou, P.; Derkatch, I.L.; Liebman, S.W. The relationship between visible intracellular aggregates that appear after overexpression of Sup35 and the yeast prion-like elements [PSI(+)] and [PIN(+)]. Mol. Microbiol. 2001, 39, 37–46. [Google Scholar] [CrossRef] [PubMed]
- Tyedmers, J. Patterns of [PSI+] aggregation allow insights into cellular organization of yeast prion aggregates. Prion 2012, 6, 191–200. [Google Scholar] [CrossRef] [PubMed]
- Serio, T.R.; Cashikar, A.G.; Moslehi, J.J.; Kowal, A.S.; Lindquist, S.L. Yeast prion [psi+] and its determinant, Sup35p. Methods Enzymol. 1999, 309, 649–673. [Google Scholar] [CrossRef]
- Danilov, L.G.; Matveenko, A.G.; Ryzhkova, V.E.; Belousov, M.V.; Poleshchuk, O.I.; Likholetova, D.V.; Sokolov, P.A.; Kasyanenko, N.A.; Kajava, A.V.; Zhouravleva, G.A.; et al. Design of a new [PSI+]-no-more mutation in SUP35 with strong inhibitory effect on the [PSI+] prion propagation. Front. Mol. Neurosci. 2019, 12, 274. [Google Scholar] [CrossRef] [PubMed]
- Malinovska, L.; Kroschwald, S.; Munder, M.C.; Richter, D.; Alberti, S. Molecular chaperones and stress-inducible protein-sorting factors coordinate the spatiotemporal distribution of protein aggregates. Mol. Biol. Cell 2012, 23, 3041–3056. [Google Scholar] [CrossRef]
- Malcova, I.; Farkasovsky, M.; Senohrabkova, L.; Vasicova, P.; Hasek, J. New integrative modules for multicolor-protein labeling and live-cell imaging in Saccharomyces cerevisiae. FEMS Yeast Res. 2016, 16, fow027. [Google Scholar] [CrossRef]
- Gokhale, K.C.; Newnam, G.P.; Sherman, M.Y.; Chernoff, Y.O. Modulation of prion-dependent polyglutamine aggregation and toxicity by chaperone proteins in the yeast model. J. Biol. Chem. 2005, 280, 22809–22818. [Google Scholar] [CrossRef]
- Gietz, R.; Akio, S. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene 1988, 74, 527–534. [Google Scholar] [CrossRef]
- Sikorski, R.S.; Hieter, P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 1989, 122, 19–27. [Google Scholar] [CrossRef]
- Trubitsina, N.P.; Zemlyanko, O.M.; Bondarev, S.A.; Zhouravleva, G.A. Nonsense mutations in the yeast SUP35 gene affect the [PSI+] prion propagation. Int. J. Mol. Sci. 2020, 21, 1648. [Google Scholar] [CrossRef] [PubMed]
- Matveenko, A.G.; Drozdova, P.B.; Belousov, M.V.; Moskalenko, S.E.; Bondarev, S.A.; Barbitoff, Y.A.; Nizhnikov, A.A.; Zhouravleva, G.A. SFP1 -mediated prion-dependent lethality is caused by increased Sup35 aggregation and alleviated by Sis1. Genes Cells 2016, 21, 1290–1308. [Google Scholar] [CrossRef] [PubMed]
- Chernoff, Y.O.; Lindquist, S.L.; Ono, B.; Inge-Vechtomov, S.G.; Liebman, S.W. Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [PSI+]. Science 1995, 268, 880–884. [Google Scholar] [CrossRef] [PubMed]
- Drozdova, P.B.; Tarasov, O.V.; Matveenko, A.G.; Radchenko, E.A.; Sopova, J.V.; Polev, D.E.; Inge-Vechtomov, S.G.; Dobrynin, P.V. Genome sequencing and comparative analysis of Saccharomyces cerevisiae strains of the Peterhof Genetic Collection. PLoS ONE 2016, 11, e0154722. [Google Scholar] [CrossRef]
- Derkatch, I.L.; Bradley, M.E.; Liebman, S.W. Overexpression of the SUP45 gene encoding a Sup35p-binding protein inhibits the induction of the de novo appearance of the [PSI+] prion. Proc. Natl. Acad. Sci. USA 1998, 95, 2400–2405. [Google Scholar] [CrossRef]
- Newnam, G.P.; Wegrzyn, R.D.; Lindquist, S.L.; Chernoff, Y.O. Antagonistic interactions between yeast chaperones Hsp104 and Hsp70 in prion curing. Mol. Cell. Biol. 1999, 19, 1325–1333. [Google Scholar] [CrossRef]
- Agaphonov, M.; Alexandrov, A. Self-excising integrative yeast plasmid vectors containing an intronated recombinase gene. FEMS Yeast Res. 2014, 14, 1048–1054. [Google Scholar] [CrossRef]
- Okamoto, A.; Hosoda, N.; Tanaka, A.; Newnam, G.P.; Chernoff, Y.O.; Hoshino, S.I. Proteolysis suppresses spontaneous prion generation in yeast. J. Biol. Chem. 2017, 292, 20113–20124. [Google Scholar] [CrossRef]
- Chernoff, Y.O.; Derkatch, I.L.; Dagkesamanskaya, A.R.; Tikhomirova, V.L.; Ter-Avanesyan, M.D.; Inge-Vechtomov, S.G. Nonsense suppression in amplification of the gene encoding a protein translation factor. Dokl. Akad. Nauk SSSR 1988, 301, 1227–1229. [Google Scholar]
- Volkov, K.V.; Aksenova, A.Y.; Soom, M.J.; Osipov, K.V.; Svitin, A.V.; Kurischko, C.; Shkundina, I.S.; Ter-Avanesyan, M.D.; Inge-Vechtomov, S.G.; Mironova, L.N. Novel non-Mendelian determinant involved in the control of translation accuracy in Saccharomyces cerevisiae. Genetics 2002, 160, 25–36. [Google Scholar] [CrossRef]
- Ter-Avanesyan, M.D.; Dagkesamanskaya, A.R.; Kushnirov, V.V.; Smirnov, V.N. The SUP35 omnipotent suppressor gene is involved in the maintenance of the non-Mendelian determinant [psi+] in the yeast Saccharomyces cerevisiae. Genetics 1994, 137, 671–676. [Google Scholar] [CrossRef]
- Moskalenko, S.E.; Chabelskaya, S.V.; Inge-Vechtomov, S.G.; Philippe, M.; Zhouravleva, G.A. Viable nonsense mutants for the essential gene SUP45 of Saccharomyces cerevisiae. BMC Mol. Biol. 2003, 4, 2. [Google Scholar] [CrossRef] [PubMed]
- Barbitoff, Y.A.; Matveenko, A.G.; Matiiv, A.B.; Maksiutenko, E.M.; Moskalenko, S.E.; Drozdova, P.B.; Polev, D.E.; Beliavskaia, A.Y.; Danilov, L.G.; Predeus, A.V.; et al. Chromosome-level genome assembly and structural variant analysis of two laboratory yeast strains from the Peterhof Genetic Collection lineage. G3 Genes Genomes Genet. 2021, 11, jkab029. [Google Scholar] [CrossRef] [PubMed]
- Sambrook, J.; Fritsch, E.; Maniatis, T. Molecular Cloning: A Laboratory Manual, 2nd ed.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 1989. [Google Scholar]
- Kaiser, C.; Michaelis, S.; Mitchell, A. Methods in Yeast Genetics, 1994 ed.; Number 316; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 1994; p. 234. [Google Scholar]
- Eaglestone, S.S.; Ruddock, L.W.; Cox, B.S.; Tuite, M.F. Guanidine hydrochloride blocks a critical step in the propagation of the prion-like determinant [PSI+] of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 2000, 97, 240–244. [Google Scholar] [CrossRef]
- Gietz, D.R.; Woods, R.A. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 2002, 350, 87–96. [Google Scholar] [CrossRef]
- Liu, C.; Apodaca, J.; Davis, L.E.; Rao, H. Proteasome inhibition in wild-type yeast Saccharomyces cerevisiae cells. BioTechniques 2007, 42, 158–162. [Google Scholar] [CrossRef] [PubMed]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2022. [Google Scholar]
- Zhang, T.; Lei, J.; Yang, H.; Xu, K.; Wang, R.; Zhang, Z. An improved method for whole protein extraction from yeast Saccharomyces cerevisiae. Yeast 2011, 28, 795–798. [Google Scholar] [CrossRef]
- Conzelmann, A.; Riezman, H.; Desponds, C.; Bron, C. A major 125-kd membrane glycoprotein of Saccharomyces cerevisiae is attached to the lipid bilayer through an inositol-containing phospholipid. EMBO J. 1988, 7, 2233–2240. [Google Scholar] [CrossRef]
- Chabelskaya, S.; Kiktev, D.; Inge-Vechtomov, S.; Philippe, M.; Zhouravleva, G. Nonsense mutations in the essential gene SUP35 of Saccharomyces cerevisiae are non-lethal. Mol. Genet. Genom. 2004, 272, 297–307. [Google Scholar] [CrossRef]
- Patino, M.M.; Liu, J.j.; Glover, J.R.; Lindquist, S. Support for the prion hypothesis for inheritance of a phenotypic trait in yeast. Science 1996, 273, 622–626. [Google Scholar] [CrossRef]
- Satpute-Krishnan, P.; Serio, T.R. Prion protein remodelling confers an immediate phenotypic switch. Nature 2005, 437, 262–265. [Google Scholar] [CrossRef]
- Song, Y.; Wu, Y.X.; Jung, G.; Tutar, Y.; Eisenberg, E.; Greene, L.E.; Masison, D.C. Role for Hsp70 chaperone in Saccharomyces cerevisiae prion seed replication. Eukaryot. Cell 2005, 4, 289–297. [Google Scholar] [CrossRef]
- Tyedmers, J.; Treusch, S.; Dong, J.; McCaffery, J.M.; Bevis, B.; Lindquist, S. Prion induction involves an ancient system for the sequestration of aggregated proteins and heritable changes in prion fragmentation. Proc. Natl. Acad. Sci. USA 2010, 107, 8633–8638. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Nawroth, P.P.; Tyedmers, J. Prion aggregates are recruited to the Insoluble Protein Deposit (IPOD) via myosin 2-based vesicular transport. PLoS Genet. 2016, 12, e1006324. [Google Scholar] [CrossRef]
- Gong, H.; Romanova, N.V.; Allen, K.D.; Chandramowlishwaran, P.; Gokhale, K.; Newnam, G.P.; Mieczkowski, P.; Sherman, M.Y.; Chernoff, Y.O. Polyglutamine toxicity is controlled by prion composition and gene dosage in yeast. PLoS Genet. 2012, 8, e1002634. [Google Scholar] [CrossRef]
- Arslan, F.; Hong, J.Y.; Kanneganti, V.; Park, S.K.; Liebman, S.W. Heterologous aggregates promote de novo prion appearance via more than one mechanism. PLoS Genet. 2015, 11, e1004814. [Google Scholar] [CrossRef]
- Dorweiler, J.E.; Lyke, D.R.; Lemoine, N.P.; Guereca, S.; Buchholz, H.E.; Legan, E.R.; Radtke, C.M.; Manogaran, A.L. Implications of the actin cytoskeleton on the multi-step process of [PSI+] prion formation. Viruses 2022, 14, 1581. [Google Scholar] [CrossRef] [PubMed]
- Shaner, N.C.; Lin, M.Z.; McKeown, M.R.; Steinbach, P.A.; Hazelwood, K.L.; Davidson, M.W.; Tsien, R.Y. Improving the photostability of bright monomeric orange and red fluorescent proteins. Nat. Methods 2008, 5, 545–551. [Google Scholar] [CrossRef]
- Pack, C.G.; Inoue, Y.; Higurashi, T.; Kawai-Noma, S.; Hayashi, D.; Craig, E.; Taguchi, H. Heterogeneous interaction network of yeast prions and remodeling factors detected in live cells. BMB Rep. 2017, 50, 478–483. [Google Scholar] [CrossRef] [PubMed]
- Greene, L.E.; Saba, F.; Silberman, R.E.; Zhao, X. Mechanisms for curing yeast prions. Int. J. Mol. Sci. 2020, 21, 6536. [Google Scholar] [CrossRef]
- Gasset-Rosa, F.; Giraldo, R. Engineered bacterial hydrophobic oligopeptide repeats in a synthetic yeast prion, [REP-PSI+]. Front. Microbiol. 2015, 6, 311. [Google Scholar] [CrossRef] [PubMed]
- Shintani, T.; Klionsky, D.J. Cargo proteins facilitate the formation of transport vesicles in the cytoplasm to vacuole targeting pathway. J. Biol. Chem. 2004, 279, 29889–29894. [Google Scholar] [CrossRef] [PubMed]
- Wilfling, F.; Lee, C.W.; Erdmann, P.S.; Zheng, Y.; Sherpa, D.; Jentsch, S.; Pfander, B.; Schulman, B.A.; Baumeister, W. A selective autophagy pathway for phase-separated endocytic protein deposits. Mol. Cell 2020, 80, 764–778.e7. [Google Scholar] [CrossRef] [PubMed]
- Speldewinde, S.H.; Doronina, V.A.; Grant, C.M. Autophagy protects against de novo formation of the [PSI+] prion in yeast. Mol. Biol. Cell 2015, 26, 4541–4551. [Google Scholar] [CrossRef]
- Dorweiler, J.E.; Obaoye, J.O.; Oddo, M.J.; Shilati, F.M.; Scheidemantle, G.M.; Coleman, T.J.; Reilly, J.A.; Smith, G.R.; Manogaran, A.L. DMSO-mediated curing of several yeast prion variants involves Hsp104 expression and protein solubilization, and is decreased in several autophagy related gene (atg) mutants. PLoS ONE 2020, 15, e0229796. [Google Scholar] [CrossRef]
- Vishveshwara, N.; Bradley, M.E.; Liebman, S.W. Sequestration of essential proteins causes prion associated toxicity in yeast. Mol. Microbiol. 2009, 73, 1101–1114. [Google Scholar] [CrossRef]
| Strain | Genotype | Background | Reference |
|---|---|---|---|
| 74-D694 | MATa ade1-14 his3-Δ200 ura3-52 leu2-3,112 trp1-289[psi-] [PIN+] | 74-D694 | [33] |
| P-74-D694 | MATa ade1-14 his3-Δ200 ura3-52 leu2-3,112 trp1-289[PSI+] [PIN+] | 74-D694 | [34] |
| 2-74-D694 | MATa ade1-14 his3-Δ200 ura3-52 leu2-3,112 trp1-289[psi-] [pin-] | 74-D694 | This work |
| OT56 | MATa ade1-14 his3-Δ200 ura3-52 leu2-3,112 trp1-289[PSI+]S[PIN+] | 74-D694 | [35,36] |
| 1-OT56 | MATa ade1-14 his3-Δ200 ura3-52 leu2-3,112 trp1-289[psi-] [PIN+] | 74-D694 | [32] |
| 2-OT56 | MATa ade1-14 his3-Δ200 ura3-52 leu2-3,112 trp1-289[psi-] [pin-] | 74-D694 | [32] |
| prb10-P-74-D694 | MATa ade1-14 his3-Δ200 ura3-52 leu2-3,112 trp1-289 prb1Δ0[PSI+] [PIN+] | 74-D694 | [37] |
| prb10-2-74-D694 | MATa ade1-14 his3-Δ200 ura3-52 leu2-3,112 trp1-289 prb1Δ0[psi-] [pin-] | 74-D694 | This work |
| yAO121 | MATa ade1-14 his3-Δ200 ura3-52 leu2-3,112 trp1-289 pep4::HIS3MX[psi-] [PIN+] | 74-D694 | [38] |
| P2.1.1-yAO121 | MATa ade1-14 his3-Δ200 ura3-52 leu2-3,112 trp1-289 pep4::HIS3MX[PSI+]2.1.1[PIN+] | 74-D694 | This work |
| 2-yAO121 | MATa ade1-14 his3-Δ200 ura3-52 leu2-3,112 trp1-289 pep4::HIS3MX[psi-] [pin-] | 74-D694 | This work |
| P1-U-1A-D1628 | MATα ade1-14 his3-Δ200 lys2 ura3-52 leu2-3,112 trp1-289 sup45::HIS3MX[pRS316-SUP45] [PSI+] | 1A-D1628 | This work |
| 3.1P.1-1B-D1606 | MATα ade1-14 his7-1 lys9-A21 ura3-52 leu2–3,112 trp1-289 gal10-1B[PSI+]3.1 | 1B-D1606 | This work |
| 1-33G-D373 | MATα ade2-144,717 pha2P-A10 (pheA10) his7-1 lys9-A21 ura3-52 leu2–3,112 trp1-289[PSI+] | 33G-D373 | [39]; this work |
| P-2V-P3982 | MATα ade1-14 his7-1 lys2-87 ura3Δ leu2-B2 thr4-B15[PSI+] | 2V-P3982 | [40] |
| U-T-PT-YAL2171 | MATa ade1-14 his3-11,-15 ura3-1 leu2-3,112 trp1-1 can1-100 sup35::hphMX sis1::kanMX[pRS314-SUP35] [YCplac33-Sis1] [PSI+] [PIN+] | W303 | This work |
| 1107-5V-H19 | MATa ade2-1 ura3-52 leu2-3,112 can1-100 SUQ5[PSI+] [PIN+] | S288C | [41] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Matveenko, A.G.; Ryzhkova, V.E.; Zaytseva, N.A.; Danilov, L.G.; Mikhailichenko, A.S.; Barbitoff, Y.A.; Zhouravleva, G.A. Processing of Fluorescent Proteins May Prevent Detection of Prion Particles in [PSI+] Cells. Biology 2022, 11, 1688. https://doi.org/10.3390/biology11121688
Matveenko AG, Ryzhkova VE, Zaytseva NA, Danilov LG, Mikhailichenko AS, Barbitoff YA, Zhouravleva GA. Processing of Fluorescent Proteins May Prevent Detection of Prion Particles in [PSI+] Cells. Biology. 2022; 11(12):1688. https://doi.org/10.3390/biology11121688
Chicago/Turabian StyleMatveenko, Andrew G., Varvara E. Ryzhkova, Natalia A. Zaytseva, Lavrentii G. Danilov, Anastasia S. Mikhailichenko, Yury A. Barbitoff, and Galina A. Zhouravleva. 2022. "Processing of Fluorescent Proteins May Prevent Detection of Prion Particles in [PSI+] Cells" Biology 11, no. 12: 1688. https://doi.org/10.3390/biology11121688
APA StyleMatveenko, A. G., Ryzhkova, V. E., Zaytseva, N. A., Danilov, L. G., Mikhailichenko, A. S., Barbitoff, Y. A., & Zhouravleva, G. A. (2022). Processing of Fluorescent Proteins May Prevent Detection of Prion Particles in [PSI+] Cells. Biology, 11(12), 1688. https://doi.org/10.3390/biology11121688






