Simple Summary
Periodontal disease represents a worldwide health problem. Human periodontal pathogens such as Treponema denticola, Porphyromonas gingivalis, Tannerella forsythia and Aggregatibacter actinomycetemcomitans are the cause of inflammatory response resulting in periodontitis. Porphyromonas gulae is mostly involved in periodontitis in dogs; however, it is not a common pathogen in humans. This study deals with the prevalence of periodontal pathogens in Slovak patients with periodontitis. Furthermore, based on the previous findings of animal-to-human transmission of periodontal pathogens, this study also assesses the possible bacterial transmission between animals and their owners. The highest prevalence in Slovak patients amongst the monitored periodontal pathogens had T. forsythia. In regard to the limited information available on T. forsythia, antibiotic sensitivity of this bacterium was evaluated. Most of the T. forsythia isolates were susceptible to antibiotics, namely amoxicillin-clavulanic acid, clindamycin and moxifloxacin, while they were resistant to metronidazole. Moreover, the transmission of P. gulae between animals and their owners was confirmed. Based on the similarity of P. gulae with human P. gingivalis, there arises the question as to whether P. gulae can also be involved in the periodontitis pathogenesis in humans. However, more studies are required for further clarification.
Abstract
Oral health and diseases are greatly influenced by oral bacteria. During dysbiosis, bacterial composition changes, which can lead to periodontitis. Periodontitis in humans is associated with periodontal pathogens such as Treponema denticola, Porphyromonas gingivalis, Tannerella forsythia and Aggregatibacter actinomycetemcomitans. Animal-to-human transmission of some of these pathogens has also been reported. The aim of this study was to evaluate the prevalence of periodontal pathogens in Slovak patients and to assess the possible risk of transmission of these pathogens from animals to their owners. The presence of periodontal pathogens in dental plaque was monitored by PCR. Amplified products were analysed using Sanger sequencing. T. forsythia isolates were assessed for the susceptibility to different antibiotics using the disk diffusion method. In humans, T. denticola, P. gingivalis, T. forsythia and A. actinomycetemcomitans were present in 69.23%, 69.23%, 100% and 84.62%, respectively. Most isolates of T. forsythia were susceptible to amoxicillin-clavulanic acid, clindamycin and moxifloxacin, but they were resistant to metronidazole. The transmission of T. forsythia from animals to their owners was not proven based on sequence analysing. On the other hand, transmission of Porphyromonas gulae was confirmed, but the risk of its involvement in the pathogenesis of periodontitis in humans must be further investigated.
1. Introduction
The oral cavity provides a habitat for a diverse bacterial community comprising hundreds of bacterial species that exist in homeostatic balance with their host [1]. Oral bacteria colonize hard and soft tissues in the oral cavity and play a significant role in influencing oral health and disease [2]. Compared to other tissues, the teeth provide a hard surface where the greatest accumulation of bacteria occurs due to better attachment, with the formation of dental plaque, also known as dental biofilm [3]. In general, various endogenous and exogenous factors are involved in the dynamics of the composition of the oral microbiota. In a state of dysbiosis, there are changes in this bacterial composition and disruption of the homeostatic balance, which can ultimately lead to oral and even systemic diseases [4]. One of the most common diseases associated with oral bacterial dysbiosis is periodontitis, which represents a worldwide health problem [5].
Periodontitis is characterized as a chronic multifactorial inflammatory disease affecting the supporting tissues surrounding the teeth. Various inflammatory manifestations can initially lead to progressive tissue destruction, followed by alveolar bone involvement and finally tooth loss [6,7]. This disease is associated with intermittent periods of remission and relapse [5]. On the basis of clinical manifestation, periodontitis was previously classified into chronic and aggressive form [8]. Due to limited differences, e.g., in the microbial composition or lack of specific immune inflammatory response patterns, these forms of periodontitis were grouped into one category under the name periodontitis [9]. It was divided into four stages based on severity and complexity of disease management and three grades according to the progression risk of disease [10].
Complex interactions between the multispecies bacterial community in dental plaque and the host’s immune response play a role in the etiopathogenesis of periodontitis [11]. The presence of specific bacterial pathogens, such as Treponema denticola, Porphyromonas gingivalis and Tannerella forsythia belonging to the red complex, or Aggregatibacter actinomycetemcomitans, is strongly associated with an increased risk of periodontitis [12]. During periodontitis, there is also a reduction or absence of some commensal bacterial species in favour of these periodontal pathogens [13]; therefore, a qualitative and quantitative bacterial difference can be observed between patients with and without periodontitis [14]. Moreover, from an epidemiological point of view, the prevalence and distribution of periodontal pathogens may be different in relation to various geographical locations and populations [15].
Periodontitis is also frequently seen in veterinary practice [16]. While it is affecting approximately 30% of adult human population, there is 80% prevalence in dogs older than 2 years [17] and 70% prevalence in cats of the same age [18]. Amongst pathogenic bacteria of companion animals there are T. denticola, Porphyromonas gulae, P. gingivalis, T. forsythia, Prevotella intermedia, Prevotella nigrescens, and A. actinomycetemcomitans [19]. It has been reported that some of these periodontal pathogens can be transmitted from companion animals to their owners, especially through close physical contact [20,21]. Approximately 16.4% of bacterial species of canine subgingival plaque were shared with the human population [22]. Such examples include P. gulae, T. forsythia, and Campylobacter rectus [23].
Based on the above, the present study was carried out to provide insights on the prevalence of periodontal pathogens in Slovak patients with periodontitis. Moreover, the risk of possible transmission of periodontal pathogens from companion animals to owners was evaluated in some patients, as well.
2. Materials and Methods
2.1. Collection of Plaque Samples from Human and Animal Population
Thirteen Slovak patients with periodontitis were included in this study. The patients were non-smokers, had more than 20 teeth and had not undergone mechanical periodontal therapy before sampling. None of them had used any nonsteroidal anti-inflammatory drugs, antibiotics or immunosuppressive drugs within the last 6 months. According to the new classification system [24], the participants of this study were assigned the appropriate stage and grade of periodontitis. Clinical examinations, assessment of periodontal status and collection of dental plaque samples were carried out by two professional practitioners. A plaque sample was taken individually from each patient from the places where the deepest periodontal pockets were present, mainly between the first and the second molars, using a sterile curette. In addition, screening of patients’ genotype was performed within three genetic variants (rs1800587, rs1143634 and rs419598) in the gene cluster for interleukin-1 (IL-1) and gene encoding the human leukocyte antigen (HLA)-DR4. Samples were taken by inserting special sampling sticks into the gingival sulcus for 10 s. These samples were sent for DentalGen genetic testing using real-time PCR (GHC GENETICS SK, Bratislava, Slovakia).
From companion animals, samples were taken from a total of five animals (1 cat and 4 dogs) that lived in the same household with some patients, using a sterile syringe needle. The sample included dental plaque from all types of teeth, especially from the upper dentes canini and dentes premolares. Before sampling, an intraoral examination was performed in non-anesthetized animals, during which the stage of periodontitis was evaluated according to Bauer et al. [25]. These examinations were carried out by a veterinarian at the Small Animal Clinic, University of Veterinary Medicine and Pharmacy in Kosice.
Dental plaque samples from humans and animals were collected into the microtubes containing 300 µL of phosphate-buffered saline (PBS). Then, samples were vortexed at maximum speed for 20 s and shaken at 1400 rpm (Thermoshaker TS-100C, BioSan, Riga, Latvia) for 5 min to homogenize the contents. A volume of 50 µL was taken from the homogenized sample into bacterial glycerol stocks (growth medium and glycerol in equal volume 1:1). Plaque samples in PBS and bacterial glycerol stocks were stored at −70 °C until processing.
2.2. Ethical Approval
The Ethics Committee of the Faculty of Medicine at the Pavol Jozef Safarik University in Kosice and Ethics Committee of Louis Pasteur University Hospital in Kosice approved the study (protocol code 2021/EK/11068). The participants were fully informed about the purposes and procedures of study and provided informed consent in accordance with the Declaration of Helsinki.
The study protocol involving companion animals was approved by the Ethics Committee of the University of Veterinary Medicine and Pharmacy in Kosice (protocol code EKVP/2022-01). The owners of the animals provided informed consent.
2.3. Extraction of Microbial DNA
Plaque samples were thawed before DNA extraction, centrifuged at 10,000× g for 10 min at 4 °C, and then the supernatant was removed. A modified protocol for phenol-chloroform DNA extraction was used [26]. In brief, 180 µL of lysis buffer (50 mM Tris-HCl, 10 mM EDTA and 1% sodium dodecyl sulfate) and 25 µL of proteinase K (Roche Diagnostics GmbH, Mannheim, Germany) were added to the pellets. The microtubes were incubated at 55 °C for 2 h, with shaking at 300 rpm (Thermoshaker TS-100C, BioSan, Riga, Latvia). Proteinase K was inactivated by heating at 95 °C for 5 min. Subsequently, they were centrifuged at 10,000× g for 5 min at 23 °C and the supernatant was transferred to the new microtubes. Phenol and chloroform were added to the supernatant in an equal volume of 1:1 and centrifuged at 10,000× g for 5 min at 23 °C. The upper aqueous phase was transferred to the new microtube. Isopropyl alcohol (0.6 volume of supernatant) and 3 M sodium acetate solution (0.1 volume of supernatant) were then added. The nucleic acids in the mixture were allowed to precipitate overnight at 4 °C. The following day, DNA was pelleted at 10,000 × g for 10 min at 4 °C, washed with 100 µL of cold 75% ethyl alcohol and dried at 35 °C for 5–10 min. The pellets were resuspended in 30 μL of TE buffer (50 mM Tris-HCl and 10 mM EDTA). The quality and quantity of the extracted DNA were analysed by spectrophotometry using NanoDrop 1000 (Thermo Fisher Scientific, Waltham, MA, USA). According to the measured DNA concentration, samples were diluted with molecular grade water to a concentration of 50 ng/µL template DNA, and the thus diluted samples were stored at −70 °C until use.
2.4. Molecular Detection of Periodontal Pathogens
PCR amplification was carried out in a thermal cycler (TProfessional Basic, Biometra GmbH, Göttingen, Germany). The reaction mixture (50 µL) contained 2 µL (100 ng) of sample (DNA template), OneTaq 2X Master Mix with standard buffer (New England Biolabs, Foster City, CA, USA), molecular water and primers at concentration of 33 µM. The primers used for PCR amplification of Treponema species, T. denticola, P. gingivalis, P. gulae, T. forsythia and A. actinomycetemcomitans, and thermocycling conditions are listed in Table 1. Negative controls (RNAse-free water) were included for each set of primers in each of the PCR reactions.

Table 1.
Primers and PCR conditions used to detect periodontal pathogens.
Amplification products were subjected to electrophoresis in 2% agarose gel and visualized with GelRed (Biotium, Inc., Hayward, CA, USA) under UV light [31]. A 100 bp DNA ladder was used as a molecular weight marker (New England Biolabs, Foster City, CA, USA).
2.5. Sequencing and Data Processing
The amplification products of all PCR reactions except A. actinomycetemcomitans were sent for Sanger sequencing from one or both directions (Microsynth, Wien, Austria). Homology searches with sequences were performed by online nucleotide BLAST (Basic Local Alignment Search Tool) algorithm (https://blast.ncbi.nlm.nih.gov/Blast.cgi, accessed on 4 August 2022). In the case of flaB2 gene, the nr/nt database was used, and for the 16S rRNA gene, both the nr/nt and 16S rRNA databases were used. The chromatograms of sequences were edited and aligned using Geneious alignment in Geneious 8.0.5 (Biomatters, Auckland, New Zealand), and phylogenetic trees were constructed for P. gingivalis, P. gulae and T. forsythia using MUSCLE Alignment and Neighbor-joining in Geneious 8.0.5. The nucleotide sequences were deposited in GenBank under accession numbers OP678047–OP678083.
2.6. Disk Diffusion Susceptibility Test
In vitro antibiotic susceptibility tests for T. forsythia were performed due to the limited information available on this bacterium. From bacterial glycerol stocks containing dental plaque from humans and animals, suspensions in a volume of 25–50 µL (based on the amount of dental plaque acquired) were inoculated onto Tryptic Soy Agar (TSA) supplemented with 5% sterile horse blood, N-acetyl muramic acid (NAM, 1 mg/L; Sigma-Aldrich, St. Louis, MO, USA) and hemin (5 mg/L; Sigma-Aldrich, St. Louis, MO, USA). Plates were incubated anaerobically (BBL GasPak™ Plus, Becton, Dickinson and Co., Franklin Lakes, NJ, USA) for 7 days at 37 °C [1]. Based on typical morphology and Gram staining, colonies were selected and cultured in an anaerobic environment for 72 h at 37 °C. For final identification of T. forsythia, DNA was isolated from pure bacterial cultures through DNAzol Direct (Molecular Research Center Inc., Cincinnati, OH, USA) according to the manufacturer’s instructions and verified by the previously mentioned PCR. Antibiotic sensitivity was determined by disk diffusion on TSA-NAM. Bacterial suspensions were prepared from pure cultures and turbidity of suspensions was adjusted to 0.5 McFarland in saline solution at 565 nm wavelength (DEN-1 McFarland densitometer, Biosan, Riga, Latvia). Subsequently, 100 µL of suspensions were inoculated onto TSA-NAM. Four antibiotic discs (HiMedia, Mumbai, India) namely amoxicillin-clavulanic acid (30 μg), clindamycin (10 μg), moxifloxacin (5 μg) and metronidazole (5 μg) were placed onto the surface of the agar plates within 15 min of inoculation. Then, the plates were incubated anaerobically for 72 h at 37 °C. The inhibition zones were measured and interpreted according to Dubreuil et al. [32] and Nagy et al. [33].
3. Results
3.1. Baseline Characteristics of the Participants and Companion Animals
Participant’s characteristics are summarized in Table 2. A total of 13 patients in the same gender ratio were ranged in age from 34–68 years, with a mean age of 50.5 ± 10.5 years. Stage I periodontitis was present in six people, while stage II was present only in one patient and stage III in three patients. In three cases, there was periodontitis in between stages II and III. Periodontitis with grade A, grade B and grade C was present in five, six, and two patients, respectively. Most of them regularly underwent dental examinations along with dental hygiene, except patients H5 and H9. Furthermore, five of them were owners of some animal, whereas patients H12 and H13 were a married couple with one dog.

Table 2.
General information about patients and their periodontal status.
Within genotyping to reveal an increased risk of immune reactivity associated with rapid progression of periodontitis, patients H1, H2, H3, H8, H9 and H10 were found to have a positive genotype within the polymorphism in the genes IL1-A (rs1800587), IL1-B (rs1143634) and IL-1RN (rs419598). On the other hand, patients H5 and H11 had an increased risk of periodontitis progression due to the presence of polymorphism in gene encoding HLA-DR4. Lastly, patient H6 had a positive genotype within gene cluster for IL-1 and in gene encoding HLA-DR4. Patients H4, H7, H12 and H13 had a negative genotype in regards to the polymorphism in genes mentioned above.
Companion animals of different breed and age were assigned periodontal status based on evaluation of clinical signs such as gingivitis, amount of dental plaque, presence of dental calculus or tooth loss (Table 3). Most of the dogs had mild gingivitis, except for dog 4 that showed presence of only dental plaque without gingivitis. None of the animals have been castrated or sterilized and their teeth had not been previously treated before sampling.

Table 3.
Basic information and periodontal status of the companion animals.
3.2. Detection of Periodontal Pathogens in Humans
Using species-specific PCR for detection of T. denticola, this pathogen was present in 69.23% of dental plaque. Similarly, P. gingivalis was detected in 69.23%, while P. gulae was found only in 23.08%. All the plaque samples in patients were positive for the presence of T. forsythia. A. actinomycetemcomitans was confirmed in most patients (84.62%) except H5 and H7. Prevalence of periodontal pathogens in plaque samples of patients is listed in Table 4. After sequencing the amplified products of PCR using primers for Treponema sp., these primers were found to be inappropriate for the detection of T. denticola, since all the samples with amplified products were identified as Treponema vincentii with 100% homology. On the other hand, based on BLAST analysis all PCR products using primers for the detection of T. denticola were identified as T. denticola showing 100% homology. The primers targeting P. gingivalis were not found to be species-specific, since after sequencing the PCR products, there was also confirmation of other bacterial species, namely P. gulae and Capnocytophaga sp. with 100% and 97.48% homology, respectively, in patients H4 and H8. Besides that, all the other amplified products showed homology ranging from 99.67% to 100% with P. gingivalis. Using species-specific primers for P. gulae, all positive PCR products after sequencing showed 100% homology with P. gulae. Lastly, for T. forsythia all sequences homology ranged from 99.3% to 100%.

Table 4.
The presence of periodontal pathogens in human dental plaque.
3.3. Comparison of Periodontal Pathogens between Companion Animals and Their Owners
In the case of companion animals, only species-specific primers were used for the identification of T. denticola. Its presence was confirmed in one dog (H10D3). P. gulae was detected in all animals using both primer pairs for P. gulae and P. gingivalis, which was subsequently confirmed by sequencing (homology ranging from 99.58% to 100%). T. forsythia and A. actinomycetemcomitans were identified in all animals by PCR. The presence of periodontal pathogens in plaque samples of companion animals is listed in Table 5.

Table 5.
The presence of periodontal pathogens in dental plaque of companion animals.
Sequence analysis of the 16S rRNA fragment using both primer pairs for P. gulae and P. gingivalis showed identical sequences of P. gulae in patient H4 and his dog (H4D1). On the other hand, in patient H9 and his dog (H9D2), identical sequences of P. gulae were confirmed only using species-specific primers for P. gulae. Based on the detection of P. gulae in owners and their dogs, there is the possibility of transmission resulting from identical sequences of human and animal strains. Sequences were compared to known P. gulae and P. gingivalis gene sequences from the sequence database of the National Center of Biotechnology Information (NCBI, Bethesda, MD, USA). A phylogenetic tree for sequences obtained using primers for P. gingivalis is shown in Figure 1.
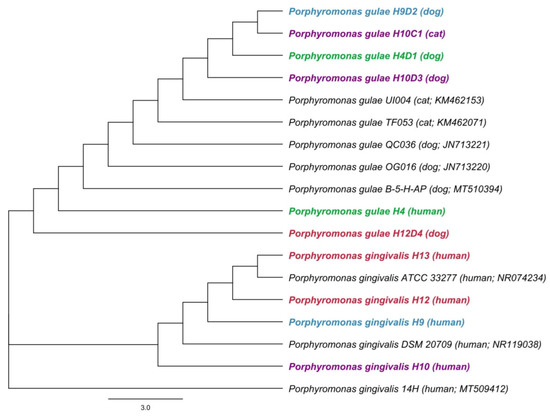
Figure 1.
The phylogenetic tree of Porphyromonas gingivalis and Porphyromonas gulae based on 16S rRNA gene sequences. A phylogenetic tree for sequences obtained using primers for P. gingivalis was constructed by the neighbor-joining method. Length of 16S rRNA gene sequence used to construct the phylogenetic tree was 240 bp. Hosts and accession numbers for 16S rRNA gene sequences obtained from the GenBank database are given in parentheses. Each color represents the owners and their animals.
A phylogenetic analysis showed differences between the sequences of the 16S rRNA gene for T. forsythia in humans and animals (Figure 2). Based on this, the transmission of T. forsythia from companion animals to their owners has not been proven.
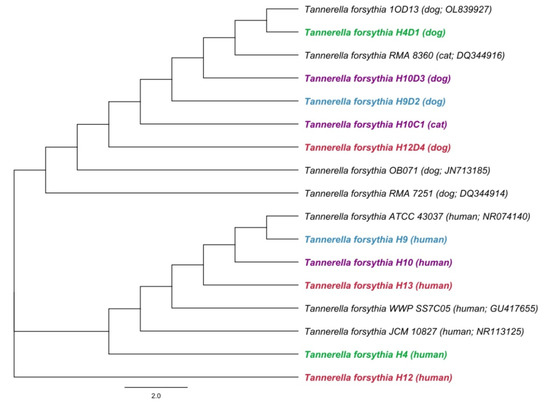
Figure 2.
A phylogenetic tree based on comparison of 16S rRNA gene sequences of Tannerella forsythia. A phylogenetic tree was constructed by the neighbor-joining method. Length of 16S rRNA gene sequence used to construct the phylogenetic tree was 480 bp. Hosts and accession numbers for 16S rRNA gene sequences obtained from the GenBank database are given in parentheses. Sequences of owners and their animals are distinguished by different colors.
3.4. Antibiotic Susceptibility of Tannerella forsythia
Regarding antibiotic sensitivity, all T. forsythia isolates were susceptible to amoxicillin-clavulanic acid and moxifloxacin (Table 6). Furthermore, all isolates were also found to be susceptible to clindamycin with the exception of H12D4a isolate. Lastly, none of the T. forsythia isolates were sensitive to metronidazole. Susceptibility for amoxicillin-clavulanic acid and moxifloxacin was defined if the diameter zone was ≥21 mm, for clindamycin it was ≥25 mm and for metronidazole the diameter zone was ≥15 mm. Despite the fact that T. forsythia was identified by PCR in all samples, T. forsythia was not isolated from patients H1, H2, H10 and companion animals H4D1, H9D2, H10D3 and H10C1.

Table 6.
The antibiotic sensitivity of Tannerella forsythia isolates.
4. Discussion
Oral diseases continue to be a major global health problem, with periodontitis being the most significant worldwide. Oral health is affected by the presence of predominantly anaerobic Gram-negative bacteria [34]. Interactions of these bacteria with the host can lead to disease; however, most of them can be present even in healthy individuals and coexist in harmony with the host due to the different virulent potential and pathogenicity of the bacterium [35]. Periodontal pathogens such as P. gingivalis, T. forsythia, T. denticola, P. intermedia and A. actinomycetemcomitans have been widely studied in terms of their virulence factors involved in periodontal pockets colonization or periodontal tissue destruction. It has been shown that the detection rate of P. gingivalis, T. forsythia and P. intermedia is significantly higher in periodontitis patients than in healthy individuals, but it does not significantly differ in between various stages of periodontal disease [36]. Similarly, determining the global distribution of bacterial species related to periodontitis may be important in terms of potential specifics associated with different ethnic and geographic factors [37]. Moreover, different detection methods can affect the capture rate of periodontal pathogens [38]. Among the available methods, PCR methods are considered to have higher sensitivity and specificity in the early detection of these periodontal bacteria [39]. For example, traditional PCR can qualitatively assess the presence of periodontal pathogens, while real-time PCR is commonly used to quantify these pathogens [40]. Using molecular methods, this study analysed the prevalence of five periodontal pathogens of humans and companion animals, namely T. denticola, P. gingivalis, P. gulae, T. forsythia and A. actinomycetemcomitans in the dental plaque of Slovak patients suffering from periodontitis and their animals.
T. denticola, being one of the most resistant bacteria frequently identified in primary dentition infection, is highly associated with periodontal diseases. It is commonly isolated from periodontal pockets of adults and it has great influence on the inflammation by stimulating the production of cytokines and it also causes disruption of tissue homeostasis [41]. Based on that, T. denticola is predominantly linked with severe periodontitis. However, based on molecular studies, other oral treponemes, including T. vincentii, are also associated with periodontal disease [42]. In our study, T. denticola was detected in 69.23% of Slovak patients. Similar findings were shown in the study by Jepsen et al. [43], in which this pathogen was present in 76.5% of German periodontitis patients. In addition, in the study by Feng et al. [44] T. denticola was present in 87.8% of Chinese patients with periodontitis and in the study by Torrungruang et al. [45] T. denticola was demonstrated in 82% of Thai patients.
Amongst the most important periodontal pathogens P. gingivalis appears to be one of the main etiological factors in the pathogenesis and development of inflammatory processes of periodontitis [46]. P. gingivalis expresses a wide array of virulence factors which include fimbriae, gingipains, collagenase, lipopolysaccharide, haemagglutinins, capsule, and superoxide dismutase [47]. These factors may cause destruction of the periodontal tissue on their own [48] or they can induce inflammatory reactions in the host and cause impairment of innate immune responses [49]. Our findings show that P. gingivalis was present in 69.23% of Slovak patients. Similar results were also presented in German [43] and Indian [50] patients, in which the prevalence was 68.2% and 66%, respectively. In the study by Rafiei et al. [51] the prevalence of P. gingivalis in periodontal disease group was 78% and in the study by Mínguez et al. [52] the prevalence in Moroccan periodontitis patients was 84.4%.
T. forsythia is also considered to be a part of periodontal oral microbiome [53] and it remains an understudied periodontal microorganism [54]. In a study by Alazemi et al. [55] T. forsythia was detected by PCR in 83.3% of Kuwait patients with severe and moderate periodontitis. In the case of the Uruguayan population with periodontitis, T. forsythia was among the most widespread species (92%) in the examined periodontal pathogens [56]. Prevalence of T. forsythia among Italian patients with periodontitis was 72.7%, with no statistical differences between geographical areas [57]. In this study, T. forsythia was detected in all Slovak patients regardless of the stages and grades of periodontitis, which probably indicates its strong association with this disease. Based on practically still limited information about the sensitivity of T. forsythia to various antimicrobial substances [54], our study also evaluated the antibacterial sensitivity of selected antibiotics. Amoxicillin-clavulanic acid, clindamycin, moxifloxacin, and metronidazole are among the most well-known drugs used as part of periodontal therapy in practice [46]. In this study, all T. forsythia isolates were sensitive to amoxicillin-clavulanic acid and moxifloxacin, while they showed resistance to metronidazole. Similarly, in the study by Ardila et al. [54], isolates of T. forsythia were completely susceptible to moxifloxacin, but the isolates showed resistance to amoxicillin and metronidazole in 25.6%. The antibacterial effect of antibiotics on T. forsythia showed resistance to metronidazole, amoxicillin and clindamycin in the study by Abdulrazzaq et al. [34]. However, in our study, 90.9% of the isolates were sensitive to clindamycin, which is contrary to the previous study. Similar to our study, T. forsythia was sensitive to clindamycin (80%) and amoxicillin-clavulanic acid (100%); however, it was found to be sensitive to metronidazole, which is in contradiction with our study [58].
A. actinomycetemcomitans is considered an oral pathogen that colonizes the oral cavity of the human population and is highly associated with rapidly progressive periodontitis [59]. Its role in the initiation and development of periodontitis as a community activist is considered essential for other periodontal pathogens, as it suppresses the initial host response, allowing their proliferation [60]. A. actinomycetemcomitans was relatively rare (16.1%) among Italian adults with periodontitis in the study by Tettamanti et al. [57]. In the study by Jensen et al. [61], the prevalence of A. actinomycetemcomitans was 13.3% in the adolescent population in Denmark. The detection frequency of A. actinomycetemcomitans in subgingival plaque samples collected from periodontitis patients living in Sweden was 26.3% [62]. In contrast to previous studies, our results show a higher rate of prevalence of this bacterium (84.62%) in Slovak patients. Similarly, in the study by Mínguez et al. [52], the detection frequency of A. actinomycetemcomitans was 79.5% in periodontitis patients in Morocco.
Host genetic predispositions within a dysregulated immune response may influence the development of periodontitis through susceptibility to oral dysbiosis and improper host response to bacterial infection [63]. Interleukin-1α plays a critical role in protecting the body from bacteria; however, in result it causes bone resorption and tissue damage, as seen in periodontal disease. Based on that, activity of IL-1 is higher in crevicular fluid obtained from inflamed gingival sites [64]. Consequently, genes encoding its production are receiving attention as potential predictors of periodontitis progression [65]. Moreover, HLA also have an importance in immune responsiveness and involvement in antigen recognition of periodontal bacteria [66]. As a result, there is a positive association of periodontitis with the presence of HLA-DR4 allele that is involved in the pathogenesis of periodontitis [67]. Individuals with positive genotype have an increased risk of periodontitis progression compared to the individuals with a negative genotype [68]. In this study, positive genotype within polymorphism in IL1-A, and IL1-B genes and IL-1RN gene encoding the antagonist receptor of IL-1 was recorded in six patients. Polymorphism in genes encoding HLA-DR4 was present in two patients. Presence of genetic variants in gene encoding HLA-DR4 and gene cluster for IL-1 was recorded in one patient. On the other hand, negative genotype of all the genes mentioned was present in four other patients.
In dogs, periodontal disease is mainly associated with the presence of P. gulae, which was originally thought to be the animal biotype of P. gingivalis; however, it was also found to colonize human oral cavity [69]. P. gulae has similar characteristics as P. gingivalis [70], which may result in the potential transmission between animals and their owners [23]. It possesses a broad spectrum of pathogenic properties such as lipopolysaccharide, haemagglutinins, fimbriae and proteases [71]. Similar proteolytic activity to P. gingivalis was also detected in P. gulae that exhibited similar virulence factors as its human counterpart [72]. Besides that, Nomura et al. [73] found that certain FimA types of fimbrial protein of canine P. gulae were phylogenetically close to those of P. gingivalis. Moreover, Fujiwara-Takahashi et al. [70] found that FimA types I, III, and IV in P. gingivalis were close to types A, B, and C, respectively, in P. gulae in dogs. Iwashita et al. [74] identified the fimA genotypes in feline P. gulae isolates, as well. In this study, the presence of P. gingivalis was confirmed in four animal owners, but the pathogen was not detected in any dental plaque samples of companion animals. Kwon et al. [75] detected P. gingivalis in 7.44% of canine subgingival plaque samples. On the other hand, in our study P. gulae was detected in two humans and their respective dogs, and based on their identical sequences, transmission between owner and its dog can be assumed. Presence of P. gulae was confirmed in all animal samples. Similar findings were described in the study by Yamasaki et al. [23], in which P. gulae was detected in 71.2% of dogs, while it was present only in 20% of the owners. Iwashita et al. [74] also detected P. gulae in approximately 90% of feline plaque samples, regardless the periodontal condition. Moreover, in this study, T. forsythia was detected in all dental plaque samples, both human and animal. However, transmission between humans and animals was not confirmed due to different taxa occurring in companion animals and their owners, which contradicts another study. Booij–Vrieling et al. [20] found T. forsythia in both cats and the owners and hypothesized that cat-to-owner transmission occurred and that cats may be a reservoir of T. forsythia. In comparison, the prevalence of T. forsythia in the study by Yamasaki et al. [23] was 77.3% in dogs, but only 30.9% in their owners. On the other hand, Kwon et al. [75] identified T. forsythia only in 34.23% of canine subgingival plaque. Further studies are needed to understand the possible transmission of periodontal pathogens.
5. Conclusions
Within the limitation of a relatively small sample size, this preliminary study clarified the prevalence of periodontal pathogens in Slovak patients with periodontitis. Based on our findings, the plaque samples analysed by molecular methods showed prevalence of T. denticola, P. gingivalis, P. gulae, T. forsythia and A. actinomycetemcomitans to be 69.23%, 69.23%, 23.08%, 100% and 84.62%, respectively. In addition, a possible owner to animal transmission of P. gulae was confirmed based on sequence analysing. However, these findings evoke additional questions for future studies regarding the risk of its involvement in the pathogenesis of periodontitis in humans. Until then, owners should be reminded that companion animals could be carriers of some periodontal pathogens; therefore, it is necessary to avoid close physical contact with animals and ensure personal hygiene compliance.
Author Contributions
Conceptualization, M.S., J.K. (Ján Kučera) and M.M.; methodology, J.K. (Ján Kučera) and J.K. (Jana Kačírová); validation, M.S., N.Š.H. and Z.K.N.; formal analysis, J.K. (Jana Kačírová) and Z.K.N.; investigation, M.S. and N.Š.H.; resources, J.K. (Ján Kučera), T.L. and M.M.; data curation, M.S., J.K. (Jana Kačírová) and N.Š.H.; writing—original draft preparation, M.S., Z.K.N. and N.Š.H.; writing—review and editing, J.K. (Ján Kučera), J.K. (Jana Kačírová) and M.M.; visualization, M.S, J.K. (Jana Kačírová) and N.Š.H.; supervision, M.M.; project administration, M.M.; funding acquisition, M.M. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Scientific Grand Agency of the Ministry of Education of the Slovak Republic (grant no. VEGA 1/0788/19).
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki and approved by Ethics Committee of the Faculty of Medicine at the Pavol Jozef Safarik University in Kosice and Ethics Committee of Louis Pasteur University Hospital in Kosice (protocol code 2021/EK/11068, 29 November 2021). The animal study protocol was approved by the Ethics Committee of the University of Veterinary Medicine and Pharmacy in Kosice (protocol code EKVP/2022-01, 14 January 2022).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Jochebed, S.R.; Jacob, C.A. Isolation, Transportation and Culture Methods of Tannerella forsythia—A Review. Int. J. Sci. Dev. Res. 2020, 5, 208–212. [Google Scholar]
- Wei, Y.; Shi, M.; Zhen, M.; Wang, C.; Hu, W.; Nie, Y.; Wu, X. Comparison of Subgingival and Buccal Mucosa Microbiome in Chronic and Aggressive Periodontitis: A Pilot Study. Front. Cell. Infect. Microbial. 2019, 9, 53. [Google Scholar] [CrossRef] [PubMed]
- Abebe, G.M. Oral Biofilm and Its Impact on Oral Health, Psychological and Social Interaction. Int. J. Oral Dent. Health 2021, 7, 127. [Google Scholar] [CrossRef]
- Li, X.; Liu, Y.; Yang, X.; Li, C.; Song, Z. The Oral Microbiota: Community Composition, Influencing Factors, Pathogenesis, and Interventions. Front. Microbiol. 2022, 13, 895537. [Google Scholar] [CrossRef]
- Boyer, E.; Martin, B.; Le Gall-David, S.; Fong, S.B.; Deugnier, Y.; Bonnaure-Mallet, M.; Meuric, V. Periodontal pathogens and clinical parameters in chronic periodontitis. Mol. Oral Microbiol. 2020, 35, 19–28. [Google Scholar] [CrossRef] [PubMed]
- de Coo, A.; Cruz, R.; Quintela, I.; Herrera, D.; Sanz, M.; Diz, P.; Rodríguez Grandío, S.; Vallcorba, N.; Ramos, I.; Oteo, A.; et al. Genome-Wide association study of stage III/IV grade C periodontitis (former aggressive periodontitis) in a Spanish population. J. Clin. Periodontol. 2021, 48, 896–906. [Google Scholar] [CrossRef] [PubMed]
- Lang, K.N.; Sculean, A.; Eick, S.; Stähli, A. A novel in vitro periodontal pocket model to evaluate the effect of root surface instrumentation on biofilm-epithelial cell interactions. Clin. Oral Investig. 2022, 26, 4021–4029. [Google Scholar] [CrossRef] [PubMed]
- Damgaard, C.; Danielsen, A.K.; Enevold, C.; Massarenti, L.; Nielsen, C.H.; Holmstrup, P.; Belstrøm, D. Porphyromonas gingivalis in saliva associates with chronic and aggressive periodontitis. J. Oral Microbiol. 2019, 11, 1653123. [Google Scholar] [CrossRef] [PubMed]
- Montenegro, S.; Retamal-Valdes, B.; Bueno-Silva, B.; Duarte, P.M.; Faveri, M.; Figueiredo, L.C.; Feres, M. Do patients with aggressive and chronic periodontitis exhibit specific differences in the subgingival microbial composition? A systematic review. J. Periodontol. 2020, 91, 1503–1520. [Google Scholar] [CrossRef] [PubMed]
- Takedachi, M.; Shimabukuro, Y.; Sawada, K.; Koshimizu, M.; Shinada, K.; Asai, H.; Mizoguchi, A.; Hayashi, Y.; Tsukamoto, A.; Miyago, M.; et al. Evaluation of periodontitis-related tooth loss according to the new 2018 classification of periodontitis. Sci. Rep. 2022, 12, 11893. [Google Scholar] [CrossRef] [PubMed]
- Schulz, S.; Porsch, M.; Grosse, I.; Hoffmann, K.; Schaller, H.G.; Reichert, S. Comparison of the oral microbiome of patients with generalized aggressive periodontitis and periodontitis-free subjects. Arch. Oral Biol. 2019, 99, 169–176. [Google Scholar] [CrossRef]
- Eick, S.; Kindblom, C.; Mizgalska, D.; Magdoń, A.; Jurczyk, K.; Sculean, A.; Stavropoulos, A. Adhesion of Porphyromonas gingivalis and Tannerella forsythia to dentin and titanium with sandblasted and acid etched surface coated with serum and serum proteins—An in vitro study. Arch. Oral Biol. 2017, 75, 81–88. [Google Scholar] [CrossRef] [PubMed]
- Abu Fanas, S.; Brigi, C.; Varma, S.R.; Desai, V.; Senok, A.; D’souza, J. The prevalence of novel periodontal pathogens and bacterial complexes in Stage II generalized periodontitis based on 16S rRNA next generation sequencing. J. Appl. Oral Sci. 2021, 29, e20200787. [Google Scholar] [CrossRef] [PubMed]
- Kumawat, R.M.; Ganvir, S.M.; Hazarey, V.K.; Qureshi, A.; Purohit, H.J. Detection of Porphyromonas gingivalis and Treponema denticola in chronic and aggressive periodontitis patients: A comparative polymerase chain reaction study. Contemp. Clin. Dent. 2016, 7, 481–486. [Google Scholar] [CrossRef]
- Lauritano, D.; Martinelli, M.; Mucchi, D.; Palmieri, A.; Lo Muzio, L.; Carinci, F. Bacterial load of periodontal pathogens among italian patients with chronic periodontitis: A comparative study of three different areas. J. Biol. Regul. Homeost. Agents 2016, 30, 149–154. [Google Scholar] [PubMed]
- Wallis, C.; Holcombe, L.J. A review of the frequency and impact of periodontal disease in dogs. J. Small Anim. Pract. 2020, 61, 529–540. [Google Scholar] [CrossRef] [PubMed]
- Albuquerque, C.; Morinha, F.; Requicha, J.; Martins, T.; Dias, I.; Guedes-Pinto, H.; Bastos, E.; Viegas, C. Canine periodontitis: The dog as an important model for periodontal studies. Vet. J. 2012, 191, 299–305. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Salcedo, L.; Laguna, E.; Sánchez, M.C.; Marín, M.J.; O’Connor, A.; González, I.; Sanz, M.; Herrera, D. Molecular identification of black-pigmented bacteria from subgingival samples of cats suffering from periodontal disease. J. Small Anim. Pract. 2015, 56, 270–275. [Google Scholar] [CrossRef] [PubMed]
- Özavci, V.; Erbas, G.; Parin, U.; Yüksel, H.T.; Kirkan, Ş. Molecular detection of feline and canine periodontal pathogens. Vet. Anim. Sci. 2019, 8, 100069. [Google Scholar] [CrossRef]
- Booij-Vrieling, H.E.; van der Reijden, W.A.; Houwers, D.J.; de Wit, W.E.; Bosch-Tijhof, C.J.; Penning, L.C.; van Winkelhoff, A.J.; Hazewinkel, H.A. Comparison of periodontal pathogens between cats and their owners. Vet. Microbiol. 2010, 144, 147–152. [Google Scholar] [CrossRef]
- Oh, C.; Lee, K.; Cheong, Y.; Lee, S.W.; Park, S.Y.; Song, C.S.; Choi, I.S.; Lee, J.B. Comparison of the Oral Microbiomes of Canines and Their Owners Using Next-Generation Sequencing. PLoS ONE 2015, 10, e0131468. [Google Scholar] [CrossRef] [PubMed]
- Nises, J.; Rosander, A.; Pettersson, A.; Backhans, A. The occurrence of Treponema spp. in gingival plaque from dogs with varying degree of periodontal disease. PLoS ONE 2018, 13, e0201888. [Google Scholar] [CrossRef] [PubMed]
- Yamasaki, Y.; Nomura, R.; Nakano, K.; Naka, S.; Matsumoto-Nakano, M.; Asai, F.; Ooshima, T. Distribution of periodontopathic bacterial species in dogs and their owners. Arch. Oral Biol. 2012, 57, 1183–1188. [Google Scholar] [CrossRef] [PubMed]
- Tonetti, M.S.; Greenwell, H.; Kornman, K.S. Staging and grading of periodontitis: Framework and proposal of a new classification and case definition. J. Periodontol. 2018, 89, S159–S172. [Google Scholar] [CrossRef] [PubMed]
- Bauer, A.E.; Stella, J.; Lemmons, M.; Croney, C.C. Evaluating the validity and reliability of a visual dental scale for detection of periodontal disease (PD) in non-anesthetized dogs (Canis familiaris). PLoS ONE 2018, 13, e0203930. [Google Scholar] [CrossRef] [PubMed]
- Vesty, A.; Biswas, K.; Taylor, M.W.; Gear, K.; Douglas, R.G. Evaluating the Impact of DNA Extraction Method on the Representation of Human Oral Bacterial and Fungal Communities. PLoS ONE 2017, 12, e0169877. [Google Scholar] [CrossRef]
- Brandt, S.; Apprich, V.; Hackl, V.; Tober, R.; Danzer, M.; Kainzbauer, C.; Gabriel, C.; Stanek, C.; Kofler, J. Prevalence of bovine papillomavirus and Treponema DNA in bovine digital dermatitis lesions. Vet. Microbiol. 2011, 148, 161–167. [Google Scholar] [CrossRef] [PubMed]
- Slots, J.; Ashimoto, A.; Flynn, M.J.; Li, G.; Chen, C. Detection of putative periodontal pathogens in subgingival specimens by 16S ribosomal DNA amplification with the polymerase chain reaction. Clin. Infect. Dis. 1995, 20, S304–S307. [Google Scholar] [CrossRef]
- Senhorinho, G.N.; Nakano, V.; Liu, C.; Song, Y.; Finegold, S.M.; Avila-Campos, M.J. Detection of Porphyromonas gulae from subgingival biofilms of dogs with and without periodontitis. Anaerobe 2011, 17, 257–258. [Google Scholar] [CrossRef]
- Kannosh, I.; Staletovic, D.; Toljic, B.; Radunovic, M.; Pucar, A.; Matic Petrovic, S.; Grubisa, I.; Lazarevic, M.; Brkic, Z.; Knezevic Vukcevic, J.; et al. The presence of periopathogenic bacteria in subgingival and atherosclerotic plaques—An age related comparative analysis. J. Infect. Dev. Ctries. 2018, 12, 1088–1095. [Google Scholar] [CrossRef] [PubMed]
- Huang, Q.; Baum, L.; Fu, W.L. Simple and practical staining of DNA with GelRed in agarose gel electrophoresis. Clin. Lab. 2010, 56, 149–152. [Google Scholar]
- Dubreuil, L.; Members of the CA-SFM 2019. Improvement of a disk diffusion method for antibiotic susceptibility testing of anaerobic bacteria. French recommendations revisited for 2020. Anaerobe 2020, 64, 102213. [Google Scholar] [CrossRef]
- Nagy, E.; Justesen, U.S.; Eitel, Z.; Urbán, E.; ESCMID Study Group on Anaerobic Infection. Development of EUCAST disk diffusion method for susceptibility testing of the Bacteroides fragilis group isolates. Anaerobe 2015, 31, 65–71. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, S.A.; Al-Jamell, D.S.; Tweij, T.A.R.; Hameed, S.A. Comparative study between pre and post bacterial growth of periodontal infections by treatment with extracts Rue. An in vitro study. J. Pharm. Sci. Res. 2019, 11, 104–109. [Google Scholar]
- Mahalakshmi, K.; Krishnan, P.; Chandrasekaran, S.C. Detection of Tannerella forsythia bspA and prtH genotypes among periodontitis patients and healthy subjects—A case-Control study. Arch. Oral Biol. 2018, 96, 178–181. [Google Scholar] [CrossRef]
- Puletic, M.; Popovic, B.; Jankovic, S.; Brajovic, G. Detection rates of periodontal bacteria and herpesviruses in different forms of periodontal disease. Microbiol. Immunol. 2020, 64, 815–824. [Google Scholar] [CrossRef] [PubMed]
- Al Yahfoufi, Z.; Hadchiti, W. Prevalence of Periodontal Pathogens in a Group of Participants from the Middle East and North Africa Geographic Region with Minimal Periodontal Disease. J. Int. Soc. Prev. Community Dent. 2017, 7, S30–S35. [Google Scholar] [CrossRef] [PubMed]
- Rajakaruna, G.A.; Negi, M.; Uchida, K.; Sekine, M.; Furukawa, A.; Ito, T.; Kobayashi, D.; Suzuki, Y.; Akashi, T.; Umeda, M.; et al. Localization and density of Porphyromonas gingivalis and Tannerella forsythia in gingival and subgingival granulation tissues affected by chronic or aggressive periodontitis. Sci. Rep. 2018, 8, 9507. [Google Scholar] [CrossRef] [PubMed]
- Badanian, A.; de León, E.P.; Rodriquez, L.; Bascuas, T.; Capo, C.; Battle, A.; Bueno, L.; Papone, V. Detection of periodontal pathogens in a Uruguayan population with aggressive periodontitis using conventional and molecular methods. Odontoestomatología 2018, 20, 68–77. [Google Scholar] [CrossRef]
- Menini, M.; Delucchi, F.; Bagnasco, F.; Pera, F.; Di Tullio, N.; Pesce, P. Analysis of the Subgingival Microbiota in Implant-Supported Full-Arch Rehabilitations. Dent. J. 2020, 8, 104. [Google Scholar] [CrossRef] [PubMed]
- dos Santos, P.B.R.E.; de Lima, P.M.N.; Palma, A.L.R.; Hasna, A.A.; Rossoni, R.D.; Junqueira, J.C.; de Oliveira, L.D. Review- The periodontal pathogen Treponema denticola: An atherosclerosis risk factor. Res. Soc. Dev. 2021, 10, e25810111637. [Google Scholar] [CrossRef]
- Zeng, H.; Chan, Y.; Gao, W.; Leung, W.K.; Watt, R.M. Diversity of Treponema denticola and Other Oral Treponeme Lineages in Subjects with Periodontitis and Gingivitis. Microbiol. Spectr. 2021, 9, e0070121. [Google Scholar] [CrossRef] [PubMed]
- Jepsen, K.; Falk, W.; Brune, F.; Fimmers, R.; Jepsen, S.; Bekeredjian-Ding, I. Prevalence and antibiotic susceptibility trends of periodontal pathogens in the subgingival microbiota of German periodontitis patients: A retrospective surveillance study. J. Clin. Periodontol. 2021, 48, 1216–1227. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.; Zhu, L.; Xu, L.; Meng, H.; Zhang, L.; Ren, X.; Lu, R.; Tian, Y.; Shi, D.; Wang, X. Distribution of 8 periodontal microorganisms in family members of Chinese patients with aggressive periodontitis. Arch. Oral. Biol. 2015, 60, 400–407. [Google Scholar] [CrossRef] [PubMed]
- Torrungruang, K.; Jitpakdeebordin, S.; Charatkulangkun, O.; Gleebbua, Y. Porphyromonas gingivalis, Aggregatibacter actinomycetemcomitans, and Treponema denticola/Prevotella intermedia Co-Infection are Associated with Severe Periodontitis in a Thai Population. PLoS ONE 2015, 10, e0136646. [Google Scholar] [CrossRef]
- Al-Deen, H.S.; Al-Ankoshy, A.A.M.; Al-Najhi, M.M.A.; Al-Kabsia, T.A.; AL-Haddad, K.A.; Al-Akwa, A.A.Y.; Al-Shamahy, H.A.; Al-labani, M.A. Porphyromonas gingivalis: Biofilm formation, antimicrobial susceptibility of isolates from cases of Localized Aggressive Periodontitis (LAP). Univers. J. Pharm. Res. 2021, 6, 1–7. [Google Scholar] [CrossRef]
- Jia, L.; Han, N.; Du, J.; Guo, L.; Luo, Z.; Liu, Y. Pathogenesis of Important Virulence Factors of Porphyromonas gingivalis via Toll-Like Receptors. Front. Cell. Infect. Microbiol. 2019, 9, 262. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Zhou, W.; Wang, H.; Liang, S. Roles of Porphyromonas gingivalis and its Virulence factors in periodontitis. Adv. Protein Chem. Struct. Biol. 2020, 120, 45–84. [Google Scholar] [CrossRef] [PubMed]
- Septiwidyati, T.R.; Bachtiar, E.W. The Role of Porphyromonas gingivalis Virulence Factors in Periodontitis Immunopathogenesis. Dentika Dent. J. 2020, 23, 6–12. [Google Scholar] [CrossRef]
- Joshi, V.M.; Bhat, K.G.; Kugaji, M.S.; Ingalagi, P.S. Prevalence of Porphyromonas gingivalis and its relationship with herpesvirus in Indian subjects with chronic periodontitis: A cross-sectional study. J. Int. Clin. Dent. Res. Organ. 2016, 8, 106–110. [Google Scholar] [CrossRef]
- Rafiei, M.; Kiani, F.; Sayehmiri, K.; Sayehmiri, F.; Tavirani, M.; Dousti, M.; Sheikhi, A. Prevalence of Anaerobic Bacteria (P. gingivalis) as Major Microbial Agent in the Incidence Periodontal Diseases by Meta-Analysis. J. Dent. (Shiraz) 2018, 19, 232–242. [Google Scholar]
- Mínguez, M.; Ennibi, O.K.; Perdiguero, P.; Lakhdar, L.; Abdellaoui, L.; Sánchez, M.C.; Sanz, M.; Herrera, D. Antimicrobial susceptibilities of Aggregatibacter actinomycetemcomitans and Porphyromonas gingivalis strains from periodontitis patients in Morocco. Clin. Oral Investig. 2019, 23, 1161–1170. [Google Scholar] [CrossRef] [PubMed]
- Santibáñez, R.; Rodríguez-Salas, C.; Flores-Yáñez, C.; Garrido, D.; Thomson, P. Assessment of Changes in the Oral Microbiome That Occur in Dogs with Periodontal Disease. Vet. Sci. 2021, 8, 291. [Google Scholar] [CrossRef] [PubMed]
- Ardila, C.M.; Bedoya-García, J.A. Antimicrobial resistance of Aggregatibacter actinomycetemcomitans, Porphyromonas gingivalis and Tannerella forsythia in periodontitis patients. J. Glob. Antimicrob. Resist. 2020, 22, 215–218. [Google Scholar] [CrossRef] [PubMed]
- Alazemi, A.M.; Jamal, W.; Al Khabbaz, A.; Rotimi, V.O. Prevalence of target anaerobes associated with chronic periodontitis. Access. Microbiol. 2020, 2, acmi000177. [Google Scholar] [CrossRef] [PubMed]
- Papone, V.; Verolo, C.; Zaffaroni, L.; Batlle, A.; Capo, C.; Bueno, L.; Gamonal, J.; Silvia, N.; Soria, S. Detection and prevalence of periodontal pathogens in a Uruguayan population with chronic periodontitis using conventional methodology and metagenomics. Odontoestomatología 2015, 17, 23–33. [Google Scholar]
- Tettamanti, L.; Gaudio, R.M.; Cura, F.; Mucchi, D.; Illuzzi, N.; Tagliabue, A. Prevalence of periodontal pathogens among italian patients with chronic periodontitis: A retrospective study on 2992 patients. Oral Implantol. 2017, 10, 28–36. [Google Scholar] [CrossRef] [PubMed]
- Tanner, A.C.R.; Izard, J. Tannerella forsythia, a periodontal pathogen entering the genomic era. Periodontology 2000 2006, 42, 88–113. [Google Scholar] [CrossRef]
- Belibasakis, G.N.; Maula, T.; Bao, K.; Lindholm, M.; Bostanci, N.; Oscarsson, J.; Ihalin, R.; Johansson, A. Virulence and Pathogenicity Properties of Aggregatibacter actinomycetemcomitans. Pathogens 2019, 8, 222. [Google Scholar] [CrossRef] [PubMed]
- Fine, D.H.; Patil, A.G.; Velusamy, S.K. Aggregatibacter actinomycetemcomitans (Aa) Under the Radar: Myths and Misunderstandings of Aa and Its Role in Aggressive Periodontitis. Front. Immunol. 2019, 10, 728. [Google Scholar] [CrossRef]
- Jensen, A.B.; Isidor, F.; Lund, M.; Væth, M.; Johansson, A.; Lauritsen, N.N.; Haubek, D. Prevalence of Aggregatibacter actinomycetemcomitans and Periodontal Findings among 14 to 15-Year Old Danish Adolescents: A Descriptive Cross-Sectional Study. Pathogens 2020, 9, 1054. [Google Scholar] [CrossRef] [PubMed]
- Claesson, R.; Höglund-Åberg, C.; Haubek, D.; Johansson, A. Age-Related prevalence and characteristics of Aggregatibacter actinomycetemcomitans in periodontitis patients living in Sweden. J. Oral Microbiol. 2017, 9, 1334504. [Google Scholar] [CrossRef] [PubMed]
- Borilova Linhartova, P.; Danek, Z.; Deissova, T.; Hromcik, F.; Lipovy, B.; Szaraz, D.; Janos, J.; Fassmann, A.; Bartova, J.; Drizhal, I.; et al. Interleukin Gene Variability and Periodontal Bacteria in Patients with Generalized Aggressive Form of Periodontitis. Int. J. Mol. Sci. 2020, 21, 4728. [Google Scholar] [CrossRef] [PubMed]
- Govindarajan, K.; Muthukumar, S.; Rangarao, S. Relationship between interleukin 1α levels in the gingival crevicular fluid in health and in inflammatory periodontal disease and periodontal inflamed surface area: A correlative study. J. Indian Soc. Periodontol. 2015, 19, 618–623. [Google Scholar] [CrossRef]
- Grigoriadou, M.E.; Koutayas, S.O.; Madianos, P.N.; Strub, J.R. Interleukin-1 as a genetic marker for periodontitis: Review of the literature. Quintessence Int. 2010, 41, 517–525. [Google Scholar]
- Sippert, E.Â.; de Oliveira e Silva, C.; Ayo, C.M.; Marques, S.B.; Visentainer, J.E.; Sell, A.M. HLA Haplotypes and Genotypes Frequencies in Brazilian Chronic Periodontitis Patients. Mediat. Inflamm. 2015, 2015, 481656. [Google Scholar] [CrossRef]
- Reichert, S.; Altermann, W.; Stein, J.M.; Schaller, H.G.; Machulla, H.K.; Schulz, S. Individual composition of human leukocyte antigens and periodontopathogens in the background of periodontitis. J. Periodontol. 2013, 84, 100–109. [Google Scholar] [CrossRef]
- Brodzikowska, A.; Górska, R.; Kowalski, J. Interleukin-1 Genotype in Periodontitis. Arch. Immunol. Ther. Exp. 2019, 67, 367–373. [Google Scholar] [CrossRef]
- Arastu-Kapur, S.; Nguyen, M.; Raha, D.; Ermini, F.; Haditsch, U.; Araujo, J.; De Lannoy, I.; Ryder, M.I.; Dominy, S.S.; Lynch, C.; et al. Treatment of Porphyromonas gulae infection and downstream pathology in the aged dog by lysine-gingipain inhibitor COR388. Pharmacol. Res. Perspect. 2020, 8, e00562. [Google Scholar] [CrossRef]
- Fujiwara-Takahashi, K.; Watanabe, T.; Shimogishi, M.; Shibasaki, M.; Umeda, M.; Izumi, Y.; Nakagawa, I. Phylogenetic diversity in fim and mfa gene clusters between Porphyromonas gingivalis and Porphyromonas gulae, as a potential cause of host specificity. J. Oral Microbiol. 2020, 12, 1775333. [Google Scholar] [CrossRef]
- Oba, P.M.; Carroll, M.Q.; Alexander, C.; Somrak, A.J.; Keating, S.; Sage, A.M.; Swanson, K.S. Dental chews positively shift the oral microbiota of adult dogs. J. Anim. Sci. 2021, 99, skab100. [Google Scholar] [CrossRef] [PubMed]
- Lenzo, J.C.; O’Brien-Simpson, N.M.; Orth, R.K.; Mitchell, H.L.; Dashper, S.G.; Reynolds, E.C. Porphyromonas gulae Has Virulence and Immunological Characteristics Similar to Those of the Human Periodontal Pathogen Porphyromonas gingivalis. Infect. Immun. 2016, 84, 2575–2585. [Google Scholar] [CrossRef] [PubMed]
- Nomura, R.; Shirai, M.; Kato, Y.; Murakami, M.; Nakano, K.; Hirai, N.; Mizusawa, T.; Naka, S.; Yamasaki, Y.; Matsumoto-Nakano, M.; et al. Diversity of fimbrillin among Porphyromonas gulae clinical isolates from Japanese dogs. Vet. Med. Sci. 2012, 74, 885–891. [Google Scholar] [CrossRef] [PubMed]
- Iwashita, N.; Nomura, R.; Shirai, M.; Kato, Y.; Murakami, M.; Matayoshi, S.; Kadota, T.; Shirahata, S.; Ohzeki, L.; Arai, N.; et al. Identification and molecular characterization of Porphyromonas gulae fimA types among cat isolates. Vet. Microbiol. 2019, 229, 100–109. [Google Scholar] [CrossRef] [PubMed]
- Kwon, D.; Bae, K.; Kim, H.; Kim, S.H.; Lee, D.; Lee, J.H. Treponema denticola as a prognostic biomarker for periodontitis in dogs. PLoS ONE 2022, 17, e0262859. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).