Simple Summary
Mitragyna speciosa (Kratom) is a narcotic plant indigenous to Southeast Asian countries, including Thailand. Traditionally, M. speciosa has been used as medicine to treat diarrhea and has anti-coughing, analgesic, and fever-reducing properties. Its leaves are commonly chewed by workers during physical labor for their coca-like stimulant effect to increase stamina and endurance. To identify important bioactive alkaloids with potential pharmaceutical uses, we performed a whole genome sequencing of Kratom to obtain information relating to the gene content in its genome, which will facilitate an improved understanding of the biosynthesis pathway and provide resources for assessing the genetic diversity in M. speciosa.
Abstract
Mitragyna speciosa (Kratom) is a tropical narcotic plant native to Southeast Asia with unique pharmacological properties. Here, we report the first chromosome-scale assembly of the M. speciosa genome. We employed PacBio sequencing to obtain a preliminary assembly, which was subsequently scaffolded using the chromatin contact mapping technique (Hi-C) into 22 pseudomolecules. The final assembly was 692 Mb with a scaffold N50 of 26 Mb. We annotated a total of 39,708 protein-coding genes, and our gene predictions recovered 98.4% of the highly conserved orthologs based on the BUSCO analysis. The phylogenetic analysis revealed that M. speciosa diverged from the last common ancestors of Coffea arabica and Coffea canephora approximately 47.6 million years ago. Our analysis of the sequence divergence at fourfold-degenerate sites from orthologous gene pairs provided evidence supporting a genome-wide duplication in M. speciosa, agreeing with the report that members of the genus Mitragyna are tetraploid. The STRUCTURE and principal component analyses demonstrated that the 85 M. speciosa accessions included in this study were an admixture of two subpopulations. The availability of our high-quality chromosome-level genome assembly and the transcriptomic resources will be useful for future studies on the alkaloid biosynthesis pathway, as well as comparative phylogenetic studies in Mitragyna and related species.
1. Introduction
Mitragyna speciosa (also known as Kratom in Thailand) is a tropical evergreen indigenous to Southeast Asia and is commonly grown in Thailand, Malaysia, Vietnam, and Papua New Guinea islands [1,2]. Mitragyna is a small genus belonging to the family Rubiaceae. Four species of Mitragyna (Mitragyna diversifolia, Mitragyna hirsuta, Mitragyna rotundifolia, and M. speciosa) are widely distributed in Thailand, especially from the central to southern parts of the country. Among these four, M. speciosa is the only species that is considered as a narcotic plant with specific medicinal importance [1]. Traditionally, M. speciosa has been used as medicine to treat diarrhea and has anti-coughing, analgesic, and fever-reducing properties. Its leaves are commonly chewed by workers during physical labor for their coca-like stimulant effect to increase stamina and endurance. In addition, M. speciosa has a medical use for treating chronic pain, and it is effective in relieving opioid-related withdrawal symptoms for heroin or morphine addicts [3,4].
Kiehn (1995) reported that Mitragyna species are tetraploid with a chromosome number of 2n = 4x = 44 [5]. Chromosome staining with DAPI was also carried out to confirm the chromosome number of 44 in the mitotic metaphase [6]. However, the identity of progenitor species that underwent hybridization to give rise to M. speciosa is currently unknown. The majority of published scientific literature on M. speciosa focuses mainly on its chemical composition with mitragynine, the principal compound responsible for analgesic activity, being the most studied alkaloid from this psychoactive medicinal plant [7,8,9,10]. Although mitragynine possesses potent opioid agonist properties, mediating μ- and κ-opioid receptors, its chemical structure is different from those of morphine and other narcotic painkillers [11]. In addition to mitragynine, 54 known alkaloids have successfully been isolated and identified in this species. Due to interest in a diverse range of bioactive alkaloids produced by M. speciosa, the draft genome sequence has recently been assembled using the linked-read 10× Genomics technology [6]. The reported assembly encompasses 1.12 Gb and contains 17,031 scaffolds with an N50 scaffold size of 1.02 Mb. To improve the quality and the contiguity of the genome assembly, we employed the Pacific Biosciences (PacBio) long-read single molecule real-time sequencing to generate the preliminary assembly of M. speciosa, which was subsequently scaffolded using the chromatin contact mapping (Hi-C) technology to achieve a final assembly that contains 22 pseudomolecules, corresponding to the diploid chromosome number in M. speciosa. Our high-quality, chromosome-level assembly of the M. speciosa genome provides a valuable resource for identifying bioactive alkaloids with potential pharmaceutical uses and for studying Kratom genetic diversity, population structure, and comparative genomics with related species.
2. Materials and Methods
2.1. Genome Size Estimation
To estimate the nuclear DNA content using flow cytometry, fresh leaf tissues were cut into small pieces with a sharp razor blade and analyzed using the protocol in [12]. We used the Galbraith’s buffer reported in [13] as a nuclear isolation buffer. Nuclei were stained with 50 ug/mL of propidium iodide (Thermo Fisher Scientific, Waltham, MA, USA). Tomato (Solanum lycopersicum) was used as the internal DNA reference standard.
2.2. Plant Materials and DNA/RNA Isolation
For high-molecular-weight DNA extraction (whole genome sequencing), healthy leaf tissues were collected from a mature M. speciosa tree (Figure 1; Sample name ‘RV’ in Supplementary Table S1), immediately frozen, and stored in liquid nitrogen until use. DNA was isolated following the protocol in [14]. Briefly, the frozen tissue was ground in liquid nitrogen and CTAB buffer was added. DNA was extracted from the aqueous phase using 25:24:1 phenol:chloroform:isoamyl alcohol and precipitated in 100% ethanol. DNA pellets were washed twice with 70% ethanol, air-dried, and resuspended in 10 mM Tris-HCl pH 8.0. After purification with the Ampure PB beads (Pacific Biosciences, Menlo Park, CA, USA), DNA integrity was evaluated using Pippin Pulse Electrophoresis System (Sage Science, Beverly, MA, USA). For the genetic diversity study, we collected leaf tissues from a total of 85 M. speciosa accessions from 15 provinces in Thailand (Supplementary Figure S1 and Table S1). DNA was isolated using the Qiagen DNeasy Plant Mini Kit following the manufacturer’s protocol (Qiagen, Hilden, Germany). For transcriptome sequencing, leaf, stem, and flower tissues were collected from the same individual used for whole genome sequencing. Total RNA was extracted using the CTAB buffer and 25:24:1 phenol:chloroform:isoamyl alcohol and precipitated overnight in ¼ vol 8M LiCl. RNA pellets were washed twice with 70% ethanol, air-dried, and resuspended in RNase-free water.

Figure 1.
Morphology of M. speciosa. (A) Tree, (B) leaves, (C) flower, and (D) seeds.
2.3. Genome and Isoform Sequencing (Iso-seq) Library Preparation
For whole genome sequencing, a SMRTbell library with an insert size of 12,000 nt was prepared from the high molecular weight DNA template using SMRTbell Express Template Prep Kit 2.0 and sequenced on the PacBio Sequel system (Pacific Biosciences, Menlo Park, CA, USA). Sequencing was performed with the Sequel Binding Kit 2.0 using a 20-h movie collection time following the manufacturer’s protocol (Pacific Biosciences, Menlo Park, CA, USA). For transcriptome sequencing, Iso-seq libraries were prepared using the NEBNext Single Cell/Low Input cDNA Synthesis and Amplification Module (New England Biolabs, Ipswich, MA, USA), Iso-Seq Express Oligo Kit, and SMRTbell Express Template Prep Kit 2.0 (Pacific Biosciences, Menlo Park, CA, USA). Iso-seq was performed as described above.
2.4. Hi-C Library Preparation and Sequencing
A chromosome conformation capturing technique (Hi-C) was conducted by Biomarker Technologies (Beijing, China) to scaffold the preliminary assembly into a chromosome-level assembly. For Hi-C library preparation, the M. speciosa sample was fixed with formaldehyde, and fixed DNA was digested with HindIII restriction endonuclease. The overhanging ends were filled with biotinylated nucleotides, and DNA fragments were circularized by blunt-end ligation. After the cross-linking was reversed, DNA was purified and sheared into fragments of 300–700 bp. Biotinylated fragments were captured with streptavidin beads. Purified fragments were used to construct a sequencing library, which was subsequently sequenced on the Illumina platform (PE150; Illumina, San Diego, CA, USA) to produce 398,879,554 read pairs.
2.5. PacBio Draft Assembly and Hi-C Scaffolding
A total of 4,049,699 PacBio raw reads totaling 36.8 Gb were subjected to read correction, trimming, overlap detection and de novo assembly by Canu v1.9 [15] using the following parameters: genomeSize = 700 m, correctedErrorRate = 0.040. For other parameters, default settings were used. An estimated genome size of 700 Mb was assumed according to our flow cytometry results (see the ‘Results’ section below). The polishing was carried out using the GenomicConcensus package in the SMRT Link v6.0 (https://github.com/PacificBiosciences/GenomicConsensus, accessed on 6 September 2022). The PacBio draft assembly (used as an input for the Hi-C scaffolding) was cut into 50-kb fragments and reassembled based on Hi-C data. Contigs in the preliminary assembly that could not be recovered in the corrected assembly were regarded as potentially misassembled regions (in those regions, positions with low Hi-C coverage were defined as error sites). The corrected contigs were further scaffolded into a chromosome-level assembly by LACHESIS [16] with the following parameter setting: CLUSTER_MIN_RE_SITES = 21; CLUSTER_MAX_LINK_DENSITY = 2; ORDER_MIN_N_RES_IN_TRUNK = 15; ORDER_MIN_N_RES_IN_SHREDS = 15. After this step, placement and orientation errors exhibiting obvious discrete chromatin interaction patterns were manually adjusted. After the manual adjustment, a total of 672 Mb sequences were anchored onto 22 chromosomes.
2.6. Genome Assembly Evaluation
The quality of the final genome assembly was assessed by aligning short-read DNA sequences (RADseq data) and transcriptome (Iso-seq) data from this study using BWA version 0.7.17-r1188 for DNA sequence alignment and HISAT2 version 2.2.0 for RNA alignment. We also tested for the presence and completeness of the orthologs using the Benchmarking Universal Single-Copy Orthologues (BUSCO) version 4.0.5 [17] and the Embryophyta OrthoDB release 10 [18].
To identify repetitive element families in the genome assembly, RepeatModeler version 2.0.3 (http://www.repeatmasker.org/RepeatModeler/, last accessed on 16 August 2022) was used to construct a de novo repeat library. This pipeline employed two distinct repeat discovery algorithms, RECON (version 1.08) and RepeatScout (version 1.0.6), to identify the boundaries of repetitive elements and build consensus models of interspersed repeats [19,20]. We aligned repeat sequences in the library to GenBank’s nr protein database using BLASTX (e-value cutoff = 10−6) to ensure that they did not contain large families of protein-coding sequences.
To identify protein-coding sequences in the unmasked assembly, we used EvidenceModeler (EVM) version 1.1.1 to combine evidences from RNA-based predictions, homology-based predictions, and ab initio predictions [21]. For RNA-based prediction, we used evidence from PacBio Iso-seq data obtained from leaf, stem, and flower tissues. Full-length transcripts were mapped to the final assembly using the genomic mapping and alignment program (GMAP) version 2020-09-12 [22]. Protein sequences from Coffea arabica, Coffea canephora, S. lycopersicum, M. speciosa, and Arabidopsis thaliana available on the public databases were aligned to the unmasked genome using AAT [23]. Protein-coding gene predictions were obtained with Augustus version 3.2.1 [24] trained with C. arabica, C. canephora, S. lycopersicum, and M. speciosa PASA transcriptome alignment assembly using M. speciosa alignment files as inputs. All gene predictions were integrated by EVM to generate consensus gene models using the following weights for each evidence type: PASA2—1, GMAP—0.5, AAT—0.3 and Augustus—0.3. Any predicted genes that had more than 20% overlapping sequence with repetitive sequences or had no RNA-seq support were excluded from the list of annotated genes.
2.7. Phylogenetic Analyses and Comparative Genomics
OrthoFinder version 2.4.0 [25] was used to identify orthologous groups in M. speciosa, Amborella trichopoda, A. thaliana, C. arabica, C. canephora, Erythranthe guttata, Populus trichocarpa, Prunus persica, Ricinus communis, S. lycopersicum, Theobroma cacao, and Vitis vinifera. We constructed a phylogenetic tree based on protein sequences from single-copy orthologous groups using RAxML-NG program version 1.0.2 [26]. We first aligned protein sequences in each single-copy orthologous group with MUSCLE [27] and removed alignment gaps with trimAI version 1.4 rev15 [28] using the automated1 heuristic method. We subsequently concatenated alignment blocks using catsequences program (https://github.com/ChrisCreevey/catsequences, last accessed on 22 August 2022), and the substitution model for each block was estimated using the ModelTest-NG program version 0.1.7 [29]. The outputs were used to compute a maximum-likelihood phylogenetic tree. Divergence times were estimated using the MCMCtree software version 4.0 (PAML 4 package) [30] using the relaxed-clock model with the known divergence time between P. trichocarpa and R. communis, which was estimated at 105–120 million years ago (MYA) [31,32].
2.8. Genome Synteny Analysis
McscanX [33] was used to analyze the collinearity within the M. speciosa genome and between M. speciosa–C. arabica and M. speciosa–C. robusta genomes. M. speciosa amino acid sequences were aligned against themselves, C. arabica, or C. canephora using BLASTP (with an e-value cutoff of 10−10) in order to identify putative paralogs. Intragenic homeologous blocks were defined as regions of ten or more genes with colinear or nearly colinear runs of paralogs elsewhere in the genome with fewer than six intervening genes. These intragenic homeologous blocks were visualized using CIRCOS version 0.69.8 [34]. Similarly, we also performed pairwise comparisons of input protein sequences from M. speciosa, C. canephora, and C. arabica. Clustering was carried out using OrthoMCL software version 2.0.9 [35] based on a Markov clustering algorithm (MCL). Syntenic blocks between M. speciosa, C. canephora, and C. arabica were identified by MCscanX and plotted with CIRCOS using the criteria mentioned above (at least ten colinear genes and fewer than six intervening genes allowed).
2.9. Population Structure and Genetic Diversity Analyses
Short-read data (RAD-seq) from 85 M. speciosa accessions collected in Thailand were used to analyze the population structure. We first mapped the sequencing reads to the final genome assembly using BWA v0.7.17, and single nucleotide polymorphism (SNP) markers were identified using GATK HaplotypeCaller 3.8 [36]. A set of 2492 SNP markers at fourfold-degenerate sites with the following criteria were used for the analyses: (I) depth coverage between 20× and 200×, (II) minor allele frequency > 0.05, and (III) less than 10% missing data. A maximum-likelihood tree with 1000 bootstrap replicates was constructed using the analysis of phylogenetic and evolution (APE) software version 5.5 in the R package [37]. To evaluate the population structure, we employed the STRUCTURE program version 2.3.4 [38] using the same set of SNP markers (for the phylogenetic tree) and 10,000 iterations with the number of clusters (K) of 2–4. The level of genetic diversity in the population investigated was estimated by calculating the gene diversity (GD, or expected heterozygosity), polymorphism information content (PIC), observed heterozygosity (HO), and minor allele frequency (MAF) using the PowerMarker software version 3.25 [39]. Principal component analysis (PCA) was performed based on genetic distances (2492 SNP markers) among 85 M. speciosa accessions computed with TASSEL version 5.2.77 [40]. Principal components were generated using the covariance method, and eigenvalues were generated to determine the proportion of variation explained by each principal component. The first and second principal components were plotted using R software package ggplot2 version 3.3.4 [41].
3. Results
3.1. M. speciosa Genome Assembly and Annotation
To obtain a preliminary draft assembly of M. speciosa, whole-genome shotgun sequencing was carried out using PacBio long-read technology, and 36.8 Gb of sequencing data (4,049,699 reads) were generated. Based on the estimated genome size of 727 Mb obtained from the DNA flow cytometry (Supplementary Figure S2), these raw sequencing data represented ~54× coverage of the genome size. A preliminary PacBio assembly yielded a draft genome of 692 Mb in 4259 scaffolds with an N50 length of 0.9 Mb (Table 1). This draft assembly was subsequently scaffolded using the long-range chromatin fixation technique (Hi-C) into a chromosome-level assembly containing 22 pseudomolecules > 10 Mb in length (from here on, they will be referred to as chromosomes; Figure 2, Supplementary Figure S3). The genus Mitragyna has been reported as a tetraploid with a base chromosome number of 11 (2n = 4x = 44) [5,6]. The 22 chromosomes corresponded to the diploid chromosome number and covered 602 Mb, or 87.04% of the 692-Mb assembly.

Table 1.
Assembly statistics of M. speciosa genome.
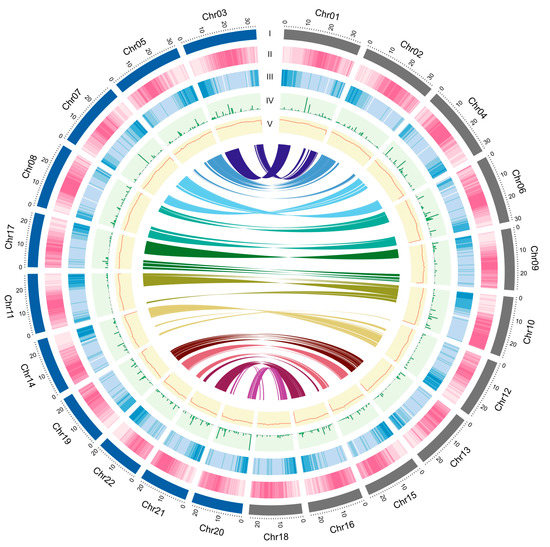
Figure 2.
Genomic landscape of M. speciosa. (I) A physical map of 22 pseudomolecules (chromosomes) numbered according to size (in Mb); (II) Content of repetitive sequences shown as proportion of genomic regions covered by repetitive sequences in 250-kb windows; (III) Gene density represented by the number of genes in 250-kb windows; (IV) SNP density represented by number of SNP markers in 250-kb windows; (V) GC content represented by the percentage of G + C bases in 250-kb windows. Syntenic blocks in the genome are displayed by colored lines.
To evaluate the quality of our assembly, short-read genomic DNA data (from RAD-seq) were aligned to the genome sequence, and 99.62% of the reads could be mapped to the M. speciosa genome. In addition, Iso-seq reads were aligned to the assembly, and 99.72% of the transcripts were mapped to the genome. An additional assessment of the gene space completeness was carried out using the BUSCO software and a plant-specific database of 1440 genes [17]. Predicted gene models in our M. speciosa assembly covered as much as 98.4% of the highly conserved orthologs in the Embryophyta lineage (81.6% classified as ‘complete and single-copy’, 15.4% as ‘complete and duplicated’, 1.4% as ‘fragmented’; Table 1). Results from the alignment and BUSCO analysis suggested that our chromosome-scale M. speciosa assembly is of high quality.
To annotate the genome assembly, three approaches were employed: ab initio prediction, homology-based search, and transcript-based evidence. A total of 42,873 predicted gene models and 39,708 protein-coding genes (92.6% of predicted gene models; Supplementary Table S2) were identified. We were able to assign gene ontology (GO) to 35,485 protein-coding genes (82.7% of predicted gene models; Supplementary Figure S4). The most prevalent GO terms associated with cellular components were the integral components of membrane, nucleus, and cytoplasm, while the largest categories of genes associated with molecular function and biological processes were ATP binding and protein phosphorylation, respectively. In addition, 70.3, 38.2 and 26.2% of the predicted gene models could be functionally annotated using the Swissprot, EC, and KEGG databases, respectively (Supplementary Table S3). The average GC content in the M. speciosa genome was 34.59% (Table 1), which was close to the average GC content in introns (33.8%; Supplementary Table S2), whereas the average GC content in exons was much higher at 43.3% (Supplementary Table S2).
We identified a number of genes involved in the specialized metabolic pathways that lead to the production of strictosidine, the intermediate in the monoterpene indole alkaloid biosynthesis (Supplementary Figure S5). Several gene families contain multiple homoeologs including those encoding key enzymes, such as loganic acid methyltransferase (LAMT), secologanin synthase (SLS), and strictosidine synthase (STR). The presence of duplicated genes is likely the consequence of the tetraploidization event. Interestingly, some of the genes encoding enzymes in the methylerythriol phosphate (MEP) pathway are present in single copies (Supplementary Figure S5), suggesting that duplicated genes may have been lost or degraded into non-functional genes after the polyploidization [42].
3.2. Identification of Repetitive Elements in the M. speciosa Genome
The M. speciosa genome assembly contained a total of 354.61 Mb of repetitive elements, representing 51.21% of the assembly (Table 2). A genome-wide distribution plot showed that DNA transposons and retrotransposons were located in proximity to the centromeric regions (Figure 2). Retrotransposons (122.38 Mb) represented the majority of the repetitive elements identified, comprising 17.83% of the assembly and 34.51% of the total repeated content. The most abundant retrotransposon types were the long terminal repeat (LTR) Copia and Gypsy, occupying 6.36% and 8.48% of the genome assembly.

Table 2.
Repeat contents in M. speciosa genome.
3.3. Comparative Genomics and Phylogenetic Analyses
To perform comparative genomics analyses, we clustered a total of 452,592 proteins (out of 469,904 input proteins from 12 species) into 28,228 orthologous groups. We then constructed a maximum-likelihood phylogenetic tree using the sequence information from single-copy orthologous genes and calculated the divergence time based on the topology and the branch length. M. speciosa diverged from the last common ancestors of C. arabica and C. canephora approximately 47.6 MYA (Figure 3A), and the last common ancestor of M. speciosa, C. arabica, and C. canephora diverged from S. lycopersicum ~106 MYA.
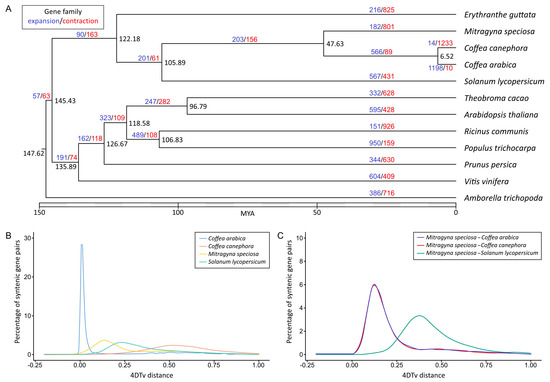
Figure 3.
Comparative genomics of M. speciosa and related species. (A) A maximum-likelihood tree of M. speciosa, two Coffea species, and nine other plant species constructed based on single-copy orthologous protein sequences. Numbers at each node represent the estimated divergence time in million years ago (MYA). Numbers in blue and red represent the numbers of gene families that have expanded or contracted, respectively, relative to their ancestors. (B) The distribution of fourfold synonymous third-codon transversion position (4DTv) distances between paralogous genes in M. speciosa, C. arabica, C. canephora, and S. lycopersicum. (C) The distribution of 4DTv distances between orthologous genes in M. speciosa and C. arabica, C. canephora and S. lycopersicum. Peaks of intraspecific and interspecific 4DTv distributions indicate whole-genome duplication and speciation events, respectively.
We determined the accumulated sequence divergence at fourfold synonymous third-codon transversion rate (4DTv) between the paralogous gene pairs in M. speciosa, C. Arabica, C. canephora, and S. lycopersicum to estimate their relative ages (Figure 3B). After correcting for multiple substitutions, we observed a sharp peak in 4DTv value at 0.012 synonymous transversions per site in C. arabica. This corresponded to the period between 1.08 and 0.54 million years ago, during which the hybridization of two progenitor species (C. canephora and Coffea eugenioides) led to the emergence of C. arabica [43]. Comparisons between the 12,198 pairs of paralogous genes residing within 418 duplicated colinear blocks within the M. speciosa genome revealed a noticeable peak at 0.139, suggesting that M. speciosa has undergone a recent whole-genome duplication (Figure 3B). This was also evidenced by the presence of intragenomic syntenic blocks throughout the genome (Figure 2) and the syntenic blocks between M. speciosa and the tetraploid C. arabica (Supplementary Figure S6). Interestingly, this peak overlapped with the 4DTv distance of M. speciosa–C. arabica and M. speciosa–C. canephora, suggesting that the genome-wide duplication event in M. speciosa took place in the time frame during which M. speciosa diverged from the last common ancestor of the two Coffea species (Figure 3C).
3.4. Genetic Diversity and Population Structure in M. speciosa
We collected and sequenced 85 M. speciosa accessions from different regions in Thailand in order to evaluate their genetic diversity and population structure. A total of 2492 SNP loci at fourfold-degenerate sites were used to assess the population structure using the Bayesian model-based clustering approach [44]. We selected the best K value based on the ΔK parameter proposed by [45], and K = 2 seemed to be the best fit for our data (Supplementary Figure S7). The STRUCTURE and PCA analyses demonstrated that the 85 M. speciosa accessions sampled in this work represented an admixture of two subpopulations (Figure 4) that appeared to originate from different geographical locations. One subpopulation consisted primarily of individuals collected from Western and Southern Thailand, whereas the other comprised individuals collected from Central Thailand (Supplementary Table S1).
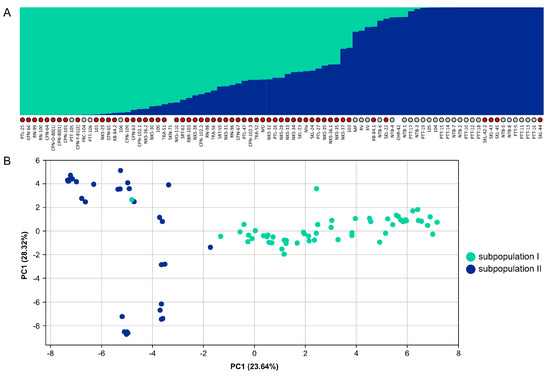
Figure 4.
Population structure and genetic diversity of M. speciosa. (A) Population structure of 85 M. speciosa accessions. Each accession is represented by a vertical bar, and the length of each color reflects the proportion contributed by ancestral populations. Accessions collected in Western and Southern Thailand are indicated by red dots while those collected in Central Thailand are indicated by grey dots. (B) Principal component analysis (PCA) of 85 M. speciosa accessions based on 2492 SNP markers. Each accession is represented by a single dot with colors indicating subpopulations.
The genetic diversity analysis of 85 M. speciosa accessions revealed that the GD ranged from 0.1 to 0.5 with an average of 0.36, while the PIC varied from 0.09 to 0.38 with an average of 0.28 (Figure 5). For the majority of SNPs analyzed, the GD and PIC values were ~0.5 and ~0.4, respectively. The HO values varied from 0.06 to 1 with most of the SNPs possessing HO values of 0.1. The mean values for HO and MAF were 0.5 and 0.28, respectively.
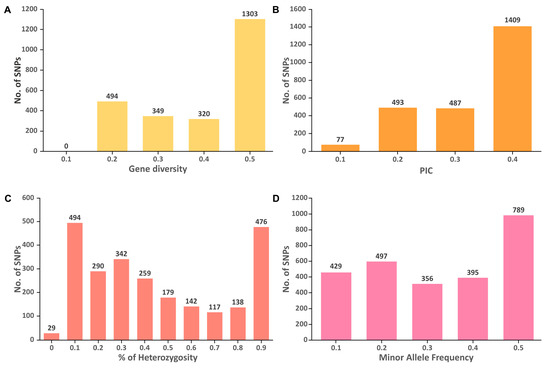
Figure 5.
Distribution of (A) gene diversity (GD), (B) polymorphism information content (PIC), (C) percentage of heterozygosity, and (D) minor allele frequency for 2492 SNP markers among the 85 M. speciosa accessions.
4. Discussion
M. speciosa, or Kratom, is a tropical plant indigenous to Southeast Asia that produces multiple biologically active phytochemicals with unique pharmacological properties. Here, we present a de novo assembly of the M. speciosa genome using the combination of PacBio long-read SMRT technology and the long-range proximity ligation technique (Hi-C). The preliminary assembly obtained from the PacBio sequencing was 692 Mb, slightly smaller than our estimated genome size of 727 Mb (by DNA flow cytometry) and a previously reported genome size of 780 Mb [6]. We subsequently employed the Hi-C method to identify chromosomal interactions using chromosome conformation capture. The Hi-C data provided long-range linkage information of up to tens of megabases that could be used to generate chromosome-level scaffolds. Our final assembly contained 22 pseudomolecules covering 602 Mb or ~87% of the 692-Mb assembly. The genus Mitragyna has been reported as a tetraploid with a base chromosome number of 11 (2n = 4x = 44) [5,6]. We believed that M. speciosa is an allotetraploid deriving from the hybridization of two currently unknown progenitors since our Hi-C assembly was phased into 22 chromosomes instead of the haploid chromosome number. We evaluated the representation of highly conserved orthologs using the BUSCO tool and demonstrated that our gene prediction recovered as much as 98.4% of the conserved orthologs in the Embryophyta lineage, suggesting that our genome assembly is more complete than the previously reported assembly, which covered only 88.5% of the BUSCO orthologs [6]. In addition, the contiguity of our chromosome-level assembly (N50 = 26 Mb and contains 2888 scaffolds) is also superior to that of the published assembly (N50 = 1 Mb and contains 17,031 scaffolds) [6]. We also noticed a huge discrepancy between the sizes of our assembly (692 Mb) and the previously published one (1122 Mb) [6]. The dot plot alignment of scaffolds/contigs from these two assemblies revealed several duplicated regions in the previously published assembly (examples shown in Supplementary Figure S8), suggesting that there were residual haplotigs remaining in the previous assembly. Those uncollapsed contigs provided the most likely explanation as to why the previous assembly was significantly larger than both the one presented here and the estimated genome size by DNA flow cytometry.
The analysis of intragenomic synteny and the 4DTv analysis of the paralogous gene pairs in M. speciosa revealed the evidence of whole-genome duplication. There appeared to be a one-to-one synteny between 11 pairs of homeologous chromosomes (Figure 1). Interestingly, our 4DTv analyses suggested that the genome-wide duplication in M. speciosa occurred during the same period in which M. speciosa diverged from the last common ancestor of those two Coffea species (Figure 3B). Our evaluation of the M. speciosa population structure showed that the 85 individuals sampled in this work could be grouped into two subpopulations. The clustering appeared to be associated with the geographical regions from which the samples were collected, with individuals from Western and Southern Thailand in one subpopulation and those from Central Thailand in another. We also assessed the level of genetic diversity of the population studied and found that among the 85 M. speciosa accessions, most of the SNP markers were moderately informative with an average PIC value of 0.28. These molecular markers will be useful for evaluating the genetic diversity of other M. speciosa populations and also for future marker-assisted breeding programs.
5. Conclusions
With M. speciosa emerging as a new cash crop, there has been interest in developing elite cultivars producing bioactive alkaloids with potential pharmaceutical uses. The availability of this high-quality reference genome assembly, the transcriptomic data along with the genomic variation information from the population will enable future studies on gene expression profiling that can improve our understanding of the biosynthetic pathways, studies on phylogeny, and comparative genomics in Rubiaceae family.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/biology11101492/s1, Figure S1: A map illustrating the locations of 85 M. speciosa accessions in Thailand collected for the population structure study; Figure S2: Genome size estimation by DNA flow cytometry. Histograms of relative DNA contents obtained after analysis of nuclei isolated from leaf tissues of M. speciosa and tomato plants (Solanum lycopersicum; used as a reference standard); Figure S3: Hi-C interaction matrix maps of M. speciosa chromosomes. The contact density is indicated by the color scale on the right from dark red (high density) to white (low density); Figure S4: Gene Ontology (GO) annotation of M. speciosa genes in the assembly; Figure S5: Mitragynine biosynthesis pathway and the list of genes in the pathway that were identified in the genome assembly; Figure S6: Synteny between M. speciosa and (A) C. canephora (B) C. arabica; Figure S7: Delta K values for STRUCTURE analysis of 85 M. speciosa accessions. Delta K is plotted against the number of modeled gene pools (K); Figure S8: Examples of the dot plot alignment of scaffolds/contigs from our genome assembly (X-axis) and the previously published one (Y-axis). Red circles indicated duplicated regions in the assembly published by Brose et al. (2021). Table S1: List of sequenced M. speciosa accessions; Table S2: Annotation statistics for M. speciosa; Table S3: Functional annotations of M. speciosa protein-coding genes.
Author Contributions
Conceptualization, W.P., S.C. and S.T.; methodology, T.Y., N.J., S.C. and P.A.; formal analysis, W.P., W.K., C.N. and C.S.; investigation, W.P, T.Y., N.J., S.C., P.A. and S.T.; writing—original draft preparation, W.P.; writing—review and editing, W.P.; visualization, W.K.; supervision, W.P. and S.T.; project administration, S.C.; funding acquisition, S.C. and S.T. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Science and Technology Development Agency, Thailand, grant numbers P2051919 and 1000221.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
M. speciosa genome assembly and Iso-seq data have been submitted to the DDBJ/EMBL/GenBank databases under Bioproject PRJNA849364 and the following accession numbers: JAMWEH000000000 (genome assembly), SRR20982408 (Iso-seq; leaf), SRR20982407 (Iso-seq; stem) and SRR20982406 (Iso-seq; flower).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Davis, A. Rubiaceae of Thailand—A pictorial guide to indigenous and cultivated genera. Bot. J. Linn. Soc. 2006, 152, 131–132. [Google Scholar] [CrossRef]
- Suwanlert, S. A study of kratom eaters in Thailand. Bull. Narc. 1975, 27, 21–27. [Google Scholar]
- Cinosi, E.; Martinotti, G.; Simonato, P.; Singh, D.; Demetrovics, Z.; Roman-Urrestarazu, A.; Bersani, F.S.; Vicknasingam, B.; Piazzon, G.; Li, J.-H. Following “the roots” of Kratom (Mitragyna speciosa): The evolution of an enhancer from a traditional use to increase work and productivity in Southeast Asia to a recreational psychoactive drug in western countries. BioMed Res. Int. 2015, 968786. [Google Scholar] [CrossRef]
- Grundmann, O. Patterns of kratom use and health impact in the US—Results from an online survey. Drug Alcohol Depend. 2017, 176, 63–70. [Google Scholar] [CrossRef] [PubMed]
- Kiehn, M. Chromosome Survey of the Rubiaceae. Ann. Mo. Bot. Gard. 1995, 82, 398–408. [Google Scholar] [CrossRef]
- Brose, J.; Lau, K.H.; Dang, T.T.T.; Hamilton, J.P.; Martins, L.d.V.; Hamberger, B.; Hamberger, B.; Jiang, J.; O’Connor, S.E.; Buell, C.R. The Mitragyna speciosa (Kratom) Genome: A resource for data-mining potent pharmaceuticals that impact human health. G3 Genes 2021, 11, jkab058. [Google Scholar] [CrossRef]
- Flores-Bocanegra, L.; Raja, H.A.; Graf, T.N.; Augustinović, M.; Wallace, E.D.; Hematian, S.; Kellogg, J.J.; Todd, D.A.; Cech, N.B.; Oberlies, N.H. The Chemistry of Kratom [Mitragyna speciosa]: Updated Characterization Data and Methods to Elucidate Indole and Oxindole Alkaloids. J. Nat. Prod. 2020, 83, 2165–2177. [Google Scholar] [CrossRef] [PubMed]
- Beckett, A.; Shellard, E.; Tackie, A. THE MITRAGYNA SPECIES OF ASIA–Part IV. The alkaloids of the leaves of Mitragyna speciosa Korth.. Isolation of Mitragynine and Speciofoline1. Planta Med. 1965, 13, 241–246. [Google Scholar] [CrossRef]
- Takayama, H.; Ishikawa, H.; Kurihara, M.; Kitajima, M.; Aimi, N.; Ponglux, D.; Koyama, F.; Matsumoto, K.; Moriyama, T.; Yamamoto, L.T.; et al. Studies on the Synthesis and Opioid Agonistic Activities of Mitragynine-Related Indole Alkaloids: Discovery of Opioid Agonists Structurally Different from Other Opioid Ligands. J. Med. Chem. 2002, 45, 1949–1956. [Google Scholar] [CrossRef]
- Karunakaran, T.; Ngew, K.Z.; Zailan, A.A.D.; Mian Jong, V.Y.; Abu Bakar, M.H. The Chemical and Pharmacological Properties of Mitragynine and Its Diastereomers: An Insight Review. Front. Pharmacol. 2022, 13, 805986. [Google Scholar] [CrossRef] [PubMed]
- Gibbons, S.; Arunotayanun, W. Chapter 14—Natural Product (Fungal and Herbal) Novel Psychoactive Substances. In Novel Psychoactive Substances; Dargan, P.I., Wood, D.M., Eds.; Academic Press: Boston, MA, USA, 2013; pp. 345–362. [Google Scholar]
- Dolezel, J.; Bartos, J. Plant DNA flow cytometry and estimation of nuclear genome size. Ann. Bot. 2005, 95, 99–110. [Google Scholar] [CrossRef]
- Galbraith, D.W.; Harkins, K.R.; Maddox, J.M.; Ayres, N.M.; Sharma, D.P.; Firoozabady, E. Rapid Flow Cytometric Analysis of the Cell Cycle in Intact Plant Tissues. Science 1983, 220, 1049–1051. [Google Scholar] [CrossRef]
- Pootakham, W.; Naktang, C.; Sonthirod, C.; Kongkachana, W.; Narong, N.; Sangsrakru, D.; Maknual, C.; Jiumjamrassil, D.; Chumriang, P.; Tangphatsornroung, S. Chromosome-level genome assembly of the Indian mangrove (Ceriops tagal) revealed a genome-wide duplication event predating the divergence of Rhizophoraceae mangroves. Plant Genome 2022, 15, e20217. [Google Scholar] [CrossRef]
- Koren, S.; Walenz, B.P.; Berlin, K.; Miller, J.R.; Bergman, N.H.; Phillippy, A.M. Canu: Scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 2017, 27, 722–736. [Google Scholar] [CrossRef] [PubMed]
- Burton, J.N.; Adey, A.; Patwardhan, R.P.; Qiu, R.; Kitzman, J.O.; Shendure, J. Chromosome-scale scaffolding of de novo genome assemblies based on chromatin interactions. Nat. Biotechnol. 2013, 31, 1119–1125. [Google Scholar] [CrossRef] [PubMed]
- Simão, F.A.; Waterhouse, R.M.; Ioannidis, P.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 2015, 31, 3210–3212. [Google Scholar] [CrossRef]
- Kriventseva, E.V.; Tegenfeldt, F.; Petty, T.J.; Waterhouse, R.M.; Simao, F.A.; Pozdnyakov, I.A.; Ioannidis, P.; Zdobnov, E.M. OrthoDB v8: Update of the hierarchical catalog of orthologs and the underlying free software. Nucleic Acids Res. 2015, 43, D250–D256. [Google Scholar] [CrossRef]
- Price, A.L.; Jones, N.C.; Pevzner, P.A. De novo identification of repeat families in large genomes. Bioinformatics 2005, 21, i351–i358. [Google Scholar] [CrossRef] [PubMed]
- Bao, Z.; Eddy, S.R. Automated de novo identification of repeat sequence families in sequenced genomes. Genome Res. 2002, 12, 1269–1276. [Google Scholar] [CrossRef]
- Haas, B.J.; Salzberg, S.L.; Zhu, W.; Pertea, M.; Allen, J.E.; Orvis, J.; White, O.; Buell, C.R.; Wortman, J.R. Automated eukaryotic gene structure annotation using EVidenceModeler and the program to assemble spliced alignments. Genome Biol. 2008, 9, R7. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.; Watanabe, C. GMAP: A genomic mapping and alignment program for mRNA and EST sequences. Bioinformatics 2005, 21, 1859–1875. [Google Scholar] [CrossRef]
- Huang, X.; Adams, M.D.; Zhou, H.; Kerlavage, A.R. A tool for analyzing and annotating genomic sequences. Genomics 1997, 46, 37–45. [Google Scholar] [CrossRef] [PubMed]
- Stanke, M.; Steinkamp, R.; Waack, S.; Morgenstern, B. AUGUSTUS: A web server for gene finding in eukaryotes. Nucleic Acids Res. 2004, 32, W309–W312. [Google Scholar] [CrossRef] [PubMed]
- Emms, D.M.; Kelly, S. OrthoFinder: Phylogenetic orthology inference for comparative genomics. Genome Biol. 2019, 20, 238. [Google Scholar] [CrossRef] [PubMed]
- Stamatakis, A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 2006, 22, 2688–2690. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Capella-Gutiérrez, S.; Silla-Martínez, J.M.; Gabaldón, T. trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 2009, 25, 1972–1973. [Google Scholar] [CrossRef] [PubMed]
- Darriba, D.; Posada, D.; Kozlov, A.M.; Stamatakis, A.; Morel, B.; Flouri, T. ModelTest-NG: A new and scalable tool for the selection of DNA and protein evolutionary models. Mol. Biol. Evol. 2020, 37, 291–294. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007, 24, 1586–1591. [Google Scholar] [CrossRef] [PubMed]
- Xi, Z.; Ruhfel, B.R.; Schaefer, H.; Amorim, A.M.; Sugumaran, M.; Wurdack, K.J.; Endress, P.K.; Matthews, M.L.; Stevens, P.F.; Mathews, S.; et al. Phylogenomics and a posteriori data partitioning resolve the Cretaceous angiosperm radiation Malpighiales. Proc. Natl. Acad. Sci. USA 2012, 109, 17519–17524. [Google Scholar] [CrossRef] [PubMed]
- Davis, C.C.; Webb, C.O.; Wurdack, K.J.; Jaramillo, C.A.; Donoghue, M.J. Explosive Radiation of Malpighiales Supports a Mid-Cretaceous Origin of Modern Tropical Rain Forests. Am. Nat. 2005, 165, E36–E65. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [PubMed]
- Krzywinski, M.I.; Schein, J.E.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef]
- Li, L.; Stoeckert, C.J., Jr.; Roos, D.S. OrthoMCL: Identification of ortholog groups for eukaryotic genomes. Genome Res. 2003, 13, 2178–2189. [Google Scholar] [CrossRef]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef]
- Paradis, E.; Claude, J.; Strimmer, K. APE: Analyses of Phylogenetics and Evolution in R language. Bioinformatics 2004, 20, 289–290. [Google Scholar] [CrossRef]
- Falush, D.; Stephens, M.; Pritchard, J.K. Inference of population structure using multilocus genotype data: Linked loci and correlated allele frequencies. Genetics 2003, 164, 1567–1587. [Google Scholar] [CrossRef]
- Liu, K.; Muse, S.V. PowerMarker: An integrated analysis environment for genetic marker analysis. Bioinformatics 2005, 21, 2128–2129. [Google Scholar] [CrossRef]
- Bradbury, P.J.; Zhang, Z.; Kroon, D.E.; Casstevens, T.M.; Ramdoss, Y.; Buckler, E.S. TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics 2007, 23, 2633–2635. [Google Scholar] [CrossRef]
- Villanueva, R.A.M.; Chen, Z.J. ggplot2: Elegant Graphics for Data Analysis (2nd ed.). Meas. Interdiscip. Res. Perspect. 2019, 17, 160–167. [Google Scholar] [CrossRef]
- Edger, P.P.; Pires, J.C. Gene and genome duplications: The impact of dosage-sensitivity on the fate of nuclear genes. Chromosome Res. 2009, 17, 699–717. [Google Scholar] [CrossRef] [PubMed]
- Bawin, Y.; Ruttink, T.; Staelens, A.; Haegeman, A.; Stoffelen, P.; Mwanga Mwanga, J.-C.I.; Roldán-Ruiz, I.; Honnay, O.; Janssens, S.B. Phylogenomic analysis clarifies the evolutionary origin of Coffea arabica. J. Syst. Evol. 2021, 59, 953–963. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef] [PubMed]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).