Humic Lake Exhibits Higher Microbial Functional Gene Diversity and Weaker Gene Interaction Efficiency than a Common Alkaline Lake
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling and Water Properties Determination
2.2. DNA Extraction and GeoChip Analysis
2.3. Data Preprocessing
2.4. Network Construction
2.5. Statistical Analyses
3. Results
3.1. Environmental Characteristics
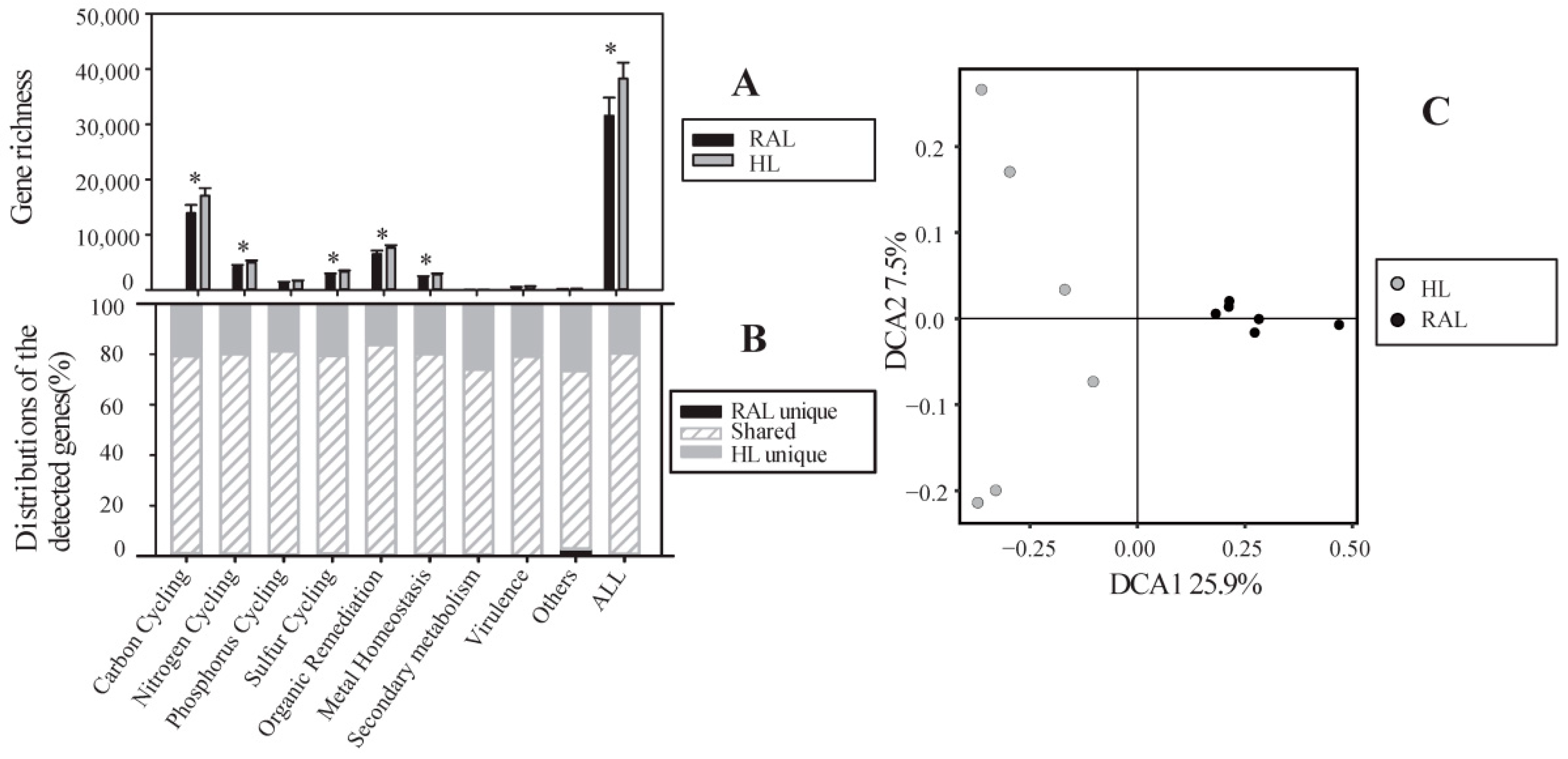
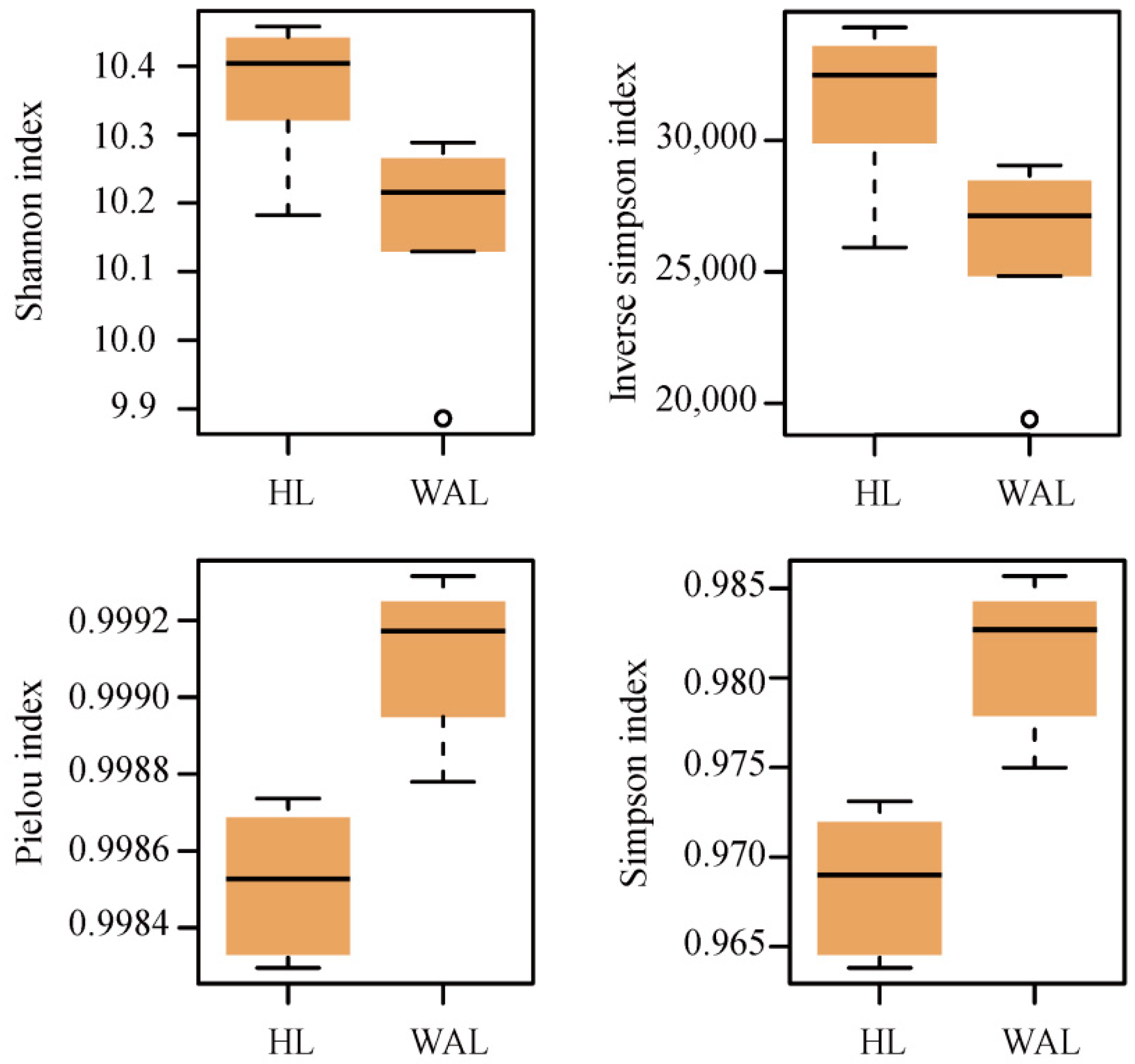
3.2. Overview of Microbial Functional Gene Composition and Structure
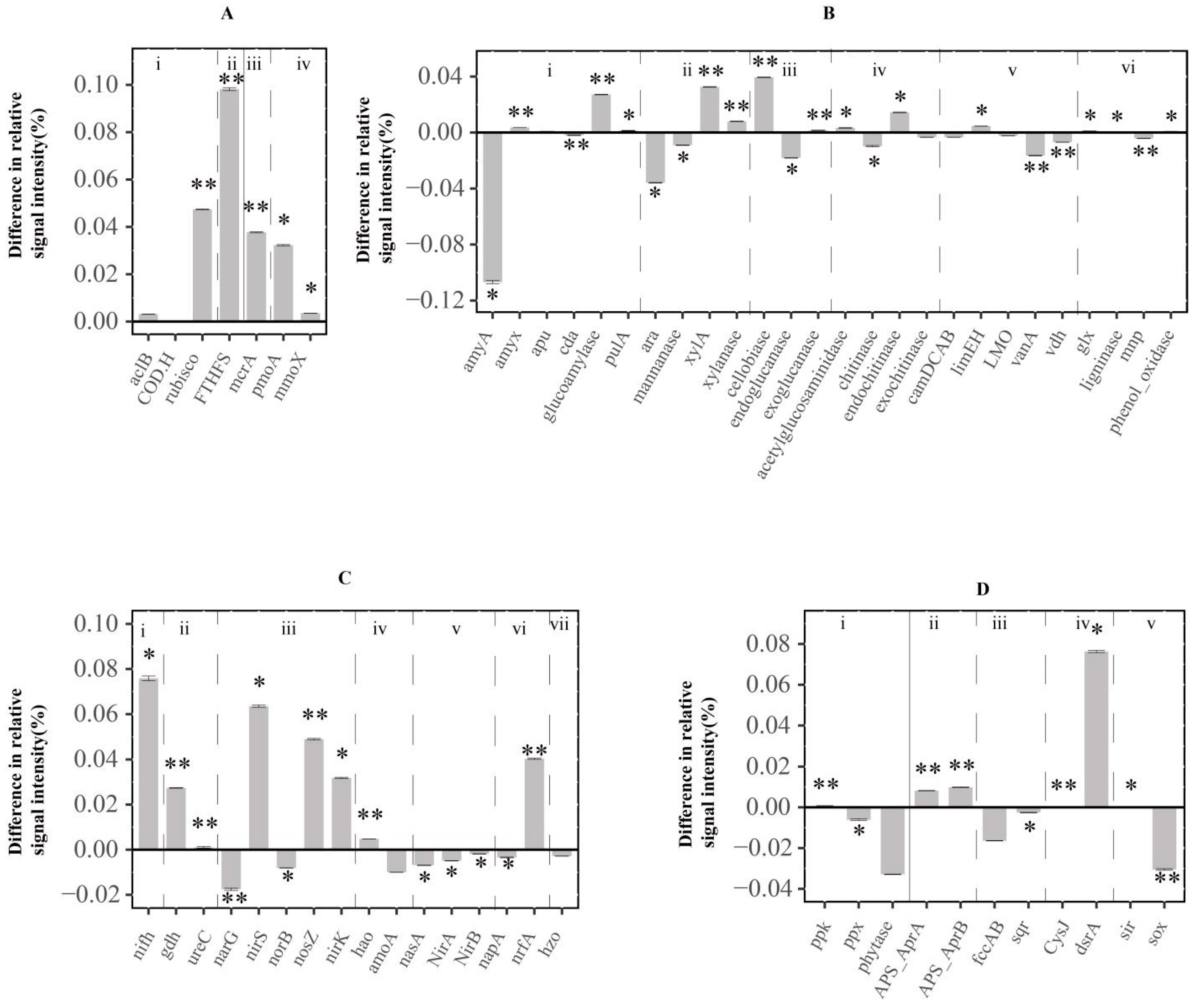
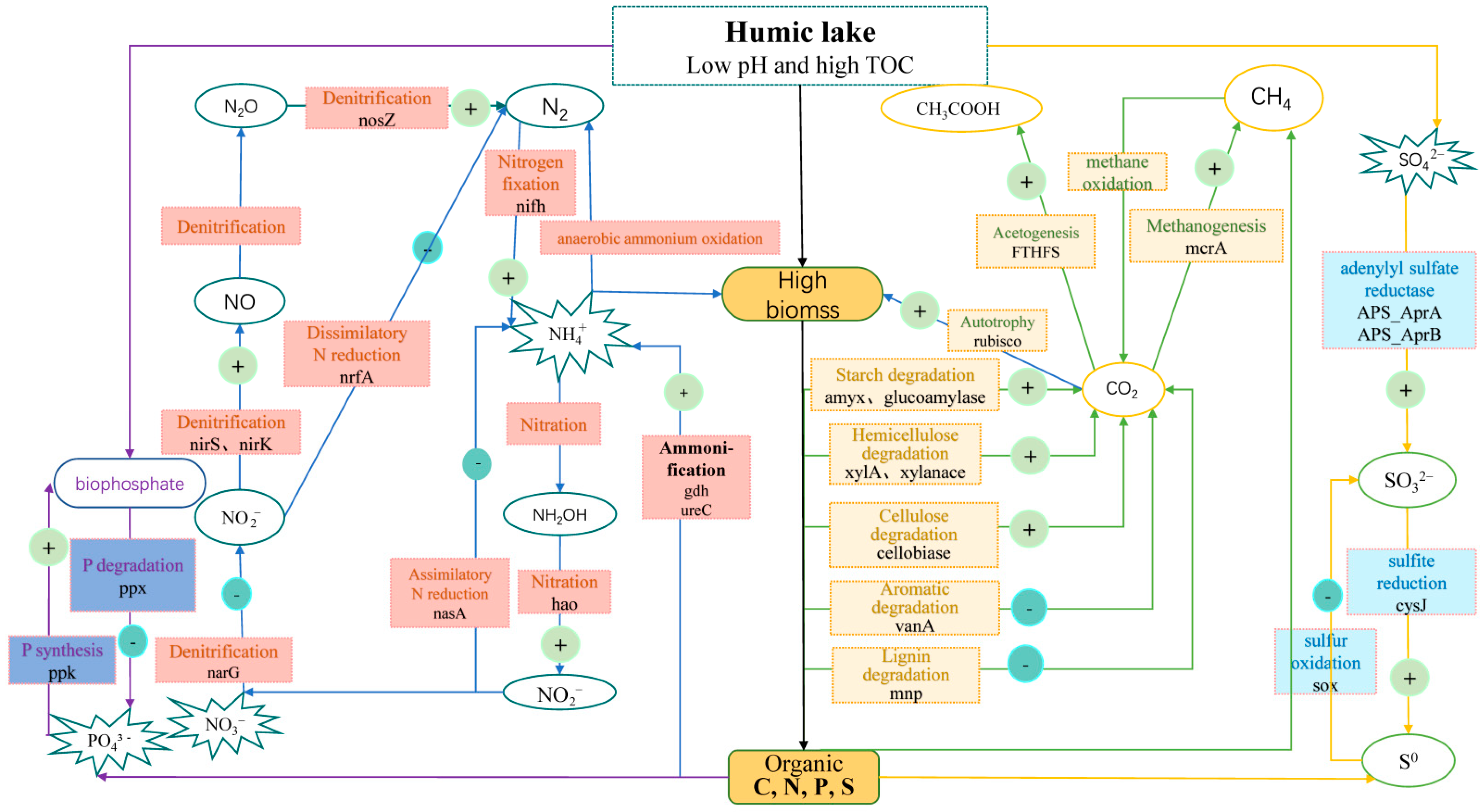
3.3. Functional Genes Involved in the C Cycle
3.4. Functional Genes Involved in N Cycle
3.5. Functional Genes Involved in Phosphorus and Sulfur Cycle
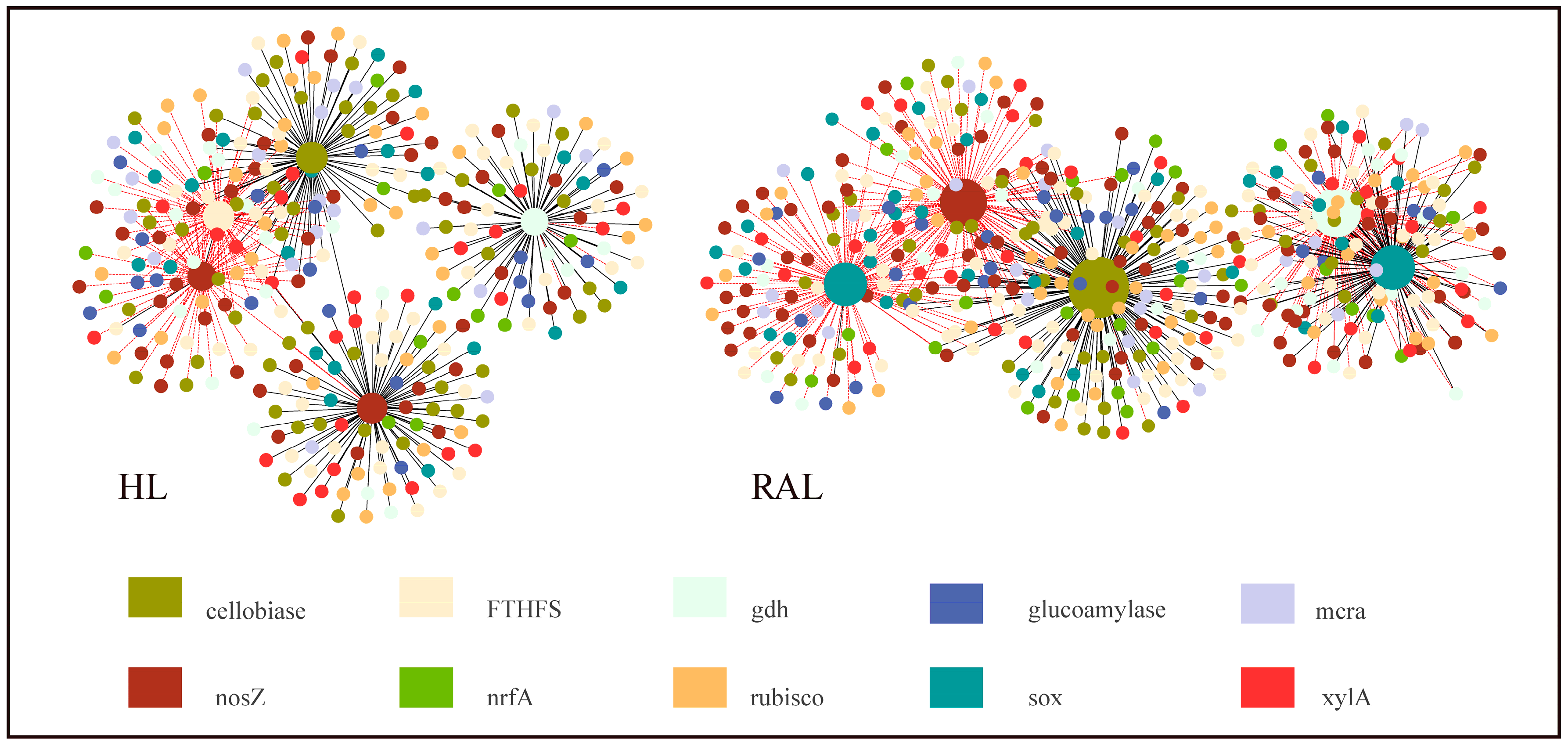
3.6. Microbial Functional Gene Network Analyses
4. Discussion
4.1. Differences in Microbial Functional Gene Compositions and Structures between the HL and RAL
4.2. Differences in Metabolic Potentials between the HL and the RAL
4.3. Potential Interactions between Functional Genes
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hessen, D.; Tranvik, L. Aquatic Humic Substances: Ecology and Biogeochemistry, 1st ed.; Springer: Berlin/Heidelberg, Germany, 1998; pp. 1–37. [Google Scholar]
- Ozoliņš, D.; Skuja, A.; Jēkabsone, J.; Kokorite, I.; Avotins, A.; Poikane, S. How to assess the ecological status of highly humic lakes? Development of a new method based on benthic invertebrates. Water 2021, 13, 223. [Google Scholar] [CrossRef]
- Song, X.Y.; Zhang, C.H.; Su, X.Y.; Zhu, L.J.; Wei, Z.M.; Zhao, Y. Characteristics of humic substance in lake sediments: The case of lakes in northeastern China. J. Hydrol. 2021, 603, 127079. [Google Scholar] [CrossRef]
- McDonald, S.; Bishop, A.G.; Prenzler, P.D.; Robards, K. Analytical chemistry of freshwater humic substances. Anal. Chim. Acta 2004, 527, 105–124. [Google Scholar] [CrossRef]
- Chupakov, A.V.; Chupakova, A.A.; Moreva, O.Y.; Shirokova, L.S.; Zabelina, S.A.; Vorobieva, T.Y.; Klimov, S.I.; Brovko, O.S.; Pokrovsky, O.S. Allochthonous and autochthonous carbon in deep, organic-rich and organic-poor lakes of the European Russian subarctic. Boreal. Environ. Res. 2017, 22, 213–230. [Google Scholar]
- Solomon, C.; Jones, S.; Weidel, B.; Buffam, I.; Fork, M.L.; Karlsson, J.; Larsen, S.; Lennon, J.T.; Read, J.S.; Sadro, S.; et al. Ecosystem consequences of changing inputs of terrestrial dissolved organic matter to lakes: Current knowledge and future challenges. Ecosystems 2015, 18, 376–389. [Google Scholar] [CrossRef]
- Kent, A.D.; Jones, S.E.; Yannarell, A.C.; Graham, J.M.; Lauster, G.H.; Kratz, T.K.; Triplett, E.W. Annual patterns in bacterioplankton community variability in a humic lake. Microb. Ecol. 2004, 48, 550–560. [Google Scholar] [CrossRef] [PubMed]
- Peura, S.; Buck, M.; Aalto, S.L.; Morales, S.E.; Nykänen, H.; Eiler, A. Novel autotrophic organisms contribute significantly to the internal carbon cycling potential of a boreal lake. mBio 2018, 9, e00916-18. [Google Scholar] [CrossRef]
- Newton, R.J.; Kent, A.D.; Triplett, E.W.; McMahon, K.D. Microbial community dynamics in a humic lake: Differential persistence of common freshwater phylotypes. Environ. Microbiol. 2006, 8, 956–970. [Google Scholar] [CrossRef]
- Rissanen, A.; Peura, S.; Mpamah, P.; Taipale, S.; Tiirola, M.; Biasi, C.; Mäki, A.; Nykänen, H. Vertical stratification of bacteria and archaea in sediments of a small boreal humic lake. FEMS Microbiol. Lett. 2019, 366, fnz044. [Google Scholar] [CrossRef]
- Rosenstock, B.; Simon, M. Consumption of dissolved amino acids and carbohydrates by limnetic bacterioplankton according to molecular weight fractions and proportions bound to humic matter. Microb. Ecol. 2003, 45, 433–443. [Google Scholar] [CrossRef]
- Kulikova, N.A.; Irina, V.P. Interactions between humic substances and microorganisms and their implications for nature-like bioremediation technologies. Molecules 2021, 26, 2706. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.L.; Ding, J.; Li, C.Y.; Yan, Z.Z.; He, J.Z.; Hu, H.W. Microbial functional attributes, rather than taxonomic attributes, drive top soil respiration, nitrification and denitrification processes. Sci. Total Environ. 2020, 734, 139479. [Google Scholar] [CrossRef] [PubMed]
- He, Z.; Deng, Y.; Van Nostrand, J.; Tu, Q.C.; Xu, M.Y.; Hemme, C.L.; Li, X.Y.; Wu, L.Y.; Gentry, T.J.; Yin, Y.F.; et al. GeoChip 3.0 as a high-throughput tool for analyzing microbial community composition, structure and functional activity. ISME J. 2010, 4, 1167–1179. [Google Scholar] [CrossRef] [PubMed]
- Low, A.; Ng, C.; He, J. Identification of antibiotic resistant bacteria community and a GeoChip based study of resistome in urban watersheds. Water Res. 2016, 106, 330–338. [Google Scholar] [CrossRef]
- Kang, S.; Van Nostrand, J.D.; Gough, H.L.; He, Z.; Hazen, T.C.; Stahl, D.A.; Zhou, J. Functional gene array-based analysis of microbial communities in heavy metals-contaminated lake sediments. FEMS Microbiol. Ecol. 2013, 86, 200–214. [Google Scholar] [CrossRef]
- Wang, X.; Xia, Y.; Wen, X.; Yang, Y.; Zhou, J. Microbial community functional structures in wastewater treatment plants as characterized by Geochip. PLoS ONE 2014, 9, e93422. [Google Scholar] [CrossRef]
- Kritzberg, E.S.; Langenheder, S.; Lindström, E.S. Influence of dissolved organic matter source on lake bacterioplankton structure and function–implications for seasonal dynamics of community composition. FEMS Microbiol. Ecol. 2006, 5, 406–417. [Google Scholar] [CrossRef][Green Version]
- Xu, M.; Zhang, Q.; Xia, C.; Zhong, Y.; Sun, G.; Guo, J.; Yuan, T.; Zhou, J.; He, Z. Elevated nitrate enriches microbial functional genes for potential bioremediation of complexly contaminated sediments. ISME J. 2014, 8, 1932–1944. [Google Scholar] [CrossRef]
- Ren, L.; Liu, Y.; Lauridsen, T.L.; Søndergaard, M.; Han, B.; Wang, J.; Jeppesen, E.; Zhou, J.; Wu, Q.L. Warming exacerbates the impact of nutrient enrichment on microbial functional potentials important to the nutrient cycling in shallow lake mesocosms. Limnol. Oceanogr. 2021, 66, 2481–2495. [Google Scholar] [CrossRef]
- Paver, S.F.; Kent, A.D. Temporal patterns in glycolate-utilizing bacterial community composition correlate with phytoplankton population dynamics in humic lakes. Microb. Ecol. 2010, 60, 406–418. [Google Scholar] [CrossRef]
- Lipczynska-Kochany, E. Humic substances, their microbial interactions and effects on biological transformations of organic pollutants in water and soil: A review. Chemosphere 2018, 202, 420–437. [Google Scholar] [CrossRef] [PubMed]
- Søndergaard, M.; Jeppesen, E.; Mortensen, E.; Dall, E.; Kristensen, P.; Sortkjær, O. Phytoplankton biomass reduction after planktivorous fish reduction in a shallow, eutrophic lake: A combined effect of reduced internal P-loading and increased zooplankton grazing. Hydrobiologia 1990, 200, 229–240. [Google Scholar] [CrossRef]
- Wu, Q.; Zwart, G.; Agterveld, M.; Liu, S.; Hahn, M. Submersed macrophytes play a key role in structuring bacterioplankton community composition in the large, shallow, subtropical Taihu Lake, China. Environ. Microbiol. 2007, 9, 2765–2774. [Google Scholar] [CrossRef] [PubMed]
- Yan, Q.; Bi, Y.; Deng, Y.; He, Z.; Wu, L.; Van Nostrand, J.; Shi, Z.; Li, J.; Wang, X.; Hu, Z.; et al. Impacts of the three gorges dam on microbial structure and potential function. Sci. Rep. 2015, 5, 8605. [Google Scholar] [CrossRef]
- Chan, Y.; Van Nostrand, J.; Zhou, J.; Pointing, S.; Farrell, R. Functional ecology of an antarctic dry valley. Proc. Natl. Acad. Sci. USA 2013, 110, 8990–8995. [Google Scholar] [CrossRef]
- Zhou, J.; Deng, Y.; Luo, F.; He, Z.; Tu, Q.; Zhi, X. Functional molecular ecological networks. mBio 2010, 1, e00169-10. [Google Scholar] [CrossRef]
- Luo, F.; Yang, Y.; Zhong, J.; Gao, H.; Khan, L.; Thompson, D.K.; Zhou, J. Constructing gene co-expression networks and predicting functions of unknown genes by random matrix theory. BMC Bioinform. 2007, 8, 299. [Google Scholar] [CrossRef]
- Deng, Y.; Jiang, Y.H.; Yang, Y.; He, Z.; Luo, F.; Zhou, J. Molecular ecological network analyses. BMC Bioinform. 2012, 13, 113. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; et al. vegan: Community Ecology Package. R Package Version 2.5-7. 2020. Available online: https://CRAN.R-project.org/package=vegan (accessed on 1 October 2021).
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2021; Available online: https://www.R-project.org/ (accessed on 1 October 2021).
- Alfreider, A.; Baumer, A.; Bogensperger, T.; Posch, T.; Salcher, M.M.; Summerer, M. CO2 assimilation strategies in stratified lakes: Diversity and distribution patterns of chemolithoautotrophs. Environ. Microbiol. 2017, 19, 2754–2768. [Google Scholar] [CrossRef]
- Zhang, L.; Fang, W.; Li, X.; Gao, G.; Jiang, J. Linking bacterial community shifts with changes in the dissolved organic matter pool in a eutrophic lake. Sci. Total Environ. 2020, 719, 137387. [Google Scholar] [CrossRef] [PubMed]
- Kraiem, K.; Wahab, M.A.; Kallali, H.; Fra-vazquez, A.; Pedrouso, A.; Mosquera-Corral, A.; Jedidi, N. Effects of short- and long-term exposures of humic acid on the Anammox activity and microbial community. Environ. Sci. Pollut. Res. 2019, 26, 19012–19024. [Google Scholar] [CrossRef]
- Zwart, G.; Crump, B.C.; Kamst-van Agterveld, M.P.; Hagen, F.; Han, S.K. Typical freshwater bacteria: An analysis of available 16S rRNA gene sequences from plankton of lakes and rivers. Aquat. Microb. Ecol. 2002, 28, 141–155. [Google Scholar] [CrossRef]
- Zhu, L.; Wei, Z.; Yang, T.; Zhao, X.; Dang, Q.; Chen, X.; Wu, J.; Zhao, Y. Core microorganisms promote the transformation of DOM fractions with different molecular weights to improve the stability during composting. Bioresour. Technol. 2020, 299, 122575. [Google Scholar] [CrossRef] [PubMed]
- Do, Y.S.; Schmidt, T.M.; Zahn, J.A.; Boyd, E.S.; Dispirito, A.A. Role of rhodobacter sp. strain ps9, a purple non-sulfur photosynthetic bacterium isolated from an anaerobic swine waste lagoon, in odor remediation. Appl. Environ. Microbiol. 2003, 69, 1710–1720. [Google Scholar] [CrossRef][Green Version]
- Sobek, S.; Söderbäck, B.; Karlsson, S.; Andersson, E.; Brunberg, A.K. A carbon budget of a small humic lake: An example of the importance of lakes for organic matter cycling in boreal catchments. Ambio 2006, 35, 469–475. [Google Scholar] [CrossRef]
- Lipczynska-Kochany, E. Effect of climate change on humic substances and associated impacts on the quality of surface water and groundwater: A review. Sci. Total Environ. 2018, 640, 1548–1565. [Google Scholar] [CrossRef]
- Assié, A.; Leisch, N.; Meier, D.V.; Gruber-Vodicka, H.; Tegetmeyer, H.E.; Meyerdierks, A.; Kleiner, M.; Hinzke, T.; Joye, S.; Saxton, M.; et al. Horizontal acquisition of a patchwork Calvin cycle by symbiotic and free-living Campylobacterota (formerly Epsilonproteobacteria). ISME J. 2020, 14, 104–122. [Google Scholar] [CrossRef]
- van Grinsven, S.; Oswald, K.; Wehrli, B.; Jegg, C.; Zopfi, J.; Lehmann, M.F.; Schubert, C.J. Methane oxidation in the waters of a humic-rich boreal lake stimulated by photosynthesis, nitrite, Fe(III) and humics. Biogeosciences 2021, 18, 3087–3101. [Google Scholar] [CrossRef]
- Smith, G.J.; Angle, J.C.; Solden, L.M.; Borton, M.A.; Morin, T.H.; Daly, R.A.; Johnston, M.D.; Stefanik, K.C.; Wolfe, R.; Gil, B.; et al. Members of the genus Methylobacter are inferred to account for the majority of aerobic methane oxidation in oxic soils from a freshwater wetland. mBio 2018, 9, e00815-18. [Google Scholar] [CrossRef]
- Zhang, Y.; Dong, J.; Yang, Z.; Zhang, S.; Wang, Y.-S. Phylogenetic diversity of nitrogen-fixing bacteria in mangrove sediments assessed by PCR–denaturing gradient gel electrophoresis. Arch. Microbiol. 2008, 190, 19–28. [Google Scholar] [CrossRef] [PubMed]
- Dong, S.; Li, M.; Chen, Y. Inherent humic substance promotes microbial denitrification of landfill leachate via shifting bacterial community, improving enzyme activity and up-regulating gene. Sci. Rep. 2017, 7, 12215. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y. Geochemical cycle and environmental effects of sulfur in lakes. In IOP Conference Series: Materials Science and Engineering, 5 July 2018; IOP Publishing: Bristol, UK, 2018; Volume 394, p. 052039. [Google Scholar]
- Kim, J.; Kim, T.H. Distribution of humic fluorescent dissolved organic matter in lake shihwa: The role of the redox condition. Estuaries Coasts 2020, 43, 578–588. [Google Scholar] [CrossRef]
- Malcolm, R.E.; Vaughan, D. Humic substances and phosphatase activities in plant tissues. Soil Biol. Biochem. 1979, 11, 253–259. [Google Scholar] [CrossRef]
- Pospíšil, F.; Hrubcová, M. The influence of humic acids on phytase activity isolated from wheat seeds. Biol. Plant. 1975, 17, 468–474. [Google Scholar] [CrossRef]
- Layeghifard, M.; Hwang, D.M.; Guttman, D.S. Disentangling interactions in the microbiome: A network perspective. Trends Microbiol. 2017, 25, 217–228. [Google Scholar] [CrossRef]
- Yuan, M.M.; Guo, X.; Wu, L.; Zhang, Y.A.; Xiao, N.; Ning, D.; Shi, Z.; Zhou, X.; Wu, L.; Yang, Y.; et al. Climate warming enhances microbial network complexity and stability. Nat. Clim. Chang. 2021, 11, 343–348. [Google Scholar] [CrossRef]
- Coyte, K.Z.; Schluter, J.; Foster, K.R. The ecology of the microbiome: Networks, competition, and stability. Science 2015, 350, 663–666. [Google Scholar] [CrossRef]
- Hoke, A.; Woodhouse, J.; Zoccarato, L.; McCarthy, V.; de Eyto, E.; Calderó-Pascual, M.; Geffroy, E.; Dillane, M.; Grossart, H.-P.; Jennings, E. Impacts of extreme weather events on bacterial community composition of a temperate humic Lake. Water 2020, 12, 2757. [Google Scholar] [CrossRef]
- Zhang, T.; Wei, N.; Wu, G. Autotrophic nitrogen removal and potential microbial interactions in anammox systems with different ammonia and organic carbon concentrations. J. Water Process Eng. 2020, 37, 101493. [Google Scholar] [CrossRef]
- Chiciudean, I.; Russo, G.; Bogdan, D.F.; Levei, E.A.; Faur, L.; Hillebrand-Voiculescu, A.; Teodora Moldovan, O.; Banciu, H.L. Competition-cooperation in the chemoautotrophic ecosystem of Movile Cave-first metagenomic approach on sediments. bioRxiv 2022. [Google Scholar] [CrossRef] [PubMed]
- Peura, S.; Nykänen, H.; Kankaala, P.; Eiler, A.; Tiirola, M.; Jones, R.I. Enhanced greenhouse gas emissions and changes in plankton communities following an experimental increase in organic carbon loading to a humic lake. Biogeochemistry 2014, 118, 177–194. [Google Scholar] [CrossRef]
| Lake | pH | TOC (mg·L−1) | TP (mg·L−1) | TFe (mg·L−1) | TN (mg·L−1) | NOX− (mg·L−1) | Conductivity (μS·cm−1) | Area (km2) |
|---|---|---|---|---|---|---|---|---|
| HL | 5.16 | 31.9 | 0.17 | 0.87 | 2.04 | 0.02 | 297 | 0.03 |
| RAL | 8.3 | 4.01 | 0.09 | 0.21 | 0.75 | 0 | 274 | 0.15 |
| Empirical Networks | Random Networks ‡ | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lake | No. of Original Genes * | Similarity Threshold (St) | Network Size (n) † | No. of Links | R Square of Power-Law | Average Connectivity (avgK) | Harmonic Geodesic Distance (HD) | Average Clustering Coefficient (avgCC) | Modularity (No. of Modules) | Harmonic Geodesic Distance (HD ± SD) | Average Clustering Coefficient (avgCC ± SD) | Average Modularity (M ± SD) |
| HL | 3275 | 0.99 | 2005 | 5551 | 0.88 | 5.537 | 4.96 | 0.21 | 0.842 (279) | 3.668 ± 0.012 | 0.019 ± 0.002 | 0.396 ± 0.003 |
| RAL | 2584 | 0.99 | 1703 | 4667 | 0.852 | 5.481 | 4.585 | 0.297 | 0.834 (172) | 3.401 ± 0.014 | 0.052 ± 0.003 | 0.396 ± 0.003 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
He, D.; Liu, Y.; Wu, Q.L.; Peng, Y.; Ren, L. Humic Lake Exhibits Higher Microbial Functional Gene Diversity and Weaker Gene Interaction Efficiency than a Common Alkaline Lake. Biology 2022, 11, 1448. https://doi.org/10.3390/biology11101448
He D, Liu Y, Wu QL, Peng Y, Ren L. Humic Lake Exhibits Higher Microbial Functional Gene Diversity and Weaker Gene Interaction Efficiency than a Common Alkaline Lake. Biology. 2022; 11(10):1448. https://doi.org/10.3390/biology11101448
Chicago/Turabian StyleHe, Dan, Yuanyuan Liu, Qinglong L. Wu, Yuyang Peng, and Lijuan Ren. 2022. "Humic Lake Exhibits Higher Microbial Functional Gene Diversity and Weaker Gene Interaction Efficiency than a Common Alkaline Lake" Biology 11, no. 10: 1448. https://doi.org/10.3390/biology11101448
APA StyleHe, D., Liu, Y., Wu, Q. L., Peng, Y., & Ren, L. (2022). Humic Lake Exhibits Higher Microbial Functional Gene Diversity and Weaker Gene Interaction Efficiency than a Common Alkaline Lake. Biology, 11(10), 1448. https://doi.org/10.3390/biology11101448







