Exaptation of Retroviral Syncytin for Development of Syncytialized Placenta, Its Limited Homology to the SARS-CoV-2 Spike Protein and Arguments against Disturbing Narrative in the Context of COVID-19 Vaccination
Simple Summary
Abstract
1. Introduction
2. Types of Mammalian Placenta
3. Exaptation of Retroviral Genes for Placental Function
4. Endogenous Retroviral Gene Function in the Placenta
5. Other Syncytins
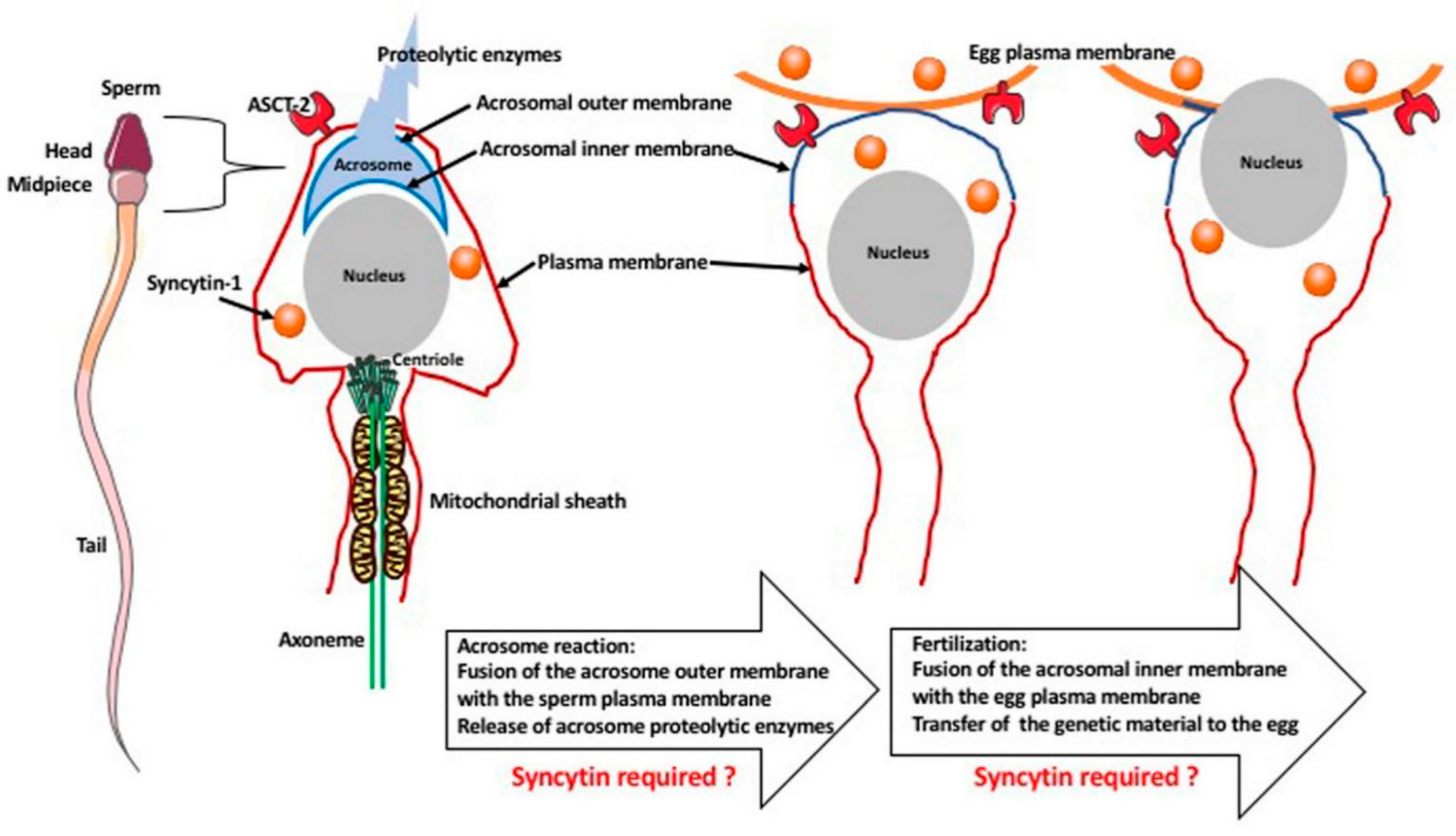
6. Hypothetical Role of Syncytin-1 in Fertilization
7. Syncytin Role in Cancer Cell Fusion
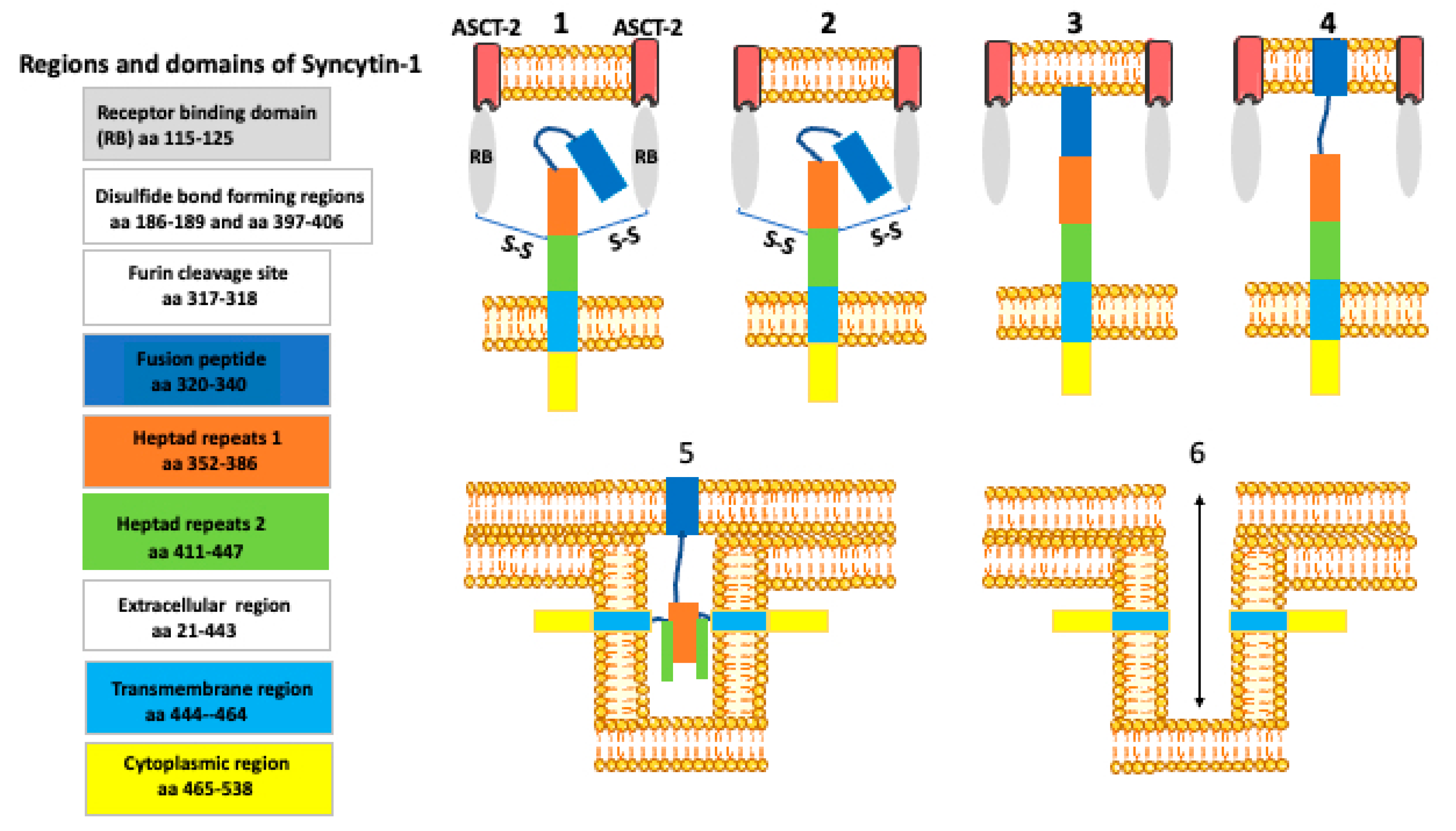
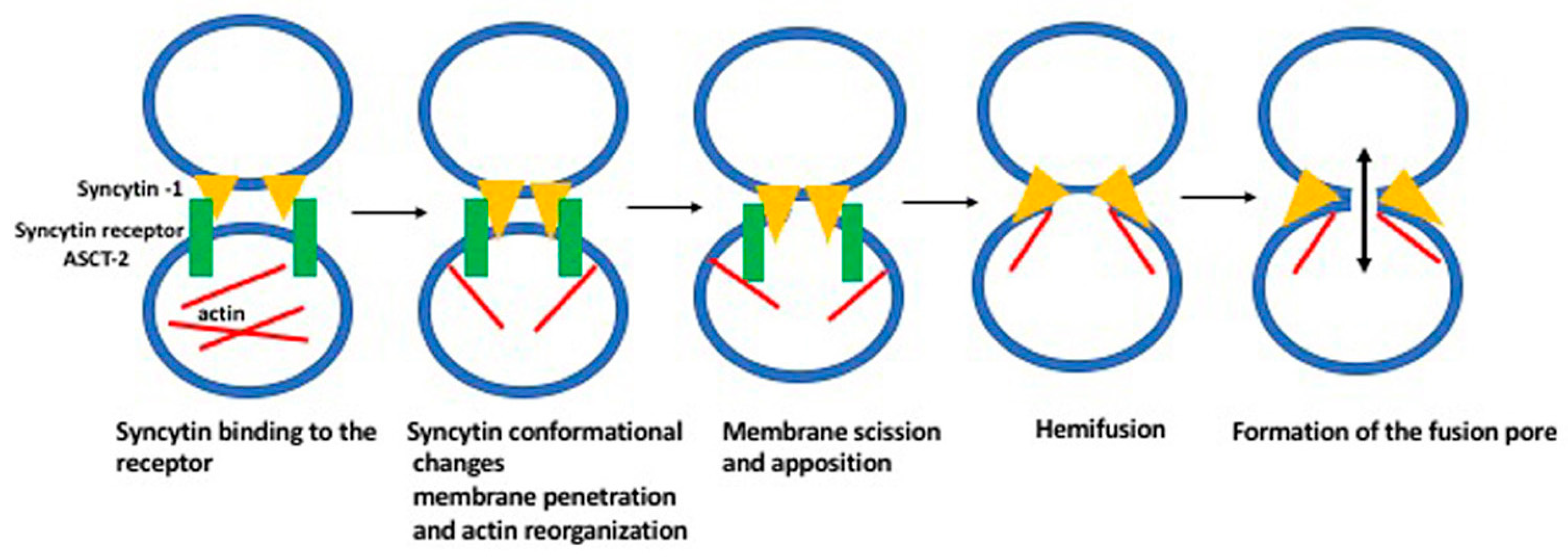
8. How Syncytin Facilitates Cell Membrane Fusion?
9. Syncytin Functions beyond Cell Fusion—The Immunomodulatory Functions
10. The Limited Similarity of Syncytin to SARS-CoV-2 Spike Protein
11. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Roberts, R.M.; Green, J.A.; Schulz, L.C. The Evolution of the Placenta. Reproduction 2016, 152, R179–R189. [Google Scholar] [CrossRef] [PubMed]
- Blackburn, D.G. Evolution of vertebrate viviparity and specializations for fetal nutrition: A quantitative and qualitative analysis. J. Morphol. 2015, 276, 961–990. [Google Scholar] [CrossRef]
- Wourms, J.P.; Lombardi, J. Reflections on the Evolution of Piscine Viviparity. Am. Zool. 1992, 32, 276–293. [Google Scholar] [CrossRef]
- Hamlett, W.C.; Eulitt, A.M.; Jarrell, R.L.; Kelly, M.A. Uterogestation and Placentation in Elasmobranchs. J. Exp. Zool. 1993, 266, 347–367. [Google Scholar] [CrossRef]
- Carcupino, M.; Baldacci, A.; Mazzini, M.; Franzoi, P. Functional significance of the male brood pouch in the reproductive strategies of pipefishes and seahorses: A morphological and ultrastructural comparative study on three anatomically different pouches. J. Fish Biol. 2002, 61, 1465–1480. [Google Scholar] [CrossRef]
- Savage, J.M. The Amphibians and Reptiles of Costa Rica: A Herpetofauna between Two Continents, between Two Seas; University of Chicago Press: Chicago, IL, USA; London, UK, 2002; ISBN 0-226-73537-0. [Google Scholar]
- Del Pino, E.M. The extraordinary biology and development of marsupial frogs (Hemiphractidae) in comparison with fish, mammals, birds, amphibians and other animals. Mech. Dev. 2018, 154, 2–11. [Google Scholar] [CrossRef] [PubMed]
- Del Pino, E.M. Embryogenesis of Marsupial Frogs (Hemiphractidae), and the Changes that Accompany Terrestrial Development in Frogs. Results Probl. Cell Differ. 2019, 68, 379–418. [Google Scholar] [CrossRef]
- Stewart, J.R. Placental specializations in lecithotrophic viviparous squamate reptiles. J. Exp. Zool. Part B Mol. Dev. Evol. 2015, 324, 549–561. [Google Scholar] [CrossRef]
- Blackburn, D.G.; Flemming, A.F. Invasive implantation and intimate placental associations in a placentotrophic african lizard, Trachylepis ivensi (scincidae). J. Morphol. 2012, 273, 137–159. [Google Scholar] [CrossRef]
- Thompson, M.B.; Speake, B.K. Energy and nutrient utilisation by embryonic reptiles. Comp. Biochem. Physiol. Part A—Mol. Integr. Physiol. 2002, 133, 529–538. [Google Scholar] [CrossRef]
- Enders, A.C. Anatomy of the placenta and its relationship to function. Mead Johnson Symp. Perinat. Dev. Med. 1981, 3–7. Available online: https://europepmc.org/article/med/6210810 (accessed on 18 March 2021).
- Huppertz, B. The anatomy of the normal placenta. J. Clin. Pathol. 2008, 61, 1296–1302. [Google Scholar] [CrossRef]
- Renfree, M.B. Review: Marsupials: Placental mammals with a difference. Placenta 2010, 31, S21–S26. [Google Scholar] [CrossRef]
- Renfree, M.B. Implantation and Placentation Vol 2, Reproduction in Mammals: Embryonic and Fetal Development; Austin, C.R., Short, R.V., Eds.; Cambridge University Press: Cambridge, UK, 1982; pp. 26–69. [Google Scholar]
- Wooding, F.B. Current topic: The synepitheliochorial placenta of ruminants: Binucleate cell fusions and hormone production. Placenta 1992, 13, 101–113. [Google Scholar] [CrossRef]
- Enders, A.C.; Carter, A.M. What can comparative studies of placental structure tell us?–A review. Placenta 2004, 25 (Suppl. A), S3–S9. [Google Scholar] [CrossRef] [PubMed]
- Funk, M. Identification and Characterization of Two Novel Syncytin-Like Retroviral Envelope Genes, Captured for a Possible role in the Atypical Structure of the Hyena Placenta and in the Emergence of the Non-Mammalian Mabuya Lizard Placenta a. Virology; 〈NNT: 2018SACLS106〉; Université Paris Saclay (COmUE): Gif-sur-Yvette, France, 2018. [Google Scholar]
- Funk, M.; Cornelis, G.; Vernochet, C.; Heidmann, O.; Dupressoir, A.; Conley, A.; Glickman, S.; Heidmann, T. Capture of a Hyena-Specific Retroviral Envelope Gene with Placental Expression Associated in Evolution with the Unique Emergence among Carnivorans of Hemochorial Placentation in Hyaenidae. J. Virol. 2019, 93, e01811-18. [Google Scholar] [CrossRef]
- Wildman, D.E.; Chen, C.; Erez, O.; Grossman, L.I.; Goodman, M.; Romero, R. Evolution of the mammalian placenta revealed by phylogenetic analysis. Proc. Natl. Acad. Sci. USA 2006, 103, 3203–3208. [Google Scholar] [CrossRef]
- Renegar, R.H.; Bazer, F.W.; Roberts, R.M. Placental Transport and Distribution of Uteroferrin in the Fetal. Pig. Biol. Reprod. 1982, 27, 1247–1260. [Google Scholar] [CrossRef]
- Lavialle, C.; Cornelis, G.; Dupressoir, A.; Esnault, C.; Heidmann, O.; Vernochet, C.; Heidmann, T. Paleovirology of ‘syncytins’, retroviral env genes exapted for a role in placentation. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2013, 368, 20120507. [Google Scholar] [CrossRef]
- Tollis, M.; Boissinot, S. The evolutionary dynamics of transposable elements in eukaryote genomes. Genome Dyn. 2010, 7, 68–91. [Google Scholar] [CrossRef]
- Makalowski, W.; Pande, A.; Gotea, V.; Makalowska, I. Transposable elements and their identification. Methods Mol. Biol. 2012, 855, 337–359. [Google Scholar] [CrossRef]
- Kurth, R.; Bannert, N. (Eds.) Retroviruses: Molecular Biology, Genomics and Pathogenesis; Horizon Scientific: Summerville, SC, USA, 2010; ISBN 978-1-904455-55-4. [Google Scholar]
- Coffin, J.M.; Fan, H. The Discovery of Reverse Transcriptase. Annu. Rev. Virol. 2016, 3, 29–51. [Google Scholar] [CrossRef]
- Andrake, M.D.; Skalka, A.M. Retroviral Integrase: Then and Now. Annu. Rev. Virol. 2015, 2, 241–264. [Google Scholar] [CrossRef]
- Menéndez-Arias, L. Special Issue: Retroviral Enzyme. Viruses 2010, 2, 1181–1184. [Google Scholar] [CrossRef]
- de Parseval, N.; Lazar, V.; Casella, J.F.; Benit, L.; Heidmann, T. Survey of human genes of retroviral origin: Identification and transcriptome of the genes with coding capacity for complete envelope proteins. J. Virol. 2003, 77, 10414–10422. [Google Scholar] [CrossRef]
- Villesen, P.; Aagaard, L.; Wiuf, C.; Pedersen, F.S. Identification of endogenous retroviral reading frames in the human genome. Retrovirology 2004, 1, 32. [Google Scholar] [CrossRef]
- Blond, J.L.; Lavillette, D.; Cheynet, V.; Bouton, O.; Oriol, G.; Chapel-Fernandes, S.; Mandrand, B.; Mallet, F.; Cosset, F.L. An envelope glycoprotein of the human endogenous retrovirus HERV-W is expressed in the human placenta and fuses cells expressing the type D mammalian retrovirus receptor. J. Virol. 2000, 74, 3321–3329. [Google Scholar] [CrossRef]
- Mi, S.; Lee, X.; Li, X.P.; Veldman, G.M.; Finnerty, H.; Racie, L.; LaVallie, E.; Tang, X.Y.; Edouard, P.; Howes, S.; et al. Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature 2000, 403, 785–789. [Google Scholar] [CrossRef]
- Blaise, S.; de Parseval, N.; Benit, L.; Heidmann, T. Genomewide screening for fusogenic human endogenous retrovirus envelopes identifies syncytin 2, a gene conserved on primate evolution. Proc. Natl. Acad. Sci. USA 2003, 100, 13013–13018. [Google Scholar] [CrossRef]
- Gong, R.; Peng, X.; Kang, S.; Feng, H.; Huang, J.; Zhang, W.; Lin, D.; Tien, P.; Xiao, G. Structural characterization of the fusion core in syncytin, envelope protein of human endogenous retrovirus family W. Biochem. Biophys. Res. Commun. 2005, 331, 1193–1200. [Google Scholar] [CrossRef]
- Esnault, C.; Priet, S.; Ribet, D.; Vernochet, C.; Bruls, T.; Lavialle, C.; Weissenbach, J.; Heidmann, T. A placenta-specific receptor for the fusogenic, endogenous retrovirus-derived, human syncytin-2. Proc. Natl Acad. Sci. USA 2008, 105, 17532–17537. [Google Scholar] [CrossRef]
- Cohen, M. The Dark Side of Cell Fusion. Int. J. Mol. Sci. 2016, 17, 638. [Google Scholar] [CrossRef]
- de Parseval, N.; Diop, G.; Blaise, S.; Helle, F.; Vasilescu, A.; Matsuda, F.; Heidmann, T. Comprehensive search for intra- and inter-specific sequence polymorphisms among coding envelope genes of retroviral origin found in the human genome: Genes and pseudogenes. BMC Genom. 2005, 6, 117. [Google Scholar] [CrossRef]
- Dupressoir, A.; Marceau, G.; Vernochet, C.; Benit, L.; Kanellopoulos, C.; Sapin, V.; Heidmann, T. Syncytin-A and syncytin-B, two fusogenic placenta-specific murine envelope genes of retroviral origin conserved in Muridae. Proc. Natl. Acad. Sci. USA 2005, 102, 725–730. [Google Scholar] [CrossRef] [PubMed]
- Coudert, A.E.; Redelsperger, F.; Chabbi-Achengli, Y.; Vernochet, C.; Marty, C.; Decrouy, X.; Heidmann, T.; de Vernejoul, M.C.; Dupressoir, A. Role of the captured retroviral envelope syncytin-B gene in the fusion of osteoclast and giant cell precursors and in bone resorption, analyzed ex vivo and in vivo in syncytin-B knockout mice. Bone Rep. 2019, 11, 100214. [Google Scholar] [CrossRef]
- Vernochet, C.; Redelsperger, F.; Harper, F.; Souquere, S.; Catzeflis, F.; Pierron, G.; Nevo, E.; Heidmann, T.; Dupressoir, A. The captured retroviral envelope syncytin-A and syncytin-B genes are conserved in the Spalacidae together with hemotrichorial placentation. Biol. Reprod. 2014, 91, 148. [Google Scholar] [CrossRef]
- Dupressoir, A.; Vernochet, C.; Bawa, O.; Harper, F.; Pierron, G.; Opolon, P.; Heidmann, T. Syncytin-A knockout mice demonstrate the critical role in placentation of a fusogenic, endogenous retrovirus-derived, envelope gene. Proc. Natl. Acad. Sci. USA 2009, 106, 12127–12132. [Google Scholar] [CrossRef]
- Dupressoir, A.; Vernochet, C.; Harper, F.; Guégan, J.; Dessen, P.; Pierron, G.; Heidmann, T. A pair of co-opted retroviral envelope syncytin genes is required for formation of the two-layered murine placental syncytiotrophoblast. Proc. Natl. Acad. Sci. USA 2011, 108, E1164–E1173. [Google Scholar] [CrossRef]
- Cornelis, G.; Heidmann, O.; Bernard-Stoecklin, S.; Véron, G.; Reynaud, K.; Mulot, B.; Dupressoír, A.; Heidmann, T. Identification of syncytin-car-1, an endogenous retroviral envelope gene involved in placentation and conserved in Carnivora: A syncytin in a new superorder of placental mammals. Retrovirology 2011, 8, P13. [Google Scholar] [CrossRef]
- Cornelis, G.; Heidmann, O.; Degrelle, S.A.; Vernochet, C.; Lavialle, C.; Letzelter, C.; Bernard-Stoecklin, S.; Hassanin, A.; Mulot, B.; Guillomot, M.; et al. A captured syncytin in Ruminantia. PANAS 2013, 110, E828–E837. [Google Scholar] [CrossRef]
- Heidmann, O.; Vernochet, C.; Dupressoir, A.; Heidmann, T. Identification of an endogenous retroviral envelope gene with fusogenic activity and placenta-specific expression in the rabbit: A new “syncytin” in a third order of mammals. Retrovirology 2009, 6, 107. [Google Scholar] [CrossRef]
- Redelsperger, F.; Cornelis, G.; Vernochet, C.; Tennant, B.C.; Catzeflis, F.; Mulot, B.; Heidmann, O.; Heidmann, T.; Dupressoir, A. Capture of syncytin-Mar1, a Fusogenic Endogenous Retroviral Envelope Gene Involved in Placentation in the Rodentia Squirrel-Related Clade. J. Virol. 2014, 88, 7915–7928. [Google Scholar] [CrossRef] [PubMed]
- Bjerregaard, B.; Lemmen, J.G.; Petersen, M.R.; Østrup, E.; Iversen, L.H.; Almstrup, K.; Larsson, L.I.; Ziebe, S. Syncytin-1 and its receptor is present in human gametes. J. Assist. Reprod. Genet. 2014, 31, 533–539. [Google Scholar] [CrossRef]
- Soygur, B.; Sati, L. The role of syncytins in human reproduction and reproductive organ cancers. Reproduction 2016, 152, R167–R178. [Google Scholar] [CrossRef]
- Jean, C.; Haghighirad, F.; Zhu, Y.; Chalbi, M.; Ziyyat, A.; Rubinstein, E.; Gourier, C.; Yip, P.; Wolf, J.P.; Lee, J.E.; et al. JUNO, the receptor of sperm IZUMO1, is expressed by the human oocyte and is essential for human fertilisation. Hum. Reprod. 2019, 34, 118–126. [Google Scholar] [CrossRef]
- Clark, T. HAP2/GCS1: Mounting evidence of our true biological EVE? PLoS Biol. 2018, 20, e3000007. [Google Scholar] [CrossRef]
- Fedry, J.; Forcina, J.; Legrand, P.; Péhau-Arnaudet, G.; Haouz, A.; Johnson, M.; Rey, F.A.; Krey, T. Evolutionary diversificationof the HAP2 membrane insertion motifs to drive gamete fusion across eukaryotes. PLoS Biol. 2018, 16, e2006357. [Google Scholar] [CrossRef] [PubMed]
- Larsson, L.-J.; Bjerregaard, B.; Wulf-Andersen, L.; Talts, J.F. Syncytin and Cancer Cell Fusions. Sci. World J. 2007, 7, 1193–1197. [Google Scholar] [CrossRef] [PubMed]
- Larsson, L.I.; Holck, S.; Christensen, I.J. Prognostic role of syncytin expression in breast cancer. Hum. Pathol. 2007, 38, 726–731. [Google Scholar] [CrossRef]
- Brukman, N.G.; Uygur, B.; Podbilewicz, B.; Chemomordik, L.V. How cell fuse. J. Cell Biol. 2019, 218, 1436–1451. [Google Scholar] [CrossRef]
- Sapir, A.; Avinoam, O.; Podbilewicz, B.; Chernomordik, L.V. Viral and developmental cell fusion mechanisms: Conservation and divergence. Dev. Cell. 2008, 14, 11–21. [Google Scholar] [CrossRef]
- Pötgens, A.J.; Drewlo, S.; Kokozidou, M.; Kaufmann, P. Syncytin: The major regulator of trophoblast fusion? Recent developments and hypotheses on its action. Hum. Reprod. Update 2004, 10, 487–496. [Google Scholar] [CrossRef]
- Zhou, X.; Platt, J.L. Molecular and cellular mechanisms of mammalian cell fusion. Adv. Exp. Med. Biol. 2011, 713, 33–64. [Google Scholar] [CrossRef]
- Lindau, M.; de Toledo, G.A. The fusion pore, Biochimica et Biophysica Acta (BBA). Mol. Cell Res. 2003, 641, 167–173. [Google Scholar] [CrossRef]
- Slokar, G.; Hasler, G. Human Endogenous Retroviruses as Pathogenic Factors in the Development of Schizophrenia. Front. Psychiatry 2015, 6, 183. [Google Scholar] [CrossRef]
- Karlsson, H.; Schröder, J.; Bachmann, S.; Bottmer, C.; Yolken, R.H. HERV-W-related RNA detected in plasma from individuals with recent-onset schizophrenia or schizoaffective disorder. Mol. Psychiatry 2004, 9, 12–13. [Google Scholar] [CrossRef]
- Laufer, G.; Mayer, J.; Mueller, B.F.; Mueller-Lantzsch, N.; Ruprecht, K. Analysis of transcribed human endogenous retrovirus W env loci clarifies the origin of multiple sclerosis-associated retrovirus env sequences. Retrovirology 2009, 6, 37. [Google Scholar] [CrossRef]
- Wang, X.; Liu, Z.; Wang, P.; Li, S.; Zeng, J.; Tu, X.; Yan, Q.; Xiao, Z.; Pan, M.; Zhu, F. Syncytin-1, an endogenous retroviral protein, triggers the activation of CRP via TLR3 signal cascade in glial cells. Brain Behav. Immun. 2018, 67, 324–334. [Google Scholar] [CrossRef]
- Wang, X.; Huang, J.; Zhu, F. Human Endogenous Retroviral Envelope Protein Syncytin-1 and Inflammatory Abnormalities in Neuropsychological Diseases. Front. Psychiatry 2018, 9, 422. [Google Scholar] [CrossRef]
- Ponferrada, V.G.; Mauck, B.S.; Wooley, D.P. The envelope glycoprotein of human endogenous retrovirus HERV-W induces cellular resistance to spleen necrosis virus. Arch. Virol. 2003, 148, 659–675. [Google Scholar] [CrossRef] [PubMed]
- Blond, J.L.; Besème, F.; Duret, L.; Bouton, O.; Bedin, F.; Perron, H.; Mandrand, B.; Mallet, F. Molecular characterization and placental expression of HERV-W, a new human endogenous retrovirus family. J. Virol. 1999, 73, 1175–1185. [Google Scholar] [CrossRef]
- Lokossou, A.G.; Toudic, C.; Nguyen, P.T.; Elisseeff, X.; Vargas, A.; Rassart, É.; Lafond, J.; Leduc, L.; Bourgault, S.; Gilbert, C.; et al. Endogenous retrovirus-encoded Syncytin-2 contributes to exosome-mediated immunosuppression of T cells†. Biol. Reprod. 2020, 102, 185–198. [Google Scholar] [CrossRef]
- Koirala, A.; Joo, Y.J.; Khatami, A.; Chiu, C.; Britton, P.N. Vaccines for COVID-19: The current state of play. Paediatr. Respir. Rev. 2020, 35, 43–49. [Google Scholar] [CrossRef]
- Dai, L.; Gao, G.F. Viral targets for vaccines against COVID-19. Nat. Rev. Immunol. 2021, 21, 73–82. [Google Scholar] [CrossRef]
- Yan, R.; Zhang, Y.; Li, Y.; Xia, L.; Guo, Y.; Zhou, Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science 2020, 367, 1444–1448. [Google Scholar] [CrossRef]
- Kloc, M.; Ghobrial, R.M.; Kubiak, J.Z. How nicotine can inhibit cytokine storm in the lungs and prevent or lessen the severity of COVID-19 infection? Immunol. Lett. 2020, 224, 28–29. [Google Scholar] [CrossRef]
- Kloc, M.; Ghobrial, R.M.; Kubiak, J.Z. SARS-CoV-2 subversion of the antiviral interferon alfa-response of lung macrophages. J. Immunol. Sci. 2020, 4, 13–16. [Google Scholar] [CrossRef]
- Kloc, M.; Ghobrial, R.M.; Kubiak, J.Z. The role of genetic sex and mitochondria in response to COVID-19 infection. Int. Arch. Allergy Immunol. 2020, 181, 629–634. [Google Scholar] [CrossRef] [PubMed]
- Available online: https://www.uniprot.org/uniprot/Q9UQF0#sequences (accessed on 18 March 2021).
- Available online: https://www.uniprot.org/uniprot/P0DTC2#sequences (accessed on 18 March 2021).
- Herman, R.A.; Song, P.; Thirumalaiswamysekhar, A. Value of eight-amino-acid matches in predicting the allergenicity status of proteins: An empirical bioinformatic investigation. Clin. Mol. Allergy 2009, 7, 9. [Google Scholar] [CrossRef] [PubMed]
- Heath, P.T.; Le Doare, K.; Khalil, A. Inclusion of pregnant women in COVID-19 vaccine development. Lancet Infect. Dis. 2020, 20, 1007–1008. [Google Scholar] [CrossRef]
- Stafford, I.A.; Parchem, J.G.; Sibai, B.M. The coronavirus disease 2019 vaccine in pregnancy: Risks, benefits, and recommendations. Am. J. Obstet. Gynecol. 2021. [Google Scholar] [CrossRef] [PubMed]
- Available online: https://www.uchicagomedicine.org/forefront/coronavirus-disease-covid-19/mrna-covid-19-vaccine-pregnancy-breastfeeding (accessed on 18 March 2021).
- Available online: https://www.cdc.gov/coronavirus/2019-ncov/vaccines/recommendations/pregnancy.html (accessed on 18 March 2021).
- Available online: https://www.acog.org/covid-19/covid-19-vaccines-and-pregnancy-conversation-guide-for-clinicians (accessed on 18 March 2021).

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kloc, M.; Uosef, A.; Kubiak, J.Z.; Ghobrial, R.M. Exaptation of Retroviral Syncytin for Development of Syncytialized Placenta, Its Limited Homology to the SARS-CoV-2 Spike Protein and Arguments against Disturbing Narrative in the Context of COVID-19 Vaccination. Biology 2021, 10, 238. https://doi.org/10.3390/biology10030238
Kloc M, Uosef A, Kubiak JZ, Ghobrial RM. Exaptation of Retroviral Syncytin for Development of Syncytialized Placenta, Its Limited Homology to the SARS-CoV-2 Spike Protein and Arguments against Disturbing Narrative in the Context of COVID-19 Vaccination. Biology. 2021; 10(3):238. https://doi.org/10.3390/biology10030238
Chicago/Turabian StyleKloc, Malgorzata, Ahmed Uosef, Jacek Z. Kubiak, and Rafik M. Ghobrial. 2021. "Exaptation of Retroviral Syncytin for Development of Syncytialized Placenta, Its Limited Homology to the SARS-CoV-2 Spike Protein and Arguments against Disturbing Narrative in the Context of COVID-19 Vaccination" Biology 10, no. 3: 238. https://doi.org/10.3390/biology10030238
APA StyleKloc, M., Uosef, A., Kubiak, J. Z., & Ghobrial, R. M. (2021). Exaptation of Retroviral Syncytin for Development of Syncytialized Placenta, Its Limited Homology to the SARS-CoV-2 Spike Protein and Arguments against Disturbing Narrative in the Context of COVID-19 Vaccination. Biology, 10(3), 238. https://doi.org/10.3390/biology10030238







