Production of d-Tagatose by Whole-Cell Conversion of Recombinant Bacillus subtilis in the Absence of Antibiotics
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains, Plasmids, and Culture Conditions
2.2. Construction of Expression Systems
2.3. Construction of Recombinant Vectors
2.4. Construction of Expression Systems
2.5. Enzyme Activity Assay
2.6. d-tagatose Production by Whole-Cell Bioconversion
3. Results
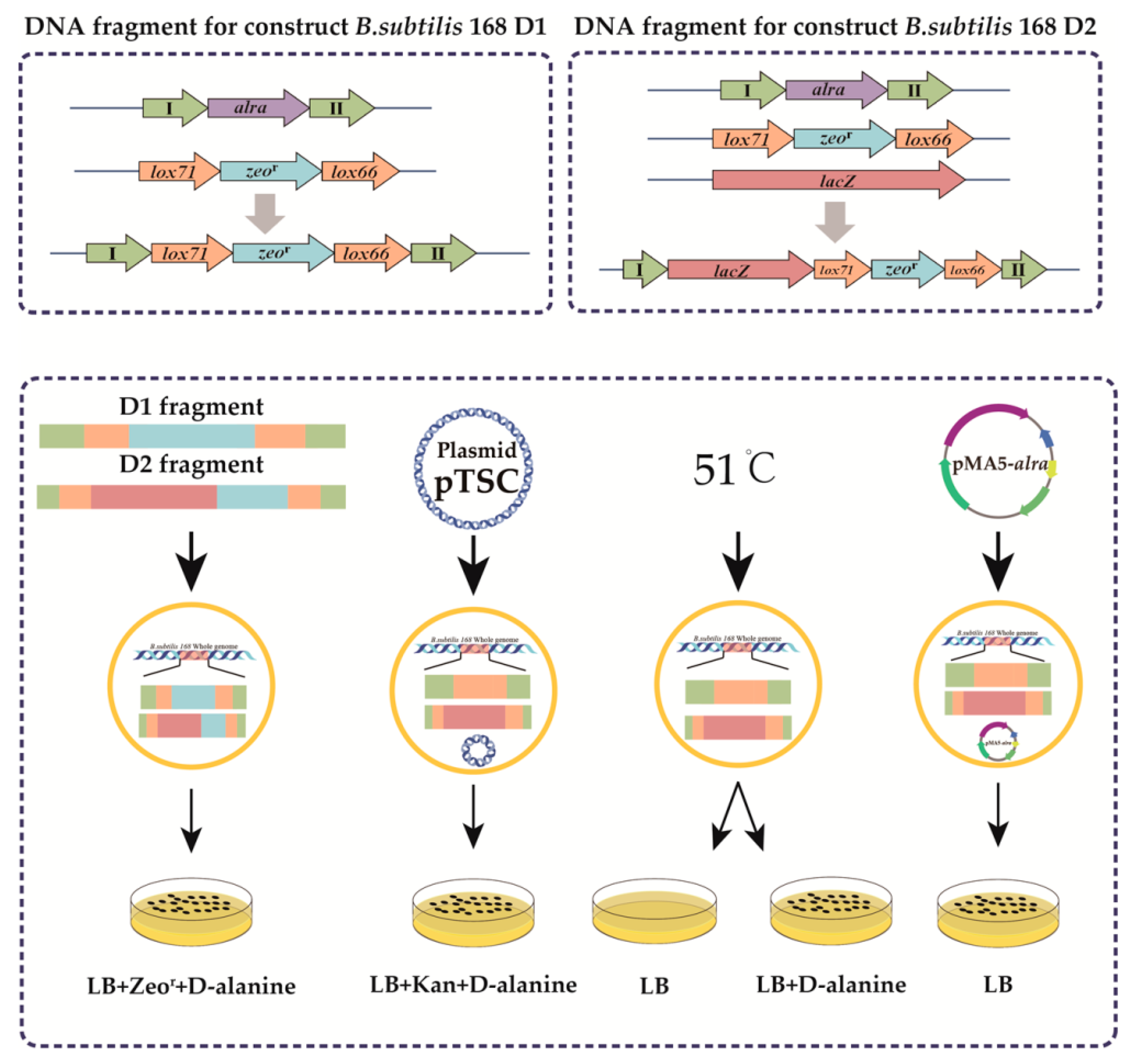
3.1. Construction of a d-alanine-deficient Plasmid Expression System
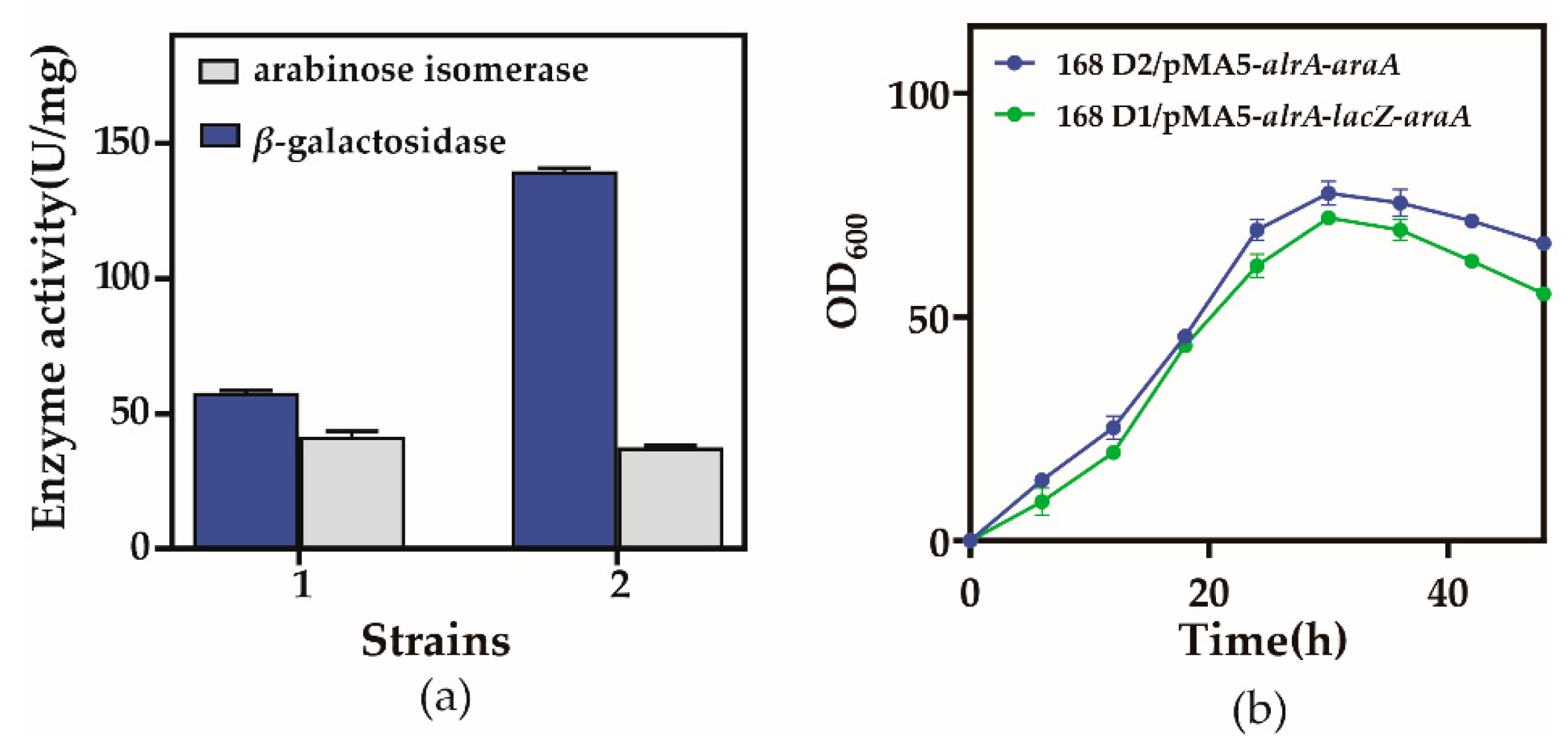
3.2. Construction of Recombinant Strains, Expression, and Enzyme Activity
3.3. Genetic Stability of the Recombinant Plasmid
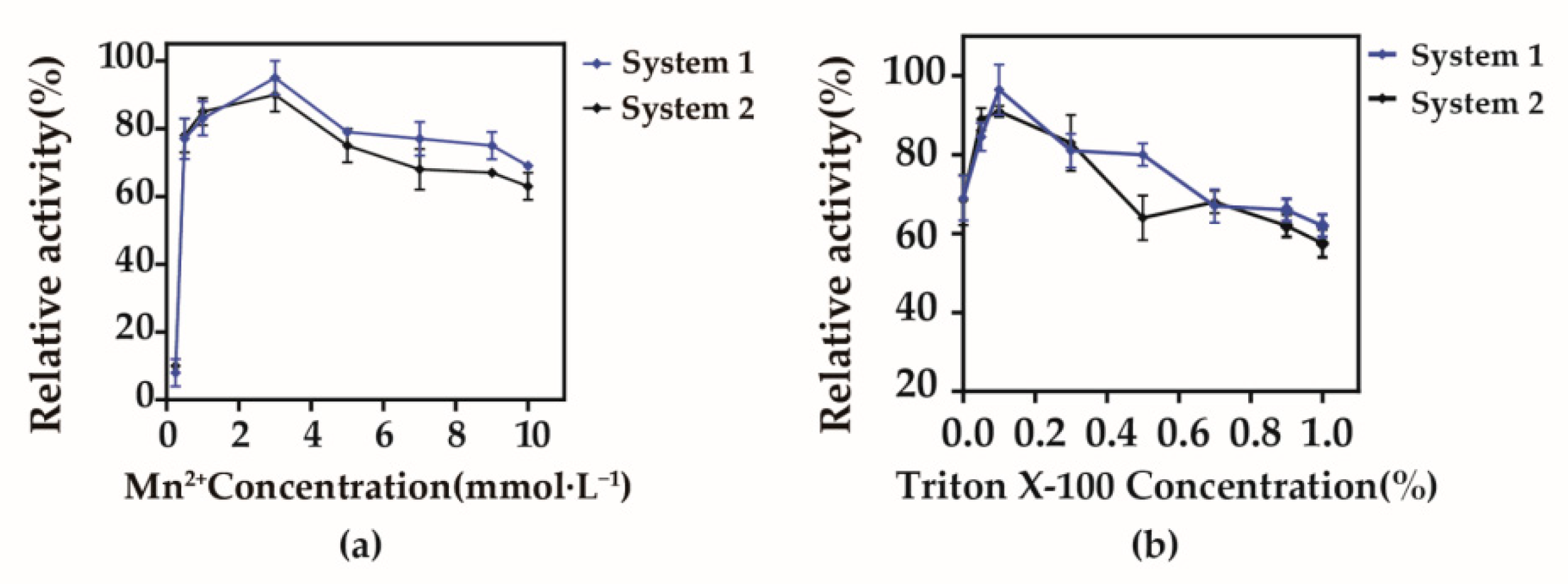
3.4. Optimization of Whole-Cell Bioconversion
3.5. d-tagatose Production
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Jayamuthunagai, J.; Gautam, P.; Srisowmeya, G.; Chakravarthy, M. Biocatalytic production of d-tagatose: A potential rare sugar with versatile applications. Crit. Rev. Food. Sci. Nutr. 2017, 57, 3430–3437. [Google Scholar] [CrossRef]
- Guo, Q.; An, Y.; Yun, J.; Yang, M.; Magocha, T.A.; Zhu, J.; Xue, Y.; Qi, Y.; Hossain, Z.; Sun, W.; et al. Enhanced d-tagatose production by spore surface-displayed l-arabinose isomerase from isolated Lactobacillus brevis PC16 and biotransformation. Bioresour. Technol. 2018, 247, 940–946. [Google Scholar] [CrossRef] [PubMed]
- Salonen, N.; Nyyssl, A.; Salonen, K.; Turunen, O. Bifidobacterium longum l-arabinose isomerase—Overexpression in Lactococcus lactis, purification, and characterization. Appl. Biochem. Biotechnol. 2012, 168, 392–405. [Google Scholar] [CrossRef] [PubMed]
- Bober, J.R.; Nair, N.U. Galactose to tagatose isomerization at moderate temperatures with high conversion and productivity. Nat. Commun. 2019, 10, 4548. [Google Scholar] [CrossRef] [Green Version]
- Wei, X.; Zhang, W.; Tao, Z.; Bo, J.; Mu, W. l-arabinose isomerases: Characteristics, modification, and application. Trends Food Sci. Technol. 2018, 78, 25–33. [Google Scholar]
- Zheng, Z.; Xie, J.; Liu, P.; Li, X.; Ouyang, J. Elegant and Efficient Biotransformation for Dual Production of d-Tagatose and Bioethanol from Cheese Whey Powder. J. Agric. Food Chem. 2019, 67, 829–835. [Google Scholar] [CrossRef]
- Saqib, S.; Akram, A.; Halim, S.A.; Tassaduq, R. Sources of β-galactosidase and its applications in food industry. Biotech 2017, 7, 79. [Google Scholar] [CrossRef] [Green Version]
- Füreder, V.; Rodriguez-Colinas, B.; Cervantes, F.V.; Fernandez-Arrojo, L.; Plou, F.J. Selective Synthesis of Galactooligosaccharides Containing β(1→3) Linkages with β-Galactosidase from Bifidobacterium bifidum (Saphera). J. Agric. Food Chem. 2020, 68, 4930–4938. [Google Scholar] [CrossRef]
- Urrutia, P.; Mateo, C.; Guisan, J.M.; Wilson, L.; Illanes, A. Immobilization of Bacillus circulans β-galactosidase and its application in the synthesis of galacto-oligosaccharides under repeated-batch operation. Biochem. Eng. J. 2013, 77, 41–48. [Google Scholar] [CrossRef]
- Sass, A.C.; Jördening, H. Immobilization of β-Galactosidase from Aspergillus oryzae on Electrospun Gelatin Nanofiber Mats for the Production of Galactooligosaccharides. Appl. Biochem. Biotechnol. 2020, 191, 1155–1170. [Google Scholar] [CrossRef] [Green Version]
- Zhan, Y.; Xu, Z.; Li, S.; Liu, X.; Xu, L.; Feng, X.; Xu, H. Coexpression of β-d-Galactosidase and l-Arabinose Isomerase in the Production of d-Tagatose: A Functional Sweetener. J. Agric. Food Chem. 2014, 62, 2412–2417. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Zabed, H.M.; Yun, J.; Yuan, J.; Qi, X. Two-stage biosynthesis of d-tagatose from milk whey powder by an engineered Escherichia coli strain expressing l-arabinose isomerase from Lactobacillus plantarum. Bioresour. Technol. 2020, 305, 123010. [Google Scholar] [CrossRef]
- Zhang, K.; Su, L.; Duan, X.; Liu, L.; Wu, J. High-level extracellular protein production in Bacillus subtilis using an optimized dual-promoter expression system. Microb. Cell Fact. 2017, 16, 32. [Google Scholar] [CrossRef] [Green Version]
- Song, Y.; Nikoloff, J.M.; Zhan, D. Improving Protein Production on the Level of Regulation of both Expression and Secretion Pathways in Bacillus subtilis. J. Microbiol. Biotechnol. 2015, 25, 963–977. [Google Scholar] [CrossRef]
- Chattopadhyay, S.; Raychaudhuri, U.; Chakraborty, R. Artificial sweeteners—A review. J. Food Sci. Technol. 2014, 51, 611–621. [Google Scholar] [CrossRef] [Green Version]
- Movahedpour, A.; Ahmadi, N.; Ghalamfarsa, F.; Ghesmati, Z.; Khalifeh, M.; Maleksabet, A.; Shabaninejad, Z.; Taheri-Anganeh, M.; Savardashtaki, A. β-Galactosidase: From its source and applications to its recombinant form. Biotechnol. Appl. Biochem. 2021. [Google Scholar] [CrossRef] [PubMed]
- Yan, X.; Yu, H.J.; Hong, Q.; Li, S.P. Cre/lox System and PCR-Based Genome Engineering in Bacillus subtilis. Appl. Environ. Microbiol. 2008, 74, 5556–5562. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferrari, E.; Henner, D.J.; Yang, M.Y. Isolation of an Alanine Racemase Gene from Bacillus subtilis and its Use for Plasmid Maintenance in B. subtilis. Nat. Biotechnol. 1985, 3, 1003–1007. [Google Scholar] [CrossRef]
- Park, C.S.; Park, C.S.; Shin, K.C.; Oh, D.K. Production of d-psicose from d-fructose by whole recombinant cells with high-level expression of d-psicose 3-epimerase from Agrobacterium tumefaciens. J. Biosci. Bioeng. 2016, 121, 186–190. [Google Scholar] [CrossRef] [PubMed]
- Laksmi, F.A.; Arai, S.; Arakawa, T.; Tsurumaru, H.; Ishibashi, M. Expression and characterization of l-arabinose isomerase from Geobacillus stearothermophilus for improved activity under acidic condition. Protein Expr. Purif. 2020, 175, 105692. [Google Scholar] [CrossRef] [PubMed]
- Hong, Y.H.; Lee, D.W.; Lee, S.J.; Choe, E.A.; Kim, S.B.; Lee, Y.H.; Cheigh, C.I.; Pyun, Y.R. Production of d-tagatose at high temperatures using immobilized Escherichia coli cells expressing l-arabinose isomerase from Thermotoga neapolitana. Biotechnol. Lett. 2007, 29, 569–574. [Google Scholar] [CrossRef] [PubMed]
- Fan, C.; Liu, K.; Zhang, T.; Zhou, L.; Xue, D.; Jiang, B.; Mu, W. Biochemical characterization of a thermostable l-arabinose isomerase from a thermoacidophilic bacterium, Alicyclobacillus hesperidum URH17-3-68. J. Mol. Catal. B Enzym. 2014, 102, 120–126. [Google Scholar] [CrossRef]
- Xu, Z.; Xu, Z.; Tang, B.; Li, S.; Gao, J.; Chi, B.; Xu, H. Construction and co-expression of polycistronic plasmids encoding thermophilic l-arabinose isomerase and hyperthermophilic β-galactosidase for single-step production of d-tagatose. Biochem. Eng. J. 2016, 109, 28–34. [Google Scholar] [CrossRef]
- Jayamuthunagai, J.; Srisowmeya, G.; Chakravarthy, M.; Gautam, P. d-tagatose production by permeabilized and immobilized Lactobacillus plantarum using whey permeate. Bioresour. Technol. 2017, 235, 250–255. [Google Scholar] [CrossRef] [PubMed]




| Generations | Stability of the Plasmid pMA5-alrA-araA (%) | Stability of the Plasmid pMA5-araA (%) | ||
|---|---|---|---|---|
| LB | LB (Add d-alanine) | LB | LB (Add d-alanine) | |
| 20 | 100 ± 0.00 | 98.00 ± 0.68 | 100 ± 0.00 | 96.00 ± 0.35 |
| 40 | 100 ± 0.00 | 96.43 ± 1.42 | 100 ± 0.00 | 94.60 ± 0.92 |
| 60 | 100 ± 0.00 | 95.00 ± 1.38 | 100 ± 0.00 | 93.48 ± 1.38 |
| 80 | 100 ± 0.00 | 93.27 ± 1.93 | 100 ± 0.00 | 92.02 ± 2.42 |
| 100 | 100 ± 0.00 | 92.30 ± 2.01 | 100 ± 0.00 | 90.54 ± 1.83 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, X.; Lu, R.; Wang, Q.; Hu, M.; Li, Z.; Xu, M.; Yang, T.; Zhang, R.; Rao, Z. Production of d-Tagatose by Whole-Cell Conversion of Recombinant Bacillus subtilis in the Absence of Antibiotics. Biology 2021, 10, 1343. https://doi.org/10.3390/biology10121343
Zhang X, Lu R, Wang Q, Hu M, Li Z, Xu M, Yang T, Zhang R, Rao Z. Production of d-Tagatose by Whole-Cell Conversion of Recombinant Bacillus subtilis in the Absence of Antibiotics. Biology. 2021; 10(12):1343. https://doi.org/10.3390/biology10121343
Chicago/Turabian StyleZhang, Xian, Ruiqi Lu, Qiang Wang, Mengkai Hu, Zhiyue Li, Meijuan Xu, Taowei Yang, Rongzhen Zhang, and Zhiming Rao. 2021. "Production of d-Tagatose by Whole-Cell Conversion of Recombinant Bacillus subtilis in the Absence of Antibiotics" Biology 10, no. 12: 1343. https://doi.org/10.3390/biology10121343
APA StyleZhang, X., Lu, R., Wang, Q., Hu, M., Li, Z., Xu, M., Yang, T., Zhang, R., & Rao, Z. (2021). Production of d-Tagatose by Whole-Cell Conversion of Recombinant Bacillus subtilis in the Absence of Antibiotics. Biology, 10(12), 1343. https://doi.org/10.3390/biology10121343








