Bta-miR-2400 Targets SUMO1 to Affect Yak Preadipocytes Proliferation and Differentiation
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals and Sample Collection
2.2. The Isolation and Induced Differentiation of Yak Preadipocytes
2.3. Cell Culture and Transfection
2.4. Cell Induced Differentiation and Oil Red O Staining
2.5. Triglyceride Assay
2.6. CCK-8 Proliferation Assay
2.7. EdU Proliferation Assay
2.8. qRT-PCR Analysis
2.9. Luciferase Reporter Assay
2.10. Western Blot Analysis
2.11. Statistical Analysis
3. Results
3.1. Expression of Bta-miR-2400 in the Yak Tissues and Adipocytes
3.2. Bta-miR-2400 Promotes the Proliferation of Yak Preadipocytes
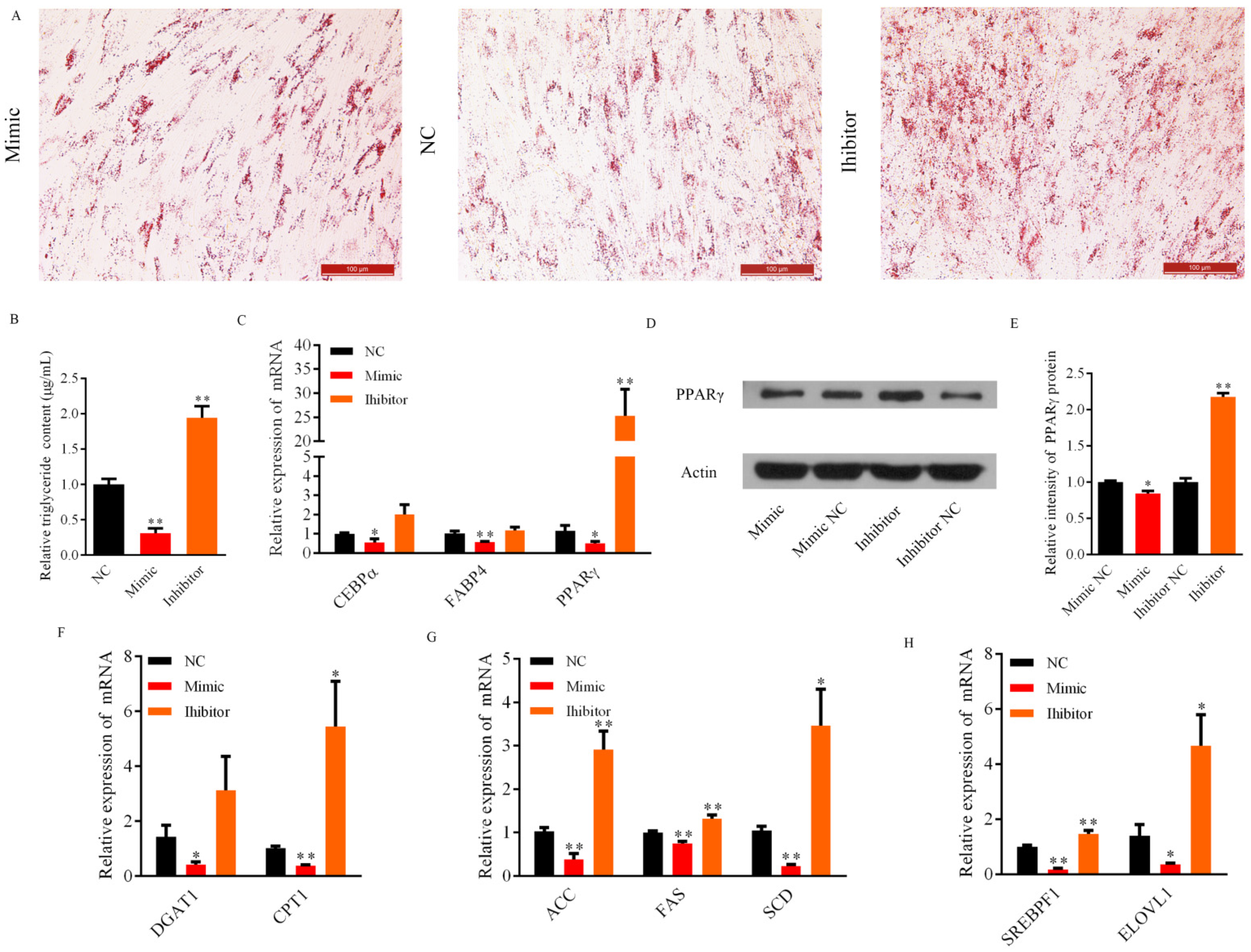
3.3. Role of Bta-miR-2400 during Yak Preadipocytes Lipogenic Differentiation
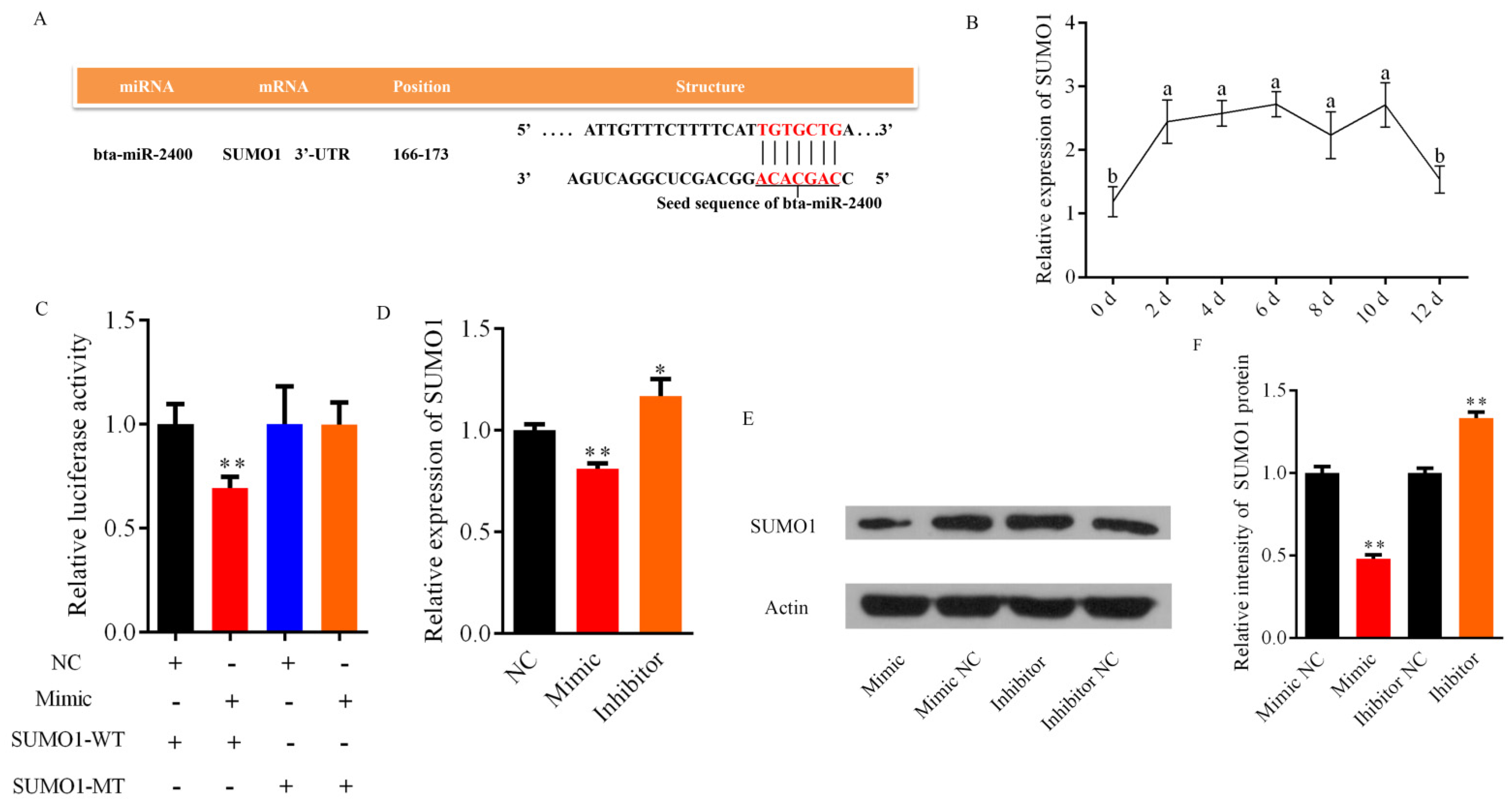
3.4. Bta-miR-2400 Directly Targets the 3′UTR of SUMO1
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Long, R.; Dong, S.; Wei, X.; Pu, X. The effect of supplementary feeds on the bodyweight of yaks in cold season. Livest. Prod. Sci. 2005, 93, 197–204. [Google Scholar] [CrossRef]
- Long, R.; Zhang, D.; Wang, X.; Hu, Z.; Dong, S. Effect of strategic feed supplementation on productive and reproductive performance in yak cows. Prev. Vet. Med. 1999, 38, 195–206. [Google Scholar] [CrossRef]
- Wiener, G.; Han, J.; Long, R. The Yak; FAO Regional office for Asia and the Pacific: Bangkok, Thailand, 2003. [Google Scholar]
- Ding, X.Z.; Guo, X.; Yan, P.; Liang, C.N.; Bao, P.J.; Chu, M. Seasonal and nutrients intake regulation of lipoprotein lipase (LPL) activity in grazing yak (Bos grunniens) in the Alpine Regions around Qinghai Lake. Livest. Sci. 2012, 143, 29–34. [Google Scholar] [CrossRef]
- Bai, X.; Zhao, X.; Zhang, Y. Dynamic changes of body weight and energy of livestock grazing on natural grassland in Qinghai-Tibetan Plateau. Anim. Husb. Vet. Med. 2005, 37, 1–4. [Google Scholar]
- Bonnet, M.; Cassar-Malek, I.; Chilliard, Y.; Picard, B. Ontogenesis of muscle and adipose tissues and their interactions in ruminants and other species. Animal 2010, 4, 1093–1109. [Google Scholar] [CrossRef] [Green Version]
- Ali, A.T.; Hochfeld, W.E.; Myburgh, R.; Pepper, M.S. Adipocyte and adipogenesis. Eur. J. Cell. Biol. 2013, 92, 229–236. [Google Scholar] [CrossRef] [PubMed]
- Li, H.X.; Xiao, L.; Wang, C.; Gao, J.L.; Zhai, Y.G. Review: Epigenetic regulation of adipocyte differentiation and adipogenesis. J. Zhejiang Univ. Sci. B. 2010, 11, 784–791. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, V.N.; Nam, J.W. Genomics of microRNA. Trends Genet. 2006, 22, 165–173. [Google Scholar] [CrossRef] [PubMed]
- Olena, A.F.; Patton, J.G. Genomic organization of microRNAs. J. Cell. Physiol. 2010, 222, 540–545. [Google Scholar] [CrossRef] [Green Version]
- Ambros, V. The functions of animal microRNAs. Nature 2004, 431, 350–355. [Google Scholar] [CrossRef]
- Karbiener, M.; Fischer, C.; Nowitsch, S.; Opriessnig, P.; Papak, C.; Ailhaud, G.; Dani, C.; Amri, E.Z.; Scheideler, M. microRNA miR-27b impairs human adipocyte differentiation and targets PPARgamma. Biochem. Biophys. Res. Commun. 2009, 390, 247–251. [Google Scholar] [CrossRef]
- Khan, R.; Raza, S.H.A.; Junjvlieke, Z.; Wang, X.; Wang, H.; Cheng, G.; Mei, C.; Elsaeid Elnour, I.; Zan, L. Bta-miR-149-5p inhibits proliferation and differentiation of bovine adipocytes through targeting CRTCs at both transcriptional and posttranscriptional levels. J. Cell Physiol. 2020, 235, 5796–5810. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Wei, D.; Cheng, G.; Li, S.; Wang, L.; Wang, Y.; Wang, X.; Zhang, S.; Wang, H.; Zan, L. Bta-miR-130a/b regulates preadipocyte differentiation by targeting PPARG and CYP2U1 in beef cattle. Mol. Cell. Probes 2018, 42, 10–17. [Google Scholar] [CrossRef]
- Mi, L.; Chen, Y.; Zheng, X.; Li, Y.; Zhang, Q.; Mo, D.; Yang, G. MicroRNA-139-5p suppresses 3T3-L1 preadipocyte differentiation through notch and IRS1/PI3K/Akt insulin signaling pathways. J. Cell. Biochem. 2015, 116, 1195–1204. [Google Scholar] [CrossRef] [PubMed]
- Qin, L.; Chen, Y.; Niu, Y.; Chen, W.; Wang, Q.; Xiao, S.; Li, A.; Xie, Y.; Li, J.; Zhao, X. A deep investigation into the adipogenesis mechanism: Profile of microRNAs regulating adipogenesis by modulating the canonical Wnt/β-catenin signaling pathway. BMC Genom. 2010, 11, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ouyang, D.; Xu, L.; Zhang, L.; Guo, D.; Tan, X.; Yu, X.; Qi, J.; Ye, Y.; Liu, Q.; Ma, Y. MiR-181a-5p regulates 3T3-L1 cell adipogenesis by targeting Smad7 and Tcf7l2. Acta Bioch. Bioph. Sin. 2016, 48, 1034–1041. [Google Scholar] [CrossRef] [Green Version]
- Glazov, E.A.; Kongsuwan, K.; Assavalapsakul, W.; Horwood, P.F.; Mitter, N.; Mahony, T.J. Repertoire of bovine miRNA and miRNA-like small regulatory RNAs expressed upon viral infection. PLoS ONE 2009, 4, e6349. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, W.W.; Tong, H.L.; Sun, X.F.; Hu, Q.; Yang, Y.; Li, S.F.; Yan, Y.Q.; Li, G.P. Identification of miR-2400 gene as a novel regulator in skeletal muscle satellite cells proliferation by targeting MYOG gene. Biochem. Biophys. Res. Commun. 2015, 463, 624–631. [Google Scholar] [CrossRef]
- Zhang, Y.; Wu, X.; Liang, C.; Bao, P.; Ding, X.; Chu, M.; Jia, C.; Guo, X.; Yan, P. MicroRNA-200a regulates adipocyte differentiation in the domestic yak Bos grunniens. Gene 2018, 650, 41–48. [Google Scholar] [CrossRef]
- Zhang, Y.; Guo, X.; Pei, J.; Chu, M.; Ding, X.; Wu, X.; Liang, C.; Yan, P. CircRNA Expression Profile during Yak Adipocyte Differentiation and Screen Potential circRNAs for Adipocyte Differentiation. Genes 2020, 11, 414. [Google Scholar] [CrossRef]
- Zhu, L.; Skoultchi, A.I. Coordinating cell proliferation and differentiation. Curr. Opin. Genet. Dev. 2001, 11, 91–97. [Google Scholar] [CrossRef]
- Moore, K.J.; Rosen, E.D.; Fitzgerald, M.L.; Randow, F.; Andersson, L.P.; Altshuler, D.; Milstone, D.S.; Mortensen, R.M.; Spiegelman, B.M.; Freeman, M.W. The role of PPAR-γ in macrophage differentiation and cholesterol uptake. Nat. Med. 2001, 7, 41–47. [Google Scholar] [CrossRef] [PubMed]
- Xue, B.; Zhao, X.; Zhang, Y. Seasonal changes in weight and body composition of yak grazing on alpine-meadow grassland in the Qinghai-Tibetan plateau of China. J. Anim. Sci. 2005, 83, 1908–1913. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ding, L.; Wang, Y.; Brosh, A.; Chen, J.; Gibb, M.; Shang, Z.; Guo, X.; Mi, J.; Zhou, J.; Wang, H. Seasonal heat production and energy balance of grazing yaks on the Qinghai-Tibetan plateau. Anim. Feed Sci. Tech. 2014, 198, 83–93. [Google Scholar] [CrossRef]
- Kiess, W.; Petzold, S.; Topfer, M.; Garten, A.; Bluher, S.; Kapellen, T.; Korner, A.; Kratzsch, J. Adipocytes and adipose tissue. Best Pr. Res. Clin. Endocrinol. Metab. 2008, 22, 135–153. [Google Scholar] [CrossRef]
- Romao, J.M.; Jin, W.; Dodson, M.V.; Hausman, G.J.; Moore, S.S.; Guan, L.L. MicroRNA regulation in mammalian adipogenesis. Exp. Biol. Med. 2011, 236, 997–1004. [Google Scholar] [CrossRef]
- Sherr, C.J.; Roberts, J.M. Inhibitors of mammalian G1 cyclin-dependent kinases. Genes Dev. 1995, 9, 1149–1163. [Google Scholar] [CrossRef] [Green Version]
- Tsai, L.-H.; Lees, E.; Faha, B.; Harlow, E.; Riabowol, K. The cdk2 kinase is required for the G1-to-S transition in mammalian cells. Oncogene 1993, 8, 1593–1602. [Google Scholar]
- Koundrioukoff, S.; Jonsson, Z.O.; Hasan, S.; de Jong, R.N.; van der Vliet, P.C.; Hottiger, M.O.; Hubscher, U. A direct interaction between proliferating cell nuclear antigen (PCNA) and Cdk2 targets PCNA-interacting proteins for phosphorylation. J. Biol. Chem. 2000, 275, 22882–22887. [Google Scholar] [CrossRef] [Green Version]
- Rosen, E.D.; MacDougald, O.A. Adipocyte differentiation from the inside out. Nat. Rev. Mol. Cell Bio. 2006, 7, 885–896. [Google Scholar] [CrossRef]
- Rosen, E.D.; Walkey, C.J.; Puigserver, P.; Spiegelman, B.M. Transcriptional regulation of adipogenesis. Genes Dev. 2000, 14, 1293–1307. [Google Scholar]
- Xie, H.; Sun, L.; Lodish, H.F. Targeting microRNAs in obesity. Expert. Opin. Tar. 2009, 13, 1227–1238. [Google Scholar]
- Peng, Y.; Li, H.; Li, X.; Yu, S.; Xiang, H.; Peng, J.; Jiang, S. MicroRNA-215 impairs adipocyte differentiation and co-represses FNDC3B and CTNNBIP1. Int. J. Biochem. Cell. Biol. 2016, 79, 104–112. [Google Scholar] [CrossRef]
- Xu, H.Y.; Shao, J.; Yin, B.Z.; Zhang, L.M.; Fang, J.C.; Zhang, J.S.; Xia, G.J. Bovine bta-microRNA-1271 Promotes Preadipocyte Differentiation by Targeting Activation Transcription Factor 3. Biochemistry 2020, 85, 749–757. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, Y.; Wang, H.; Ma, X.; Zan, L. MicroRNA-224 impairs adipogenic differentiation of bovine preadipocytes by targeting LPL. Mol. Cell. Probes 2019, 44, 29–36. [Google Scholar] [CrossRef]
- Mikkonen, L.; Hirvonen, J.; Jänne, O.A. Sumo-1 regulates body weight and adipogenesis via PPARγ in male and female mice. Endocrinology 2013, 154, 698–708. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shukla, G.C.; Singh, J.; Barik, S. MicroRNAs: Processing, maturation, target recognition and regulatory functions. Mol. Cell. Pharmacol. 2011, 3, 83. [Google Scholar] [PubMed]
- Dohmen, R.J. SUMO protein modification. Biochim. Et Biophys. Acta (BBA)-Mol. Cell. Res. 2004, 1695, 113–131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brunmeir, R.; Xu, F. Functional Regulation of PPARs through Post-Translational Modifications. Int. J. Mol. Sci. 2018, 19, 1738. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sarkar, R. Stiffening of flexible SUMO1 protein upon peptide-binding: Analysis with anisotropic network model. Math. Biosci. 2018, 295, 67–72. [Google Scholar] [CrossRef] [PubMed]
- Ohshima, T.; Koga, H.; Shimotohno, K. Transcriptional activity of peroxisome proliferator-activated receptor gamma is modulated by SUMO-1 modification. J. Biol. Chem. 2004, 279, 29551–29557. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, H.B.; Luo, J.; Yao, D.W.; Zhu, J.J.; Xu, H.F.; Shi, H.P.; Loor, J.J. Peroxisome proliferator-activated receptor-gamma stimulates the synthesis of monounsaturated fatty acids in dairy goat mammary epithelial cells via the control of stearoyl-coenzyme A desaturase. J. Dairy Sci. 2013, 96, 7844–7853. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shimizu, M.; Yamashita, D.; Yamaguchi, T.; Hirose, F.; Osumi, T. Aspects of the regulatory mechanisms of PPAR functions: Analysis of a bidirectional response element and regulation by sumoylation. Mol. Cell. Biochem. 2006, 286, 33–42. [Google Scholar] [CrossRef] [PubMed]


| Gene | Primer Sequence (5′ to 3′) | Tm/°C | Product Size/bp | Accession Numbers |
|---|---|---|---|---|
| SCD | F:CTGGTGTCCTGTTGTTGTGC R:GGTCTTGTCATAAGGGCGGT | 60.0 | 179 | XM_005892055.2 |
| PPAR γ | F:CATTTCCACTCCGCACTA R:GGGATACAGGCTCCACTT | 60.3 | 122 | XM_005902846.1 |
| C/EBPα | F:GGCATCTGCGAACACGAGA R:AGGAACTCGTCGTTGAAGGC | 60.3 | 212 | XM_005893918.2 |
| FABP4 | F:CATTAAATCCGAAAGCA R:CTCATAAACTCTGGTGGC | 60.3 | 242 | XM_014478668.1 |
| FAS | F:GCAAAGTGGTCATTCAGGTACG R:CCCAGTGATGATGTAGCTCTTG | 64.5 | 125 | NM_001012669.1 |
| ACC | F:GAGACAAACAGGGACCATT R:AGGGACTGCCGAAACAT | 61.4 | 142 | AJ132890.1 |
| CPT1 | F:TCGACCCAAACAAGTACCCC R:CGCTGGGCATTTGTCTCTGA | 60.0 | 157 | NM_001034349.2 |
| DGAT1 | F:GAGACAAACAGGGACCATT R: AGGGACTGCCGAAACAT | 64.5 | 92 | JF913457.1 |
| SUMO1 | F:CATTGGACAGGATAGCA R:CTCCATTCCCAGTTCTT | 59.0 | 172 | NM_001035458.1 |
| ELOVL1 | F:AGAAAGACGGACAGGTGAC R:GCAGGAAGTAGTATTGGGAG | 61.4 | 273 | XM_010803685.3 |
| bta-miR-2400 | F:CCAGCACAGGCAGCTCGGACTGA | 60.3 | _ | |
| SREBPF1 | F:GCTGACCGACATAGAAGACA R:CTCATCGTGGAAGGAGGTGG | 61.0 | 165 | NM_001113302.1 |
| PCNA | F:TCCGAGGGCTTCGACACTTA R:GTGCCAACGTGTCTGCGTTA | 63.3 | 142 | XM_005906528.2 |
| CDK2 | F:AGGTGGTGACTCTGTGGTA R:GCTCCGTCCATCTTCAT | 59.0 | 299 | XM_005905862.2 |
| CDK4 | F:TTGGTGTCGGTGCCTAT R:TCCAGACGCCGCAGTAA | 57.0 | 157 | XM_005901664.1 |
| U6 | F:GGAACGATACAGAGAAGATTAGC R:TGGAACGCTTCACGAATTTGCG | 60.3 | _ | |
| β-Actin | F:GCAGGTCATCACCATCGG R:CCGTGTTGGCGTAGAGGT | 60.3 | 158 | XM_005887322.2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.; Ma, L.; Gu, Y.; Chang, Y.; Liang, C.; Guo, X.; Bao, P.; Chu, M.; Ding, X.; Yan, P. Bta-miR-2400 Targets SUMO1 to Affect Yak Preadipocytes Proliferation and Differentiation. Biology 2021, 10, 949. https://doi.org/10.3390/biology10100949
Zhang Y, Ma L, Gu Y, Chang Y, Liang C, Guo X, Bao P, Chu M, Ding X, Yan P. Bta-miR-2400 Targets SUMO1 to Affect Yak Preadipocytes Proliferation and Differentiation. Biology. 2021; 10(10):949. https://doi.org/10.3390/biology10100949
Chicago/Turabian StyleZhang, Yongfeng, Lanhua Ma, Yarong Gu, Yongfang Chang, Chunnian Liang, Xian Guo, Pengjia Bao, Min Chu, Xuezhi Ding, and Ping Yan. 2021. "Bta-miR-2400 Targets SUMO1 to Affect Yak Preadipocytes Proliferation and Differentiation" Biology 10, no. 10: 949. https://doi.org/10.3390/biology10100949
APA StyleZhang, Y., Ma, L., Gu, Y., Chang, Y., Liang, C., Guo, X., Bao, P., Chu, M., Ding, X., & Yan, P. (2021). Bta-miR-2400 Targets SUMO1 to Affect Yak Preadipocytes Proliferation and Differentiation. Biology, 10(10), 949. https://doi.org/10.3390/biology10100949






