Microbial Characteristics and Safety of Dairy Manure ComPosting for Reuse as Dairy Bedding
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and Dry Matter Determination
2.2. DNA Extraction
2.3. Qualitative Analysis of the Pathogenic Bacteria Causing Mastitis
2.4. High Throughput Sequencing
2.5. Bioinformatics and Statistical Analysis
3. Results and Discussion
3.1. Dry Mand α-Diversity
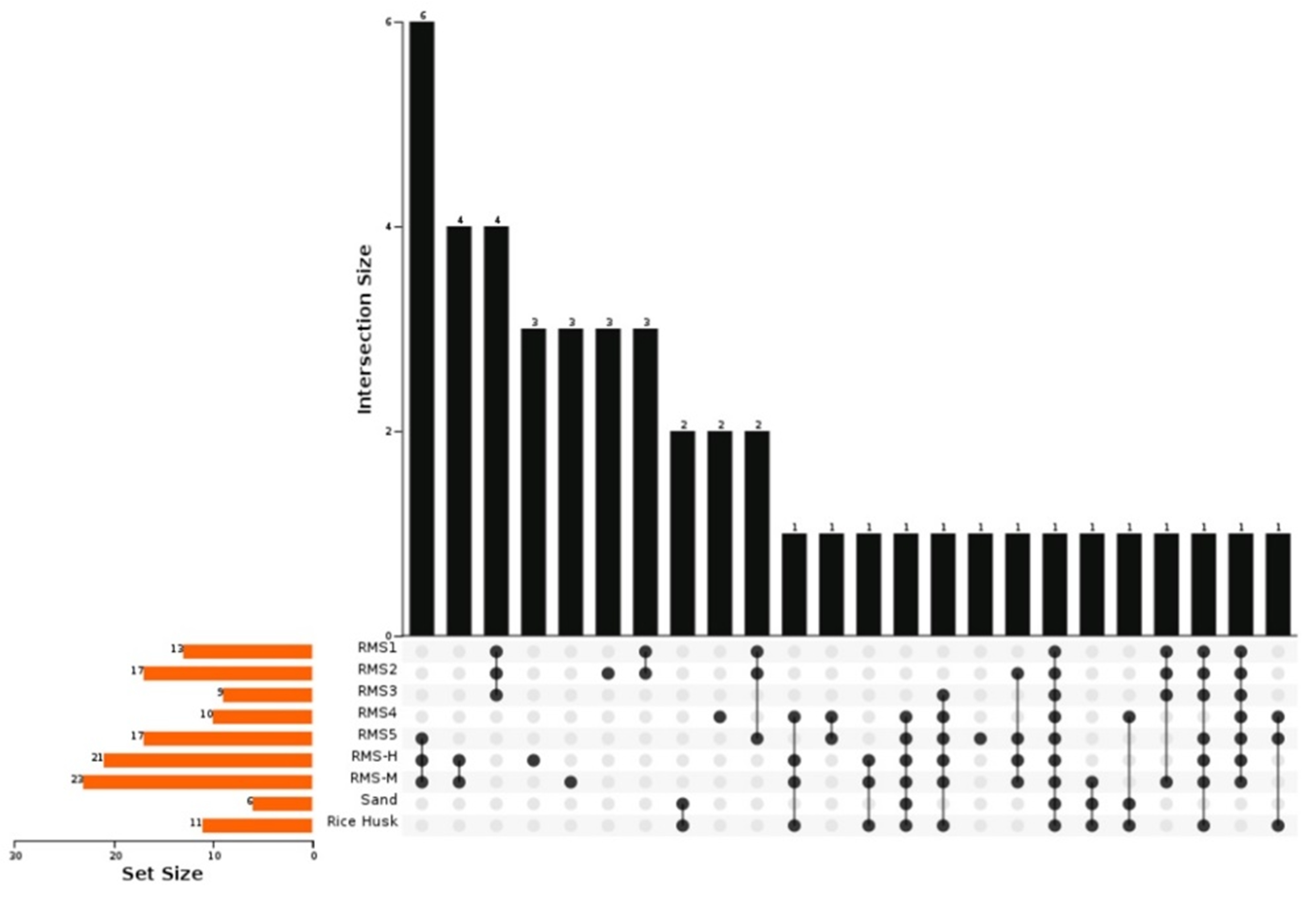
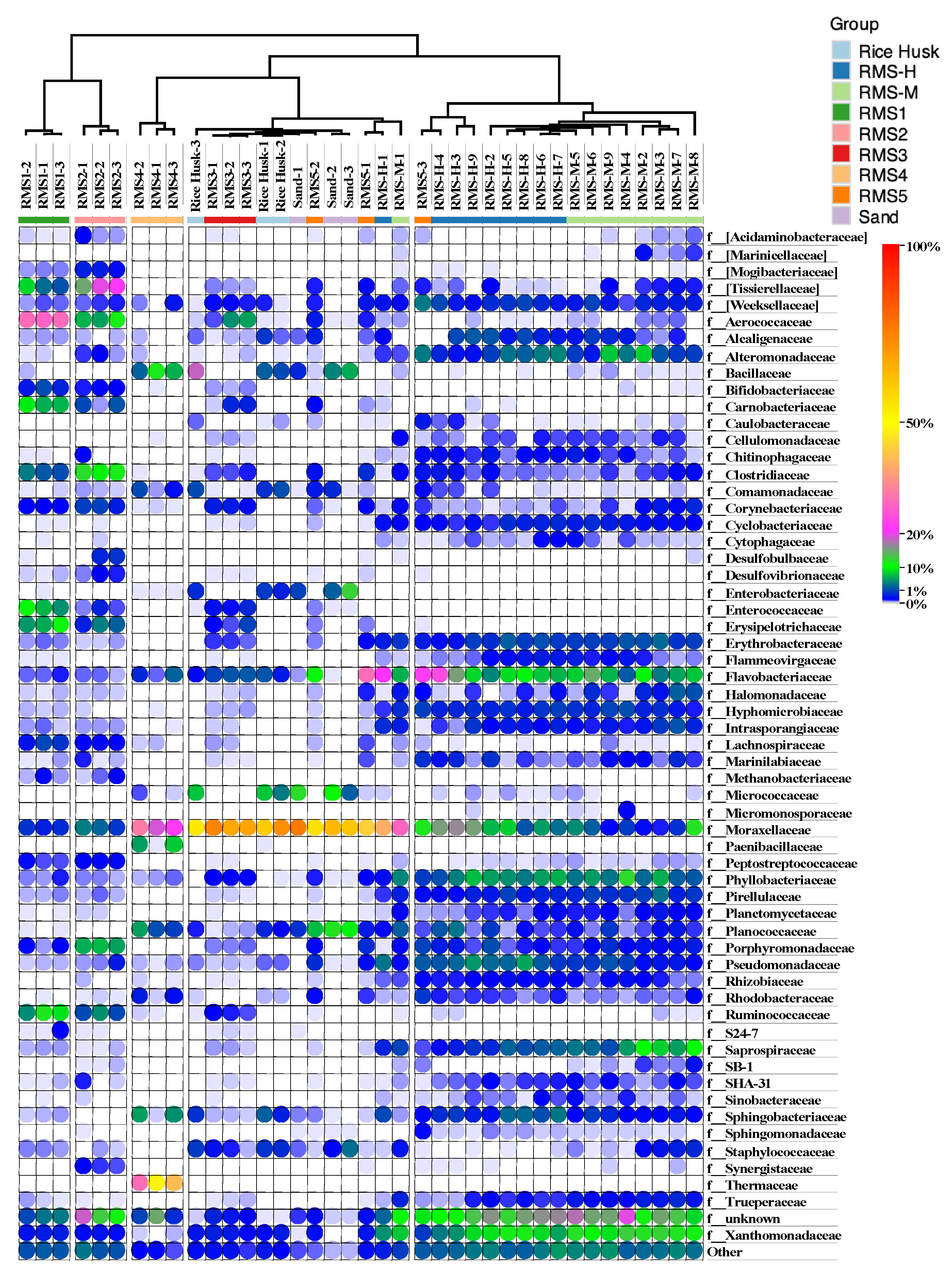
3.2. Bacterial Community Composition
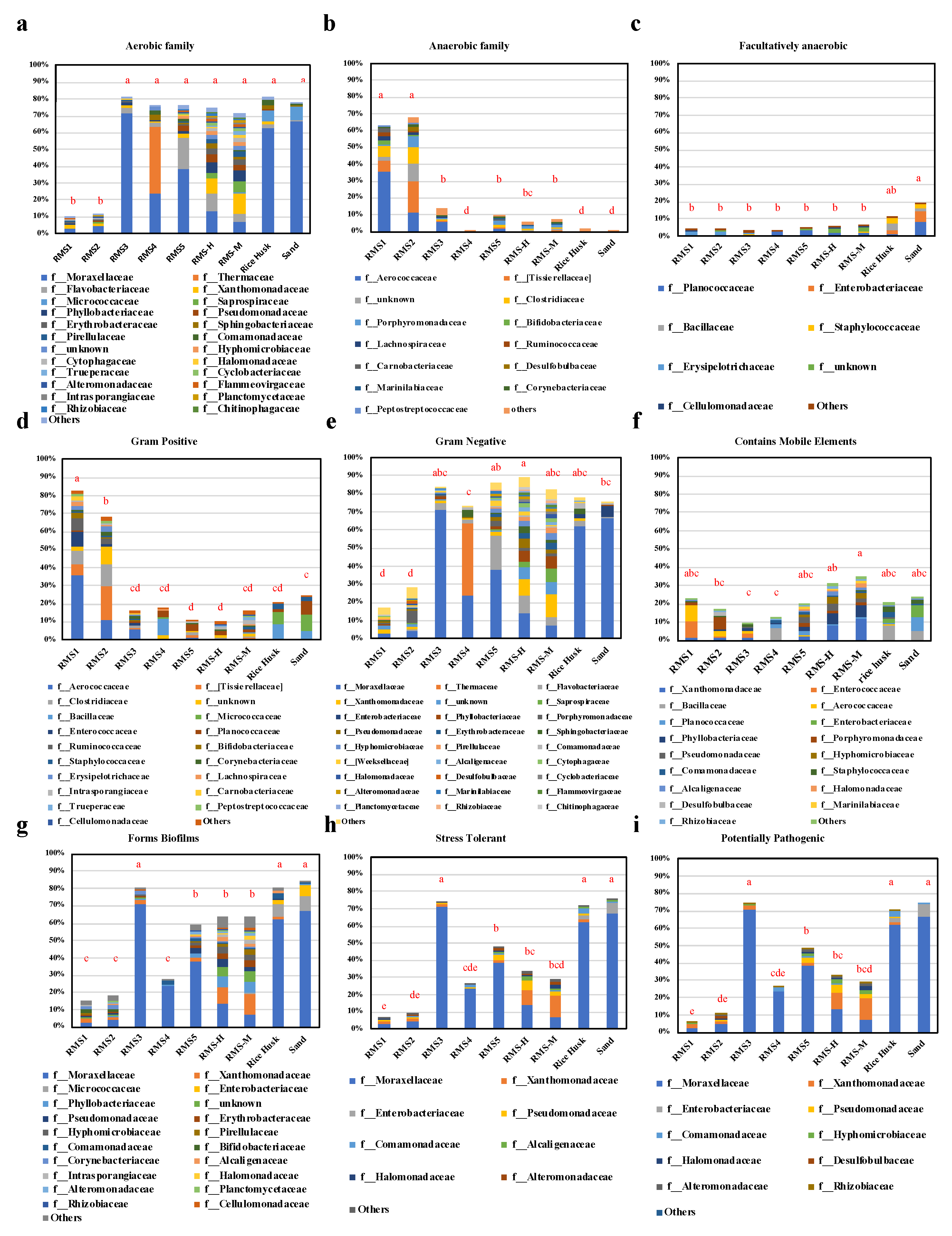
3.3. Phenotype of the Bacterial Community
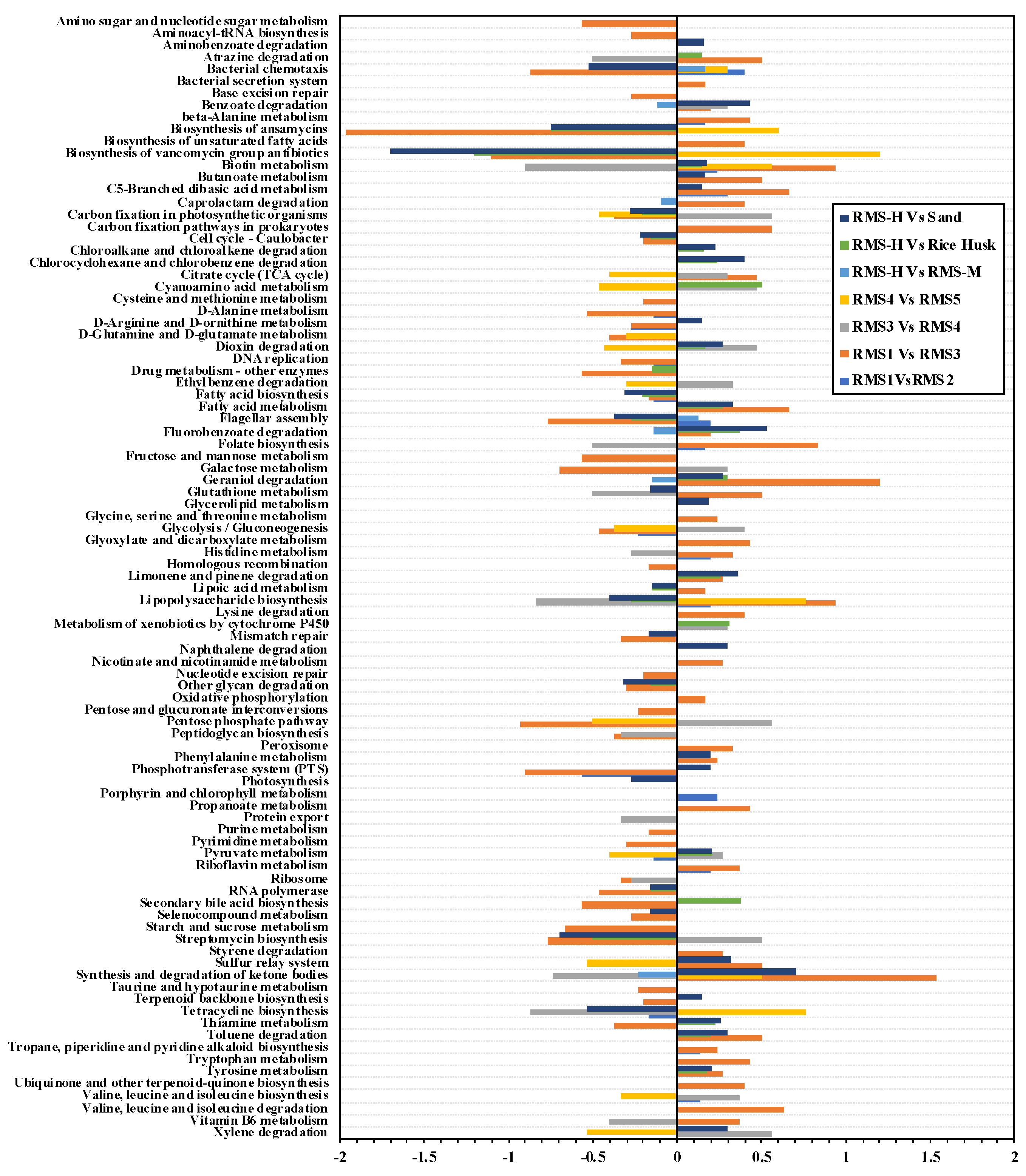
3.4. Predicted Potential Metabolic Functions of the Bacterial Community
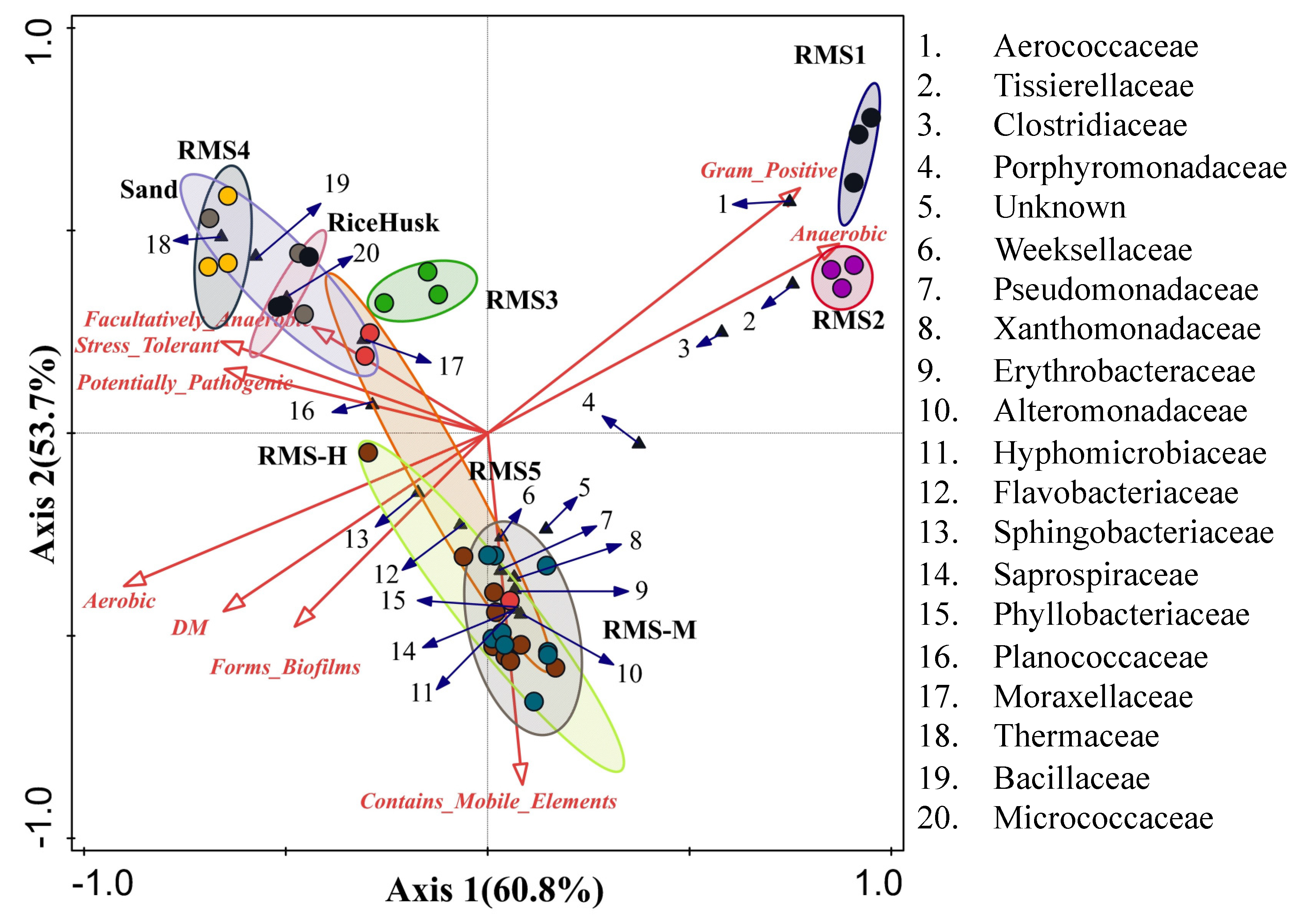
3.5. Relationships among Phenotypic Properties, Bacterial Community, and Metabolic Functions
3.6. Mastitis Pathogen Detection
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Wang, K.; Chu, C.; Li, X.; Wang, W.; Ren, N. Succession of bacterial community function in cow manure composing. Bioresour. Technol. 2018, 267, 63–70. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Yin, X.; Mao, H.; Chu, C.; Tian, Y. Changes in structure and function of fungal community in cow manure composting. Bioresour. Technol. 2018, 255, 123–130. [Google Scholar] [CrossRef] [PubMed]
- Hogan, J.; Smith, K.L. Managing environmental mastitis. Vet. Clin. Food Anim. Pract. 2012, 28, 217–224. [Google Scholar] [CrossRef] [PubMed]
- van Gastelen, S.; Westerlaan, B.; Houwers, D.J.; van Eerdenburg, F.J.C.M. A study on cow comfort and risk for lameness and mastitis in relation to different types of bedding materials. J. Dairy Sci. 2011, 94, 4878–4888. [Google Scholar] [CrossRef]
- Rowbotham, R.F.; Ruegg, P.L. Associations of selected bedding types with incidence rates of subclinical and clinical mastitis in primiparous Holstein dairy cows. J. Dairy Sci. 2016, 99, 4707–4717. [Google Scholar] [CrossRef]
- Rowbotham, R.F.; Ruegg, P.L. Bacterial counts on teat skin and in new sand, recycled sand, and recycled manure solids used as bedding in freestalls. J. Dairy Sci. 2016, 99, 6594–6608. [Google Scholar] [CrossRef]
- Husfeldt, A.W.; Endres, M.I.; Salfer, J.A.; Janni, K.A. Management and characteristics of recycled manure solids used for bedding in Midwest freestall dairy herds. J. Dairy Sci. 2012, 95, 2195–2203. [Google Scholar] [CrossRef]
- Petersen, H.H.; Nielsen, J.P.; Heegaard, P.M.H. Application of acute phase protein measurements in veterinary clinical chemistry. Vet. Res. 2004, 35, 163–187. [Google Scholar] [CrossRef]
- Tucker, C.B.; Weary, D.M. Bedding on geotextile mattresses: How much is needed to improve cow comfort? J. Dairy Sci. 2004, 87, 2889–2895. [Google Scholar] [CrossRef]
- De, L.; Laval, U.; Hamelin, L.; Fréchette, A.; Dufour, S.; Roy, D. Effect of recycled manure solids as bedding on bulk tank milk and implications for cheese microbiological quality. J. Dairy Sci. 2020, 128–140. [Google Scholar] [CrossRef]
- De Vliegher, S.; Ohnstad, I.; Piepers, S. Management and prevention of mastitis: A multifactorial approach with a focus on milking, bedding and data-management. J. Integr. Agric. 2018, 17, 1214–1233. [Google Scholar] [CrossRef]
- Li, X.; Shi, X.S.; Lu, M.Y.; Zhao, Y.Z.; Li, X.; Peng, H.; Guo, R.B. Succession of the bacterial community and functional characteristics during continuous thermophilic composting of dairy manure amended with recycled ceramsite. Bioresour. Technol. 2019, 294. [Google Scholar] [CrossRef] [PubMed]
- Bradley, A.J.; Leach, K.A. Scoping Study on the Potential Risks (and Benefits) of Using Recycled Manure Solids as Bedding for Dairy Cattle; Quality Milk Management Services Ltd.: Wells, UK, 2014. [Google Scholar]
- Fournel, S.; Godbout, S.; Ruel, P.; Fortin, A.; Létourneau, V.; Généreux, M.; Lemieux, J.; Potvin, D.; Côté, C.; Duchaine, C.; et al. Production of recycled manure solids for use as bedding in Canadian dairy farms: II. Composting methods. J. Dairy Sci. 2019, 102, 1847–1865. [Google Scholar] [CrossRef] [PubMed]
- Leach, K.A.; Archer, S.C.; Breen, J.E.; Green, M.J.; Ohnstad, I.C.; Tuer, S.; Bradley, A.J. Recycling manure as cow bedding: Potential benefits and risks for UK dairy farms. Vet. J. 2015, 206, 123–130. [Google Scholar] [CrossRef]
- Cole, K.J.; Hogan, J.S. Short communication: Environmental mastitis pathogen counts in freestalls bedded with composted and fresh recycled manure solids. J. Dairy Sci. 2016, 99, 1501–1505. [Google Scholar] [CrossRef]
- Sorter, D.E.; Kester, H.J.; Hogan, J.S. Short communication: Bacterial counts in recycled manure solids bedding replaced daily or deep packed in freestalls. J. Dairy Sci. 2014, 97, 2965–2968. [Google Scholar] [CrossRef]
- Godden, S.; Bey, R.; Lorch, K.; Farnsworth, R.; Rapnicki, P. Ability of organic and inorganic bedding materials to promote growth of environmental bacteria. J. Dairy Sci. 2008, 91, 151–159. [Google Scholar] [CrossRef]
- Bradley, A.J.; Leach, K.A.; Green, M.J.; Gibbons, J.; Ohnstad, I.C.; Black, D.H.; Payne, B.; Prout, V.E.; Breen, J.E. The impact of dairy cows’ bedding material and its microbial content on the quality and safety of milk—A cross sectional study of UK farms. Int. J. Food Microbiol. 2018, 269, 36–45. [Google Scholar] [CrossRef]
- Lasprilla-Mantilla, M.I.; Wagner, V.; Pena, J.; Frechette, A.; Thivierge, K.; Dufour, S.; Fernandez-Prada, C. Effects of recycled manure solids bedding on the spread of gastrointestinal parasites in the environment of dairies and milk. J. Dairy Sci. 2019. [Google Scholar] [CrossRef]
- Hall, M.; Beiko, R.G. 16S rRNA gene analysis with QIIME2. In Microbiome Analysis; Springer: Berlin/Heidelberg, Germany, 2018; pp. 113–129. [Google Scholar]
- Ward, T.; Larson, J.; Meulemans, J.; Hillmann, B.; Lynch, J.; Sidiropoulos, D.; Spear, J.R.; Caporaso, G.; Blekhman, R.; Knight, R.; et al. BugBase predicts organism-level microbiome phenotypes. bioRxiv 2017, 133462. [Google Scholar] [CrossRef]
- Conway, J.R.; Lex, A.; Gehlenborg, N. UpSetR: An R package for the visualization of intersecting sets and their properties. Bioinformatics 2017, 33, 2938–2940. [Google Scholar] [CrossRef] [PubMed]
- Fan, S.; Liang, Z.; Gao, Z.; Pan, Z.; Han, S.; Liu, X.; Zhao, C.; Yang, W.; Pan, Z.; Feng, W. Identification of the key genes and pathways in prostate cancer. Oncol. Lett. 2018, 16, 6663–6669. [Google Scholar] [CrossRef] [PubMed]
- Han, M.-M.; Zhu, X.-Y.; Peng, Y.-F.; Lin, H.; Liu, D.-C.; Li, L. The alterations of gut microbiota in mice with chronic pancreatitis. Ann. Transl. Med. 2019, 7, 464. [Google Scholar] [CrossRef] [PubMed]
- Zuo, T.; Lu, X.J.; Zhang, Y.; Cheung, C.P.; Lam, S.; Zhang, F.; Tang, W.; Ching, J.Y.L.; Zhao, R.; Chan, P.K.S.; et al. Gut mucosal virome alterations in ulcerative colitis. Gut 2019, 68, 1169–1179. [Google Scholar] [CrossRef]
- Zähner, M.; Schmidtko, J.; Schrade, S.; Schaeren, W. Alternative Einstreumaterialien in Liegeboxen. Available online: https://docplayer.org/42942950-Bautagung-raumberg-gumpenstein-technik-in-der-rinderhaltung-emissionen-rahmenbedingungen-fuer-die-schweinehaltung.html (accessed on 18 December 2020).
- Du, J.; Wang, T.; Zhou, Q.; Hu, X.; Wu, J.; Li, G.; Li, G.; Hou, F.; Wu, Y. Graphene oxide enters the rice roots and disturbs the endophytic bacterial communities. Ecotoxicol. Environ. Saf. 2020, 192, 110304. [Google Scholar] [CrossRef]
- Liu, H.; Li, C.; Liang, Z.; Zhang, S.; Yang, W.; Ye, Y.; Lin, Y.; Chen, R.; Zhou, H.; Su, J. Moraxellaceae and Moraxella interact with the altered airway mycobiome in asthma. bioRxiv 2019, 525113. [Google Scholar] [CrossRef]
- Vidakovics, M.L.P.; Riesbeck, K. Virulence mechanisms of moraxella in the pathogenesis of infection. Curr. Opin. Infect. Dis. 2009, 22, 279–285. [Google Scholar] [CrossRef]
- Wu, H.; Nguyen, Q.D.; Tran, T.T.M.; Tang, M.T.; Tsuruta, T.; Nishino, N. Rumen fluid, feces, milk, water, feed, airborne dust, and bedding microbiota in dairy farms managed by automatic milking systems. Anim. Sci. J. 2019, 90, 445–452. [Google Scholar] [CrossRef]
- Robles, I.; Kelton, D.F.; Barkema, H.W.; Keefe, G.P.; Roy, J.P.; Von Keyserlingk, M.A.G.; Devries, T.J. Bacterial concentrations in bedding and their association with dairy cow hygiene and milk quality. Animal 2020, 1052–1066. [Google Scholar] [CrossRef]
- Wang, L.; Luo, Z.; Zhen, Z.; Yan, Y.; Yan, C.; Ma, X.; Sun, L.; Wang, M.; Zhou, X.; Hu, A. Bacterial community colonization on tire microplastics in typical urban water environments and associated impacting factors. Environ. Pollut. 2020, 265, 114922. [Google Scholar] [CrossRef]
- Glantz, M.; Rosenlöw, M.; Lindmark-Månsson, H.; Buhelt Johansen, L.; Hartmann, J.; Höjer, A.; Waak, E.; Löfgren, R.; Hallin Saedén, K.; Svensson, C.; et al. Impact of protease and lipase activities on quality of Swedish raw milk. Int. Dairy J. 2020, 107, 104724. [Google Scholar] [CrossRef]
- Stoeckel, M.; Lidolt, M.; Achberger, V.; Glück, C.; Krewinkel, M.; Stressler, T.; von Neubeck, M.; Wenning, M.; Scherer, S.; Fischer, L.; et al. Growth of Pseudomonas weihenstephanensis, Pseudomonas proteolytica and Pseudomonas sp. in raw milk: Impact of residual heat-stable enzyme activity on stability of UHT milk during shelf-life. Int. Dairy J. 2016, 59, 20–28. [Google Scholar] [CrossRef]
- Langille, M.G.I.; Zaneveld, J.; Caporaso, J.G.; McDonald, D.; Knights, D.; Reyes, J.A.; Clemente, J.C.; Burkepile, D.E.; Vega Thurber, R.L.; Knight, R.; et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 2013, 31, 814–821. [Google Scholar] [CrossRef] [PubMed]
- Staninska-Pięta, J.; Czarny, J.; Piotrowska-Cyplik, A.; Juzwa, W.; Wolko, Ł.; Nowak, J.; Cyplik, P. Heavy metals as a factor increasing the functional genetic potential of bacterial community for polycyclic aromatic hydrocarbon biodegradation. Molecules 2020, 25, 319. [Google Scholar] [CrossRef] [PubMed]
- Ward, W.R.; Hughes, J.W.; Faull, W.B. Observational study of temperature, moisture, pH and bacteria in straw bedding, and faecal consistency, cleanliness and mastitis in cows in four dairy herds. Vet. Rec. 2002, 151, 199–206. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Liu, Y.C.; Wang, Y.; Li, H.; Wang, X.M.; Wu, Y.; Zhang, D.R.; Gao, S.; Qi, Z.L. Impact of yeast and lactic acid bacteria on mastitis and milk microbiota composition of dairy cows. AMB Express 2020, 10, 22. [Google Scholar] [CrossRef]
| DM (%) | Diversity | Evenness | |||
|---|---|---|---|---|---|
| Observed OTU | Chao 1 | Simpson | Shannon | ||
| RMS1 | 6.05 ± 0.16 d | 248 ± 14 ab | 250 ± 14 ab | 0.97 ± 0.004 ab | 6.33 ± 0.087 a |
| RMS2 | 4.52 ± 1.81 d | 281 ± 6 a | 282 ± 6 a | 0.99 ± 0 a | 7.16 ± 0.036 a |
| RMS3 | 20.20 ± 0.64 c | 159 ± 14 b | 160 ± 15 b | 0.61 ± 0.032 c | 3.34 ± 0.177 c |
| RMS4 | 47.52 ± 1.12 a | 181 ± 32 ab | 184 ± 33 ab | 0.90 ± 0.019 b | 4.80 ± 0.395 bc |
| RMS5 | 36.94 ± 3.53 b | 198 ± 31 ab | 199 ± 31 ab | 0.96 ± 0.013 ab | 5.91 ± 0.545 ab |
| RMS-H | 61.78 ± 3.017 y | 241 ± 12 x | 242 ± 12 | 0.98 ± 0.007 x | 6.71 ± 0.19 x |
| RMS-M | 68.75 ± 2.60 y | 267 ± 12 x | 268 ± 12 | 0.98 ± 0.002 x | 6.97 ± 0.122 x |
| Sand | 92.67 ± 1.20 x | 153 ± 46 y | 206 ± 85 | 0.79 ± 0.028 y | 3.37 ± 0.15 z |
| Rice Husk | 84.97 ± 2.43 x | 249 ± 16 x | 285 ± 18 | 0.84 ± 0.028 y | 4.44 ± 0.226 y |
| Target Bacteria | RMS1 (n = 3) | RMS2 (n = 3) | RMS3 (n = 3) | RMS4 (n = 3) | RMS5 (n = 3) | RMSH (n = 9) | RMS M (n = 9) | |
|---|---|---|---|---|---|---|---|---|
| Environmental pathogens | ||||||||
| Yeast (Yea) | (+ +) | 67% | 67% | 89% | 89% | |||
| (+) | 100% | 33% | 100% | 100% | 33% | 11% | 11% | |
| Enterococcus (Ensp) | (+ +) | 100% | 100% | 33% | 100% | 100% | 100% | 100% |
| (+) | 67% | |||||||
| Klebsiella (Klsp) | (+) | 100% | 100% | 33% | 33% | 100% | 100% | 100% |
| (−) | 67% | 67% | ||||||
| Escherichia coli (Ec) | (++) | 100% | 67% | 33% | 22% | |||
| (+) | 33% | 100% | 67% | 67% | 78% | 89% | ||
| (−) | 33% | 11% | ||||||
| Streptococcus uberis (Sub) | (+ + +) | 67% | 100% | 67% | 44% | |||
| (+ +) | 33% | 33% | 22% | 11% | ||||
| (+) | 67% | 11% | ||||||
| (−) | 67% | 33% | 33% | 67% | 44% | |||
| Trueperella pyogenes (Tpy) | (+ +) | 100% | 100% | 100% | ||||
| (+) | 67% | 22% | 78% | |||||
| (−) | 100% | 33% | 78% | 22% | ||||
| Serratia marcescens (Sm) | (+) | 67% | 11% | |||||
| (−) | 100% | 33% | 100% | 100% | 100% | 89% | 100% | |
| Streptococcus dysgalactiae (Sdy) | (+ + +) | 22% | ||||||
| (+) | 67% | 100% | ||||||
| (−) | 33% | 100% | 100% | 100% | 78% | 100% | ||
| Protozoan (Psp) | (−) | 100% | 100% | 100% | 100% | 100% | 100% | 100% |
| Contact pathogen | ||||||||
| Corynebacterium bovis (Cb) | (−) | 100% | 100% | 100% | 100% | 100% | 100% | 100% |
| Staphylococcus aureus (Sau) | (−) | 100% | 100% | 100% | 100% | 100% | 100% | 100% |
| Streptococcus agalactiae (Sag) | (−) | 100% | 100% | 100% | 100% | 100% | 100% | 100% |
| Mycoplasma (Mysp) | (−) | 100% | 100% | 100% | 100% | 100% | 100% | 100% |
| Mycoplasma bovis (Myb) | (−) | 100% | 100% | 100% | 100% | 100% | 100% | 100% |
| β-lactamase resistance gene (Lac) | Others | |||||||
| (−) | 100% | 100% | 100% | 100% | 100% | 100% | 100% | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, H.; Wang, Y.; Dong, L.; Hu, H.; Meng, L.; Liu, H.; Zheng, N.; Wang, J. Microbial Characteristics and Safety of Dairy Manure ComPosting for Reuse as Dairy Bedding. Biology 2021, 10, 13. https://doi.org/10.3390/biology10010013
Wu H, Wang Y, Dong L, Hu H, Meng L, Liu H, Zheng N, Wang J. Microbial Characteristics and Safety of Dairy Manure ComPosting for Reuse as Dairy Bedding. Biology. 2021; 10(1):13. https://doi.org/10.3390/biology10010013
Chicago/Turabian StyleWu, Haoming, Yang Wang, Lei Dong, Haiyan Hu, Lu Meng, Huimin Liu, Nan Zheng, and Jiaqi Wang. 2021. "Microbial Characteristics and Safety of Dairy Manure ComPosting for Reuse as Dairy Bedding" Biology 10, no. 1: 13. https://doi.org/10.3390/biology10010013
APA StyleWu, H., Wang, Y., Dong, L., Hu, H., Meng, L., Liu, H., Zheng, N., & Wang, J. (2021). Microbial Characteristics and Safety of Dairy Manure ComPosting for Reuse as Dairy Bedding. Biology, 10(1), 13. https://doi.org/10.3390/biology10010013






