Quorum Sensing and the Use of Quorum Quenchers as Natural Biocides to Inhibit Sulfate-Reducing Bacteria
Abstract
:1. Introduction
2.1. Sulfate Reducers: Phylogenetic Affiliation and Their Contributory Roles in Industrial Problems
2.2. Current Strategies to Tackle SRBs and Their Limitations
2.3. Biofilm Formation by SRB
2.4. Biofilm Formation: the Role of QS and the Possible Link to Biocorrosion by SRB
2.5. QS in SRBs: What is Known Thus Far?
2.6. QQ as a Potential Green Biocidal Approach to Tackle QS
2.7. Potential Strategies for QQ Application to Tackle SRB
3. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Lin, J.; Ballim, R. Biocorrosion control: Current strategies and promising alternatives. Afr. J. Biotechnol. 2012, 11, 15736–15747. [Google Scholar] [CrossRef]
- AlAbbas, F.M.; Williamson, C.; Bhola, S.M.; Spear, J.R.; Olson, D.L.; Mishra, B.; Kakpovbia, A.E. Influence of sulfate reducing bacterial biofilm on corrosion behavior of low-alloy, high-strength steel (api-5l x80). Int. Biodeter. Biodegr. 2013, 78, 34–42. [Google Scholar] [CrossRef]
- Williamson, R.; Javaherdashti, R.; Tan, H. Corrosion and Materials in the Oil and Gas Industries; Javaherdashti, R., Nwaoha, C., Tan, H., Eds.; CRC Press: Boca Raton, FL, USA, 2013; p. 6. [Google Scholar]
- NACE. Corrosion costs and preventive strategies in the united states. Available online: https://www.nace.org/uploadedFiles/Publications/ccsupp.pdf (accessed on 5 May 2016).
- Heitz, E.; Flemming, H.C.; Sand, W. Microbially Influenced Corrosion of Materials: Scientific and Engineering Aspects; Springer: Berlin/Heidelberg, Germany, 2012; p. 475. [Google Scholar]
- Enning, D.; Garrelfs, J. Corrosion of iron by sulfate-reducing bacteria: New views of an old problem. Appl. Environ. Microbiol. 2014, 80, 1226–1236. [Google Scholar] [CrossRef] [PubMed]
- Sawford, M.K.; Ateya, B.G.; Abdullah, A.M.; Pickering, H.W. The role of oxygen on the stability of crevice corrosion. J. Electrochem. Soc. 2002, 149, B198–B205. [Google Scholar] [CrossRef]
- Kahrilas, G.A.; Blotevogel, J.; Stewart, P.S.; Borch, T. Biocides in hydraulic fracturing fluids: A critical review of their usage, mobility, degradation, and toxicity. Environ. Sci. Technol. 2015, 49, 16–32. [Google Scholar] [CrossRef] [PubMed]
- OSHA. Osha fact sheet formaldehyde. Available online: https://www.osha.gov/OshDoc/data_General_Facts/formaldehyde-factsheet.pdf (accessed on 8 May 2016).
- Muyzer, G.; Stams, A.J.M. The ecology and biotechnology of sulphate-reducing bacteria. Nat. Rev. Microbiol. 2008, 6, 441–454. [Google Scholar] [CrossRef] [PubMed]
- Baumgartner, L.K.; Reid, R.P.; Dupraz, C.; Decho, A.W.; Buckley, D.H.; Spear, J.R.; Przekop, K.M.; Visscher, P.T. Sulfate reducing bacteria in microbial mats: Changing paradigms, new discoveries. Sediment. Geol. 2006, 185, 131–145. [Google Scholar] [CrossRef]
- Decho, A.W.; Visscher, P.T.; Ferry, J.; Kawaguchi, T.; He, L.J.; Przekop, K.M.; Norman, R.S.; Reid, R.P. Autoinducers extracted from microbial mats reveal a surprising diversity of N-acylhomoserine lactones (ahls) and abundance changes that may relate to diel pH. Environ. Microbiol. 2009, 11, 409–420. [Google Scholar] [CrossRef] [PubMed]
- Jonkers, H.; Koh, I.-O.; Behrend, P.; Muyzer, G.; de Beer, D. Aerobic organic carbon mineralization by sulfate-reducing bacteria in the oxygen-saturated photic zone of a hypersaline microbial mat. Microbiol. Ecol. 2005, 49, 291–300. [Google Scholar] [CrossRef] [PubMed]
- Decho, A.W.; Norman, R.S.; Visscher, P.T. Quorum sensing in natural environments: Emerging views from microbial mats. Trends Microbiol. 2010, 18, 73–80. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Wen, F.; Cao, Y. Progress in research of corrosion and protection by sulfate-reducing bacteria. Procedia Environ. Sci. 2011, 10, 1177–1182. [Google Scholar] [CrossRef]
- Kuang, F.; Wang, J.; Yan, L.; Zhang, D. Effects of sulfate-reducing bacteria on the corrosion behavior of carbon steel. Electrochim. Acta 2007, 52, 6084–6088. [Google Scholar] [CrossRef]
- Kato, S. Microbial extracellular electron transfer and its relevance to iron corrosion. Microbial. Biotechnol. 2016, 9, 141–148. [Google Scholar] [CrossRef] [PubMed]
- Hong, P.-Y.; Moosa, N.; Mink, J. Dynamics of microbial communities in an integrated ultrafiltration–Reverse osmosis desalination pilot plant located at the arabian gulf. Desalin. Water Treat. 2016, 57, 16310–16323. [Google Scholar] [CrossRef]
- Turkiewicz, A.; Brzeszcz, J.; Kapusta, P. The application of biocides in the oil and gas industry. Nafta-Gaz 2013, 69, 103–111. [Google Scholar] [CrossRef]
- Videla, H.A. Prevention and control of biocorrosion. Int. Biodeter. Biodegr. 2002, 49, 259–270. [Google Scholar] [CrossRef]
- Chang, Y.J.; Chang, Y.T.; Hung, C.H. The use of magnesium peroxide for the inhibition of sulfate-reducing bacteria under anoxic conditions. J. Ind. Microbiol. Biot. 2008, 35, 1481–1491. [Google Scholar] [CrossRef] [PubMed]
- Wen, J.; Xu, D.; Gu, T.; Raad, I. A green triple biocide cocktail consisting of a biocide, edds and methanol for the mitigation of planktonic and sessile sulfate-reducing bacteria. World J. Microbiol. Biot. 2012, 28, 431–435. [Google Scholar] [CrossRef] [PubMed]
- Wen, J.; Zhao, K.L.; Gu, T.Y.; Raad, I.I. A green biocide enhancer for the treatment of sulfate-reducing bacteria (SRB) biofilms on carbon steel surfaces using glutaraldehyde. Int. Biodeter. Biodegr. 2009, 63, 1102–1106. [Google Scholar] [CrossRef]
- Rasol, R.M.; Noor, N.M.; Yahaya, N.; Abdullah, A.; Abu Bakar, A.; Rashid, A.S.A. Combination effects of ultrasound wave and biocide treatment on the growth of sulfate reducing bacteria (SRB). Desalin. Water Treat. 2014, 52, 3637–3646. [Google Scholar] [CrossRef]
- Korenblum, E.; Goulart, F.R.D.; Rodrigues, I.D.; Abreu, F.; Lins, U.; Alves, P.B.; Blank, A.F.; Valoni, E.; Sebastian, G.V.; Alviano, D.S.; et al. Antimicrobial action and anti-corrosion effect against sulfate reducing bacteria by lemongrass (Cymbopogon citratus) essential oil and its major component, the citral. AMB Express 2013. [Google Scholar] [CrossRef] [PubMed]
- Lavania, M.; Sarma, P.M.; Mandal, A.K.; Cheema, S.; Lal, B. Efficacy of natural biocide on control of microbial induced corrosion in oil pipelines mediated by Desulfovibrio vulgaris and Desulfovibrio gigas. J. Environ. Sci. 2011, 23, 1394–1402. [Google Scholar] [CrossRef]
- Aiad, I.; Emam, D.; El-Deeb, A.; Abd-Alrahman, E. Novel imidazolium-based gemini surfactants: Synthesis, surface properties, corrosion inhibition and biocidal activity against sulfate-reducing bacteria. J. Surfactants Deterg. 2013, 16, 927–935. [Google Scholar] [CrossRef]
- Aiad, I.A.; Tawfik, S.M.; Shaban, S.M.; Abd-Elaal, A.A.; El-Shafie, M. Enhancing of corrosion inhibition and the biocidal effect of phosphonium surfactant compounds for oil field equipment. J. Surfactants Deterg. 2014, 17, 391–401. [Google Scholar] [CrossRef]
- Shaban, S.M.; Saied, A.; Tawfik, S.M.; Abd-Elaal, A.; Aiad, I. Corrosion inhibition and biocidal effect of some cationic surfactants based on schiff base. J. Ind. Eng. Chem. 2013, 19, 2004–2009. [Google Scholar] [CrossRef]
- Labena, A.; Hegazy, M.A.; Horn, H.; Muller, E. The biocidal effect of a novel synthesized gemini surfactant on environmental sulfidogenic bacteria: Planktonic cells and biofilms. Mater. Sci. Eng. 2015, 47, 367–375. [Google Scholar] [CrossRef] [PubMed]
- Braissant, O.; Decho, A.W.; Dupraz, C.; Glunk, C.; Przekop, K.M.; Visscher, P.T. Exopolymeric substances of sulfate-reducing bacteria: Interactions with calcium at alkaline pH and implication for formation of carbonate minerals. Geobiology 2007, 5, 401–411. [Google Scholar] [CrossRef]
- Cordas, C.M.; Guerra, L.T.; Xavier, C.; Moura, J.J. Electroactive biofilms of sulphate reducing bacteria. Electrochim. Acta 2008, 54, 29–34. [Google Scholar] [CrossRef]
- Li, H.B.; Xu, D.K.; Li, Y.C.; Feng, H.; Liu, Z.Y.; Li, X.G.; Gu, T.Y.; Yang, K. Extracellular electron transfer is a bottleneck in the microbiologically influenced corrosion of C1018 carbon steel by the biofilm of sulfate-reducing bacterium Desulfovibrio vulgaris. PLoS ONE 2015. [Google Scholar] [CrossRef] [PubMed]
- Wikieł, A.J.; Datsenko, I.; Vera, M.; Sand, W. Impact of Desulfovibrio alaskensis biofilms on corrosion behaviour of carbon steel in marine environment. Bioelectrochemistry 2014, 97, 52–60. [Google Scholar] [CrossRef] [PubMed]
- Videla, H.C.A. Manual of Biocorrosion; CRC Press: Boca Raton, FL, USA, 1996; p. 273. [Google Scholar]
- Duan, D.X.; Lin, C.G. Effect of surface free energy and electrochemical polarization on attachment of sulfate reducing bacteria. Adv. Mater. Res. 2011, 199, 1967–1972. [Google Scholar] [CrossRef]
- Elmouaden, K.; Jodeh, S.; Chaouay, A.; Oukhrib, R.; Salghi, R.; Bazzi, L.; Hilali, M. Sulfate-reducing bacteria impact on copper corrosion behavior in natural seawater environment. J. Surf. Eng. Mater. Adv. Technol. 2016. [Google Scholar] [CrossRef]
- Beech, I.B.; Sunner, J.A.; Hiraoka, K. Microbe-surface interactions in biofouling and biocorrosion processes. Int. Microbiol. 2005, 8, 157–168. [Google Scholar] [PubMed]
- Lovley, D.R. Live wires: Direct extracellular electron exchange for bioenergy and the bioremediation of energy-related contamination. Energ. Environ. Sci. 2011, 4, 4896–4906. [Google Scholar] [CrossRef]
- Xu, D.; Li, Y.; Gu, T. Mechanistic modeling of biocorrosion caused by biofilms of sulfate reducing bacteria and acid producing bacteria. Bioelectrochemistry 2016, 110, 52–58. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; He, Q.; Hemme, C.L.; Mukhopadhyay, A.; Hillesland, K.; Zhou, A.; He, Z.; van Nostrand, J.D.; Hazen, T.C.; Stahl, D.A. How sulphate-reducing microorganisms cope with stress: Lessons from systems biology. Nat. Rev. Microbiol. 2011, 9, 452–466. [Google Scholar] [CrossRef] [PubMed]
- Wall, J.; Bill Yen, H.; Drury, E.; Barton, L.; Hamilton, W. Evaluation of Stress Response in Sulphate-Reducing Bacteria through Genome Analysis; Cambridge University Press: Cambridge, UK, 2007; pp. 141–165. [Google Scholar]
- Keller, K.L.; Wall, J.D. Genetics and molecular biology of the electron flow for sulfate respiration in Desulfovibrio. Front. Microbiol. 2011. [Google Scholar] [CrossRef] [PubMed]
- Pereira, I.A.C.; Ramos, A.R.; Grein, F.; Marques, M.C.; da Silva, S.M.; Venceslau, S.S. A comparative genomic analysis of energy metabolism in sulfate reducing bacteria and archaea. Front. Microbiol. 2011. [Google Scholar] [CrossRef] [PubMed]
- Price, M.N.; Ray, J.; Wetmore, K.M.; Kuehl, J.V.; Bauer, S.; Deutschbauer, A.M.; Arkin, A.P. The genetic basis of energy conservation in the sulfate-reducing bacterium Desulfovibrio alaskensis g20. Front. Microbiol 2014. [Google Scholar] [CrossRef] [PubMed]
- Caffrey, S.M.; Park, H.S.; Been, J.; Gordon, P.; Sensen, C.W.; Voordouw, G. Gene expression by the sulfate-reducing bacterium Desulfovibrio vulgaris hildenborough grown on an iron electrode under cathodic protection conditions. Appl. Environ. Microbiol. 2008, 74, 2404–2413. [Google Scholar] [CrossRef] [PubMed]
- Krumholz, L.R.; Bradstock, P.; Sheik, C.S.; Diao, Y.; Gazioglu, O.; Gorby, Y.; McInerney, M.J. Syntrophic growth of Desulfovibrio alaskensis requires genes for H2 and formate metabolism as well as those for flagellum and biofilm formation. Appl. Environ. Microbiol. 2015, 81, 2339–2348. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Culley, D.E.; Nie, L.; Scholten, J.C. Comparative transcriptome analysis of Desulfovibrio vulgaris grown in planktonic culture and mature biofilm on a steel surface. Appl. Microbiol. Biotechnol. 2007, 76, 447–457. [Google Scholar] [CrossRef] [PubMed]
- Wegener, G.; Krukenberg, V.; Riedel, D.; Tegetmeyer, H.E.; Boetius, A. Intercellular wiring enables electron transfer between methanotrophic archaea and bacteria. Nature 2015, 526, 587–590. [Google Scholar] [CrossRef] [PubMed]
- Reguera, G.; McCarthy, K.D.; Mehta, T.; Nicoll, J.S.; Tuominen, M.T.; Lovley, D.R. Extracellular electron transfer via microbial nanowires. Nature 2005, 435, 1098–1101. [Google Scholar] [CrossRef] [PubMed]
- Gorby, Y.A.; Yanina, S.; McLean, J.S.; Rosso, K.M.; Moyles, D.; Dohnalkova, A.; Beveridge, T.J.; Chang, I.S.; Kim, B.H.; Kim, K.S.; et al. Electrically conductive bacterial nanowires produced by Shewanella oneidensis strain MR-1 and other microorganisms. Proc. Natl. Acad. Sci. USA 2006, 103, 11358–11363. [Google Scholar] [CrossRef] [PubMed]
- Clark, M.E.; He, Z.; Redding, A.M.; Joachimiak, M.P.; Keasling, J.D.; Zhou, J.Z.; Arkin, A.P.; Mukhopadhyay, A.; Fields, M.W. Transcriptomic and proteomic analyses of Desulfovibrio vulgaris biofilms: Carbon and energy flow contribute to the distinct biofilm growth state. BMC Genom. 2012, 13. [Google Scholar] [CrossRef] [PubMed]
- Defoirdt, T.; Boon, N.; Sorgeloos, P.; Verstraete, W.; Bossier, P. Quorum sensing and quorum quenching in Vibrio harveyi: Lessons learned from in vivo work. ISME J. 2008, 2, 19–26. [Google Scholar] [CrossRef] [PubMed]
- Henke, J.M.; Bassler, B.L. Three parallel quorum-sensing systems regulate gene expression in Vibrio harveyi. J. Bacteriol. 2004, 186, 6902–6914. [Google Scholar] [CrossRef] [PubMed]
- Ng, W.L.; Perez, L.J.; Wei, Y.Z.; Kraml, C.; Semmelhack, M.F.; Bassler, B.L. Signal production and detection specificity in Vibrio CqsA/CqsS quorum-sensing systems. Mol. Microbiol. 2011, 79, 1407–1417. [Google Scholar] [CrossRef] [PubMed]
- Rezzonico, F.; Duffy, B. Lack of genomic evidence of AI-2 receptors suggests a non-quorum sensing role for luxs in most bacteria. BMC Microbiol. 2008. [Google Scholar] [CrossRef] [PubMed]
- Pereira, C.S.; Thompson, J.A.; Xavier, K.B. Ai-2-mediated signalling in bacteria. FEMS Microbiol. Rev. 2013, 37, 156–181. [Google Scholar] [CrossRef] [PubMed]
- Weiland-Bräuer, N.; Kisch, M.J.; Pinnow, N.; Liese, A.; Schmitz, R.A. Highly effective inhibition of biofilm formation by the first metagenome-derived AI-2 quenching enzyme. Front. Microbiol. 2016. [Google Scholar] [CrossRef] [PubMed]
- Surette, M.G.; Miller, M.B.; Bassler, B.L. Quorum sensing in Escherichia coli, Salmonella typhimurium, and Vibrio harveyi: A new family of genes responsible for autoinducer production. Proc. Natl. Acad. Sci. USA 1999, 96, 1639–1644. [Google Scholar] [CrossRef] [PubMed]
- Aubert, D.F.; O′Grady, E.P.; Hamad, M.A.; Sokol, P.A.; Valvano, M.A. The burkholderia cenocepacia sensor kinase hybrid AtsR is a global regulator modulating quorum-sensing signalling. Environ. Microbiol. 2013, 15, 372–385. [Google Scholar] [CrossRef] [PubMed]
- Yarwood, J.M.; Schlievert, P.M. Quorum sensing in Staphylococcus infections. J. Clin. Investig. 2003, 112, 1620–1625. [Google Scholar] [CrossRef] [PubMed]
- Vuong, C.; Saenz, H.L.; Gotz, F.; Otto, M. Impact of the agr quorum-sensing system on adherence to polystyrene in Staphylococcus aureus. J. Infect. Dis. 2000, 182, 1688–1693. [Google Scholar] [CrossRef] [PubMed]
- Pratten, J.; Foster, S.J.; Chan, P.F.; Wilson, M.; Nair, S.P. Staphylococcus aureus accessory regulators: Expression within biofilms and effect on adhesion. Microbes Infect. 2001, 3, 633–637. [Google Scholar] [CrossRef]
- Nickzad, A.; Lepine, F.; Deziel, E. Quorum sensing controls swarming motility of Burkholderia glumae through regulation of rhamnolipids. PLoS ONE 2015. [Google Scholar] [CrossRef] [PubMed]
- Blus-Kadosh, I.; Zilka, A.; Yerushalmi, G.; Banin, E. The effect of pstS and phoB on quorum sensing and swarming motility in Pseudomonas aeruginosa. PLoS ONE 2013. [Google Scholar] [CrossRef] [PubMed]
- Krysciak, D.; Grote, J.; Orbegoso, M.R.; Utpatel, C.; Forstner, K.U.; Li, L.; Schmeisser, C.; Krishnan, H.B.; Streit, W.R. RNA sequencing analysis of the broad-host-range strain sinorhizobium fredii NGR234 identifies a large set of genes linked to quorum sensing-dependent regulation in the background of a trai and ngri deletion mutant. Appl. Environ. Microbiol. 2014, 80, 5655–5671. [Google Scholar] [CrossRef] [PubMed]
- Gao, R.; Krysciak, D.; Petersen, K.; Utpatel, C.; Knapp, A.; Schmeisser, C.; Daniel, R.; Voget, S.; Jaeger, K.E.; Streit, W.R. Genome-wide RNA sequencing analysis of quorum sensing-controlled regulons in the plant-associated Burkholderia glumae PG1 strain. Appl. Environ. Microbiol. 2015, 81, 7993–8007. [Google Scholar] [CrossRef] [PubMed]
- Sperandio, V.; Mellies, J.L.; Nguyen, W.; Shin, S.; Kaper, J.B. Quorum sensing controls expression of the type III secretion gene transcription and protein secretion in enterohemorrhagic and enteropathogenic Escherichia coli. Proc. Natl. Acad. Sci. USA 1999, 96, 15196–15201. [Google Scholar] [CrossRef] [PubMed]
- Sha, J.; Pillai, L.; Fadl, A.A.; Galindo, C.L.; Erova, T.E.; Chopra, A.K. The type III secretion system and cytotoxic enterotoxin alter the virulence of Aeromonas hydrophila. Infect. Immun. 2005, 73, 6446–6457. [Google Scholar] [CrossRef] [PubMed]
- Schuster, M.; Lostroh, C.P.; Ogi, T.; Greenberg, E.P. Identification, timing, and signal specificity of Pseudomonas aeruginosa quorum-controlled genes: A transcriptome analysis. J. Bacteriol. 2003, 185, 2066–2079. [Google Scholar] [CrossRef] [PubMed]
- Decho, A.W.; Kawaguchi, T.; Chen, Y.P. High Throughput in vitro Translation (Cell-Lysate Based) Assay for Detecting Quorum Sensing Signals. Patent 20140030750, 30 January 2015. [Google Scholar]
- Kawaguchi, T.; Chen, Y.P.; Norman, R.S.; Decho, A.W. Rapid screening of quorum-sensing signal N-acyl homoserine lactones by an in vitro cell-free assay. Appl. Environ. Microbiol. 2008, 74, 3667–3671. [Google Scholar] [CrossRef] [PubMed]
- Montgomery, K.; Charlesworth, J.C.; LeBard, R.; Visscher, P.T.; Burns, B.P. Quorum sensing in extreme environments. Life 2013, 3, 131–148. [Google Scholar] [CrossRef] [PubMed]
- Kimura, N. Metagenomic approaches to understanding phylogenetic diversity in quorum sensing. Virulence 2014, 5, 433–442. [Google Scholar] [CrossRef] [PubMed]
- Brady, S.F.; Chao, C.J.; Clardy, J. Long-chain N-acyltyrosine synthases from environmental DNA. Appl. Environ. Microbiol. 2004, 70, 6865–6870. [Google Scholar] [CrossRef] [PubMed]
- Williamson, L.L.; Borlee, B.R.; Schloss, P.D.; Guan, C.H.; Allen, H.K.; Handelsman, J. Intracellular screen to identify metagenomic clones that induce or inhibit a quorum-sensing biosensor. Appl. Environ. Microbiol. 2005, 71, 6335–6344. [Google Scholar] [CrossRef] [PubMed]
- Kaksonen, A.H.; Spring, S.; Schumann, P.; Kroppenstedt, R.M.; Puhakka, J.A. Desulfotomaculum alcoholivorax sp. nov., a moderately thermophilic, spore-forming, sulfate-reducer isolated from a fluidized-bed reactor treating acidic metal- and sulfate-containing wastewater. Int. J. Syst. Evol. Microbiol. 2008, 58, 833–838. [Google Scholar] [CrossRef] [PubMed]
- Mayilraj, S.; Kaksonen, A.H.; Cord-Ruwisch, R.; Schumann, P.; Spröer, C.; Tindall, B.J.; Spring, S. Desulfonauticus autotrophicus sp. nov., a novel thermophilic sulfate-reducing bacterium isolated from oil-production water and emended description of the genus Desulfonauticus. Extremophiles 2009, 13, 247–255. [Google Scholar] [CrossRef] [PubMed]
- LaRock, C.N.; Yu, J.; Horswill, A.R.; Parsek, M.R.; Minion, F.C. Transcriptome analysis of acetyl-homoserine lactone-based quorum sensing regulation in Yersinia pestis. PLoS ONE 2013. [Google Scholar] [CrossRef]
- Khan, R.; Shen, F.; Khan, K.; Liu, L.X.; Wu, H.H.; Luo, J.Q.; Wan, Y.H. Biofouling control in a membrane filtration system by a newly isolated novel quorum quenching bacterium, Bacillus methylotrophicus sp. WY. Rsc. Adv. 2016, 6, 28895–28903. [Google Scholar] [CrossRef]
- Ivanova, K.; Fernandes, M.M.; Francesko, A.; Mendoza, E.; Guezguez, J.; Burnet, M.; Tzanov, T. Quorum-quenching and matrix-degrading enzymes in multilayer coatings synergistically prevent bacterial biofilm formation on urinary catheters. ACS Appl. Mater. Interfaces 2015, 7, 27066–27077. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Lara, B.; Saucedo-Mora, M.A.; Roldan-Sanchez, J.A.; Perez-Eretza, B.; Ramasamy, M.; Lee, J.; Coria-Jimenez, R.; Tapia, M.; Varela-Guerrero, V.; Garcia-Contreras, R. Inhibition of quorum-sensing-dependent virulence factors and biofilm formation of clinical and environmental Pseudomonas aeruginosa strains by zno nanoparticles. Lett. Appl. Microbiol. 2015, 61, 299–305. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Park, S.-K.; Kwon, H.; Lee, S.H.; Lee, K.; Nahm, C.H.; Jo, S.J.; Oh, H.-S.; Park, P.-K.; Choo, K.-H. Crossing the border between laboratory and field: Bacterial quorum quenching for anti-biofouling strategy in an MBR. Environ. Sci. Technol. 2016, 50, 1788–1795. [Google Scholar] [CrossRef] [PubMed]
- Siddiqui, M.F.; Rzechowicz, M.; Harvey, W.; Zularisam, A.W.; Anthony, G.F. Quorum sensing based membrane biofouling control for water treatment: A review. J. Water Proc. Eng. 2015, 7, 112–122. [Google Scholar] [CrossRef]
- Chan, K.-G.; Liu, Y.-C.; Chang, C.-Y. Inhibiting N-acyl-homoserine lactone synthesis and quenching Pseudomonas quinolone quorum sensing to attenuate virulence. Front. Microbiol. 2015. [Google Scholar] [CrossRef] [PubMed]
- Shen, G.; Rajan, R.; Zhu, J.; Bell, C.E.; Pei, D. Design and synthesis of substrate and intermediate analogue inhibitors of S-ribosylhomocysteinase. J. Med. Chem. 2006, 49, 3003–3011. [Google Scholar] [CrossRef] [PubMed]
- Widmer, K.; Soni, K.; Hume, M.; Beier, R.; Jesudhasan, P.; Pillai, S. Identification of poultry meat-derived fatty acids functioning as quorum sensing signal inhibitors to autoinducer-2 (AI-2). J. Food Sci. 2007, 72, M363–M368. [Google Scholar] [CrossRef] [PubMed]
- Hentzer, M.; Riedel, K.; Rasmussen, T.B.; Heydorn, A.; Andersen, J.B.; Parsek, M.R.; Rice, S.A.; Eberl, L.; Molin, S.; Hoiby, N.; et al. Inhibition of quorum sensing in Pseudomonas aeruginosa biofilm bacteria by a halogenated furanone compound. Microbiol-Sgm 2002, 148, 87–102. [Google Scholar] [CrossRef] [PubMed]
- Katebian, L.; Gomez, E.; Skillman, L.; Li, D.; Ho, G.; Jiang, S.C. Inhibiting quorum sensing pathways to mitigate seawater desalination ro membrane biofouling. Desalination 2016, 393, 135–143. [Google Scholar] [CrossRef]
- Almasoud, A.; Hettiarachchy, N.; Rayaprolu, S.; Babu, D.; Kwon, Y.M.; Mauromoustakos, A. Inhibitory effects of lactic and malic organic acids on autoinducer type 2 (AI-2) quorum sensing of Escherichia coli O157:H7 and Salmonella typhimurium. LWT-Food Sci. Technol. 2016, 66, 560–564. [Google Scholar] [CrossRef]
- Ibacache-Quiroga, C.; Ojeda, J.; Espinoza-Vergara, G.; Olivero, P.; Cuellar, M.; Dinamarca, M. The hydrocarbon-degrading marine Bacterium cobetia sp. strain mm1ida2h-1 produces a biosurfactant that interferes with quorum sensing of fish pathogens by signal hijacking. Microbial Biotechnol. 2013, 6, 394–405. [Google Scholar] [CrossRef] [PubMed]
- Abbas, R.; Saudi Aramco, Dhahran, Saudi Arabia. Personal communication, 2016.
- Galloway, W.R.; Hodgkinson, J.T.; Bowden, S.; Welch, M.; Spring, D.R. Applications of small molecule activators and inhibitors of quorum sensing in gram-negative bacteria. Trends Microbiol. 2012, 20, 449–458. [Google Scholar] [CrossRef] [PubMed]
- Teasdale, M.E.; Liu, J.; Wallace, J.; Akhlaghi, F.; Rowley, D.C. Secondary metabolites produced by the marine bacterium Halobacillus salinus that inhibit quorum sensing-controlled phenotypes in gram-negative bacteria. Appl. Environ. Microbiol. 2009, 75, 567–572. [Google Scholar] [CrossRef] [PubMed]
- Golberg, K.; Pavlov, V.; Marks, R.S.; Kushmaro, A. Coral-associated bacteria, quorum sensing disrupters, and the regulation of biofouling. Biofouling 2013, 29, 669–682. [Google Scholar] [CrossRef] [PubMed]
- Liaqat, I.; Bachmann, R.T.; Edyvean, R.G.J. Type 2 quorum sensing monitoring, inhibition and biofilm formation in marine microrganisms. Curr. Microbiol. 2014, 68, 342–351. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.Y.; Song, G.J.; Zhu, Q.; Liu, S.J.; Xia, C.H. The influence of bacterial quorum-sensing inhibitors against the formation of the diatom-biofilm. Chem. Ecol. 2016, 32, 169–181. [Google Scholar] [CrossRef]
- Santhakumari, S.; Kannappan, A.; Pandian, S.K.; Thajuddin, N.; Rajendran, R.B.; Ravi, A.V. Inhibitory effect of marine cyanobacterial extract on biofilm formation and virulence factor production of bacterial pathogens causing vibriosis in aquaculture. J. Appl. Phycol. 2016, 28, 313–324. [Google Scholar] [CrossRef]
- Mai, T.; Tintillier, F.; Lucasson, A.; Moriou, C.; Bonno, E.; Petek, S.; Magre, K.; Al Mourabit, A.; Saulnier, D.; Debitus, C. Quorum sensing inhibitors from Leucetta chagosensis dendy, 1863. Lett. Appl. Microbiol. 2015, 61, 311–317. [Google Scholar] [CrossRef] [PubMed]
- Nithya, C.; Pandian, S.K. Bacillus pumilus of palk bay origin inhibits quorum-sensing-mediated virulence factors in gram-negative bacteria. Res. Microbiol. 2010, 161, 293–304. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.; Zhou, Z.; Cao, Y.; Bai, Y.; Yao, B. High yield expression of an AHL-lactonase from Bacillus sp. B546 in pichia pastoris and its application to reduce aeromonas hydrophila mortality in aquaculture. Microbial Cell Factories 2010. [Google Scholar] [CrossRef] [PubMed]
- Seo, M.J.; Lee, B.S.; Pyun, Y.R.; Park, H. Isolation and characterization of N-acylhomoserine lactonase from the thermophilic bacterium, Geobacillus caldoxylosilyticus ys-8. Biosci. Biotechnol. Biochem. 2011, 75, 1789–1795. [Google Scholar] [CrossRef] [PubMed]
- Easwaran, N.; Karthikeyan, S.; Sridharan, B.; Gothandam, K.M. Identification and analysis of the salt tolerant property of ahl lactonase (aiiatsawb) of Bacillus species. J. Basic Microbiol. 2015, 55, 579–590. [Google Scholar] [CrossRef] [PubMed]
- Hiblot, J.; Gotthard, G.; Chabriere, E.; Elias, M. Structural and enzymatic characterization of the lactonase sis lac from Sulfolobus islandicus. PLoS ONE 2012, 7, e47028. [Google Scholar] [CrossRef] [PubMed]
- Merone, L.; Mandrich, L.; Rossi, M.; Manco, G. A thermostable phosphotriesterase from the archaeon Sulfolobus solfataricus: Cloning, overexpression and properties. Extremophiles 2005, 9, 297–305. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; He, S.; Zhou, Z.; Zhang, M.; Mao, W.; Zhang, H.; Yao, B. Orally administered thermostable N-acyl homoserine lactonase from Bacillus sp. strain AI96 attenuates aeromonas hydrophila infection in zebrafish. Appl. Environ. Microbiol. 2012, 78, 1899–1908. [Google Scholar] [CrossRef] [PubMed]
- Vinoj, G.; Vaseeharan, B.; Thomas, S.; Spiers, A.J.; Shanthi, S. Quorum-quenching activity of the ahl-lactonase from Bacillus licheniformis dahb1 inhibits Vibrio biofilm formation in vitro and reduces shrimp intestinal colonisation and mortality. Marine Biotechnol. 2014, 16, 707–715. [Google Scholar] [CrossRef] [PubMed]
- Mayer, C.; Romero, M.; Muras, A.; Otero, A. Aii20j, a wide-spectrum thermostable N-acylhomoserine lactonase from the marine bacterium Tenacibaculum sp. 20j, can quench ahl-mediated acid resistance in Escherichia coli. Appl. Microbiol. Biotechnol. 2015, 99, 9523–9539. [Google Scholar] [CrossRef] [PubMed]
- Morohoshi, T.; Tominaga, Y.; Someya, N.; Ikeda, T. Characterization of a novel thermostable N-acylhomoserine lactonase from the thermophilic bacterium Thermaerobacter marianensis. J. Biosci. Bioeng. 2015, 120, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Lin, Y.; Yi, S.; Liu, P.; Shen, J.; Shao, Z.; Liu, Z. Qsdh, a novel ahl lactonase in the rnd-type inner membrane of marine Pseudoalteromonas byunsanensis strain 1a01261. PLoS ONE 2012, 7, e46587. [Google Scholar] [CrossRef] [PubMed]
- Tang, K.; Su, Y.; Brackman, G.; Cui, F.; Zhang, Y.; Shi, X.; Coenye, T.; Zhang, X.-H. Moml, a novel marine-derived N-acyl homoserine lactonase from Muricauda olearia. Appl. Environ. Microbiol. 2015, 81, 774–782. [Google Scholar] [CrossRef] [PubMed]
- Tang, K.H.; Zhang, Y.H.; Yu, M.; Shi, X.C.; Coenye, T.; Bossier, P.; Zhang, X.H. Evaluation of a new high-throughput method for identifying quorum quenching bacteria. Sci. Rep. 2013. [Google Scholar] [CrossRef] [PubMed]
- Chow, J.Y.; Xue, B.; Lee, K.H.; Tung, A.; Wu, L.; Robinson, R.C.; Yew, W.S. Directed evolution of a thermostable quorum-quenching lactonase from the amidohydrolase superfamily. J. Biol. Chem. 2010, 285, 40911–40920. [Google Scholar] [CrossRef] [PubMed]
- Fetzner, S. Quorum quenching enzymes. J. Biotechnol. 2015, 201, 2–14. [Google Scholar] [CrossRef] [PubMed]
- Roy, V.; Fernandes, R.; Tsao, C.Y.; Bentley, W.E. Cross species quorum quenching using a native AI-2 processing enzyme. ACS Chem. Biol. 2010, 5, 223–232. [Google Scholar] [CrossRef] [PubMed]
- Maqbool, T.; Khan, S.J.; Waheed, H.; Lee, C.H.; Hashmi, I.; Iqbal, H. Membrane biofouling retardation and improved sludge characteristics using quorum quenching bacteria in submerged membrane bioreactor. J. Membr. Sci. 2015, 483, 75–93. [Google Scholar] [CrossRef]
- Oh, H.S.; Yeon, K.M.; Yang, C.S.; Kim, S.R.; Lee, C.H.; Park, S.Y.; Han, J.Y.; Lee, J.K. Control of membrane biofouling in MBR for wastewater treatment by quorum quenching bacteria encapsulated in microporous membrane. Environ. Sci. Technol. 2012, 46, 4877–4884. [Google Scholar] [CrossRef] [PubMed]
- Kalia, V.C.; Wood, T.K.; Kumar, P. Evolution of resistance to quorum-sensing inhibitors. Microbial Ecol. 2014, 68, 13–23. [Google Scholar] [CrossRef] [PubMed]
- Yeon, K.M.; Lee, C.H.; Kim, J. Magnetic enzyme carrier for effective biofouling control in the membrane bioreactor based on enzymatic quorum quenching. Environ. Sci. Technol. 2009, 43, 7403–7409. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Liu, Y. Control and cleaning of membrane biofouling by energy uncoupling and cellular communication. Environ. Sci. Technol. 2010, 45, 595–601. [Google Scholar] [CrossRef] [PubMed]
| 4 H2 + SO42− + H+ = HS− + 4 H2O | ΔG0′ (KJ/rx) = −151.9 |
| CH3COO− + SO42− = 2 HCO3− + HS− | ΔG0′ (KJ/rx) = −47.6 |
| CH3CH2COO− +0.75 SO42− = CH3COO− + HCO3− 0.75 HS− + 0.25 H+ | ΔG0′ (KJ/rx) = −37.7 |
| CH₃CH₂CH₂COO− + 0.5 SO42− = 2 CH3COO− + 0.5 HS− + 0.5 H+ | ΔG0′ (KJ/rx) = −27.8 |
| CH3CHOHCOO− + 0.5 SO42− = CH3COO− + HCO3− + 0.5 HS− | ΔG0′ (KJ/rx) = −80.2 |
| Class | Biocide | Action | Dosage | Other Information | Ref. |
|---|---|---|---|---|---|
| Oxidizing biocides | Chlorine | Direct oxidation, destruction of the cell walls through modification of membrane permeability, leakage of cellular constituents, protein inactivation, damage of nucleic acid. | 0.5 ppm | They have numerous disadvantages: (i) interaction with other chemicals to result in toxic disinfectant byproducts (ii) contribute to corrosion of structural metals (iii) weaken the integrity of non-metallic components (iv) ineffective against bacteria embedded within biofilm matrix. | [1,20] |
| Bromine | 0.05–0.1 ppm | ||||
| Ozone | 0.2–0.5 ppm | ||||
| Hydrogen peroxide | 50–100 ppm | ||||
| Magnesium peroxide/ORC™ | 1%–2% MgO2: 1% MgO2 + 1% ORC | [21] | |||
| Non-oxidizing biocides | Glutaraldehyde | Reacts with proteins on the cell membrane and cytoplasm. | 10–70 ppm | Generally toxic and persistent in the environment into which they are being discarded. To reduce the dosage, in the recent past they have been tested in a cocktail with 1000-2000 ppm Ethylenediaminedisuccinate (EDDS), a chelator that increases the permeability of membranes by chelating with Mg2+ and Ca2+, and methanol or ethanol that denature the proteins of the outer membrane [22,23]. The use of ultrasound was also able to increase their efficacy [24]. | [1,20] |
| QUATS (Quaternary ammonium compounds) | Impose detergent effect on cell, dissolute lipids and thus cause loss of cellular content. | 8–35 ppm | |||
| Isothiazolones | Exhibit cytotoxicity on different types of cells. | 0.9–10 ppm | |||
| MTB (Methylene-bisthiocyanate) | Prevents cell growth by blocking essential chemical reactions that occur within the cell. | 1.5–8 ppm | |||
| THPS (tetrakishydroxymethyl phosphonium sulfate) | Cytotoxic effect, with loss of membrane integrity. Mainly used in water treatment systems and oil field operations. Low environmental toxicity. | 10–90 ppm | |||
| Natural biocides | Lemongrass essential oil and citrus | Antimicrobial effect due to membrane alteration and formation of electron-dense inclusions. Loss of ions and reduction of membrane potential will occur. | 0.17–0.84 ppm | Showed limited efficacies in large-scale operations. | [25] |
| Cow Urine | Reduces the planktonic and biofilm population in the same way. A reduction of sulfide, Fe(III), and EPS production was observed. | 25 ppm | [26] | ||
| Surfactants | Imidazolium-based Gemini Surfactants | Amphiphilic molecules create a biomolecular layer on the metal surface. Also, hydrophobic chains of surfactants can penetrate through bacterial cell membranes, leading to strong bacterial damage. Shchiff bases are usually used to synthesize other antibacterial compounds. | 5000 ppm | Applied in the oil and gas industry to reduce the action of SRB to delay the biocorrosion process. | [27] |
| Phosphonium Surfactant compounds | 50–400 ppm for 3 h | [28] | |||
| Cationic surfactants based on Schiff bases | 20–400 ppm on cultured media | [29] | |||
| Gemini Surfactant | Forms a protective film on the surface. Electrostatic interaction between the negatively charged cell membrane (lipoprotein) and the positively charged ammonium groups of the synthesized gemini surfactant. Moreover, physical disruption of the bacterial cell membrane takes place when the surfactant’s alkyl hydrophobic chain penetrates into the bacterial cell membrane. | 1 mM | [30] |
| Protein | SRB | Best Matched Protein Name in Database that was Homologous to the Listed QS Protein | Amino Acids Identity % | Query Cover % | E Value |
|---|---|---|---|---|---|
| LuxS | Desulfovibrio hydrothermalis | S-ribosylhomocysteine lyase | 33 | 100 | 3E-19 |
| Desulfovibrio salexigens | Quorum sensing AI-2, LuxS | 34 | 100 | 2E-18 | |
| Desulfotalea psychrophila | Probable q. s. AI-2 production protein, LuxS | 34 | 85 | 4E-16 | |
| LuxR | Desulfovibrio desulfuricans | 2 components transcriptional regulator | 38 | 99 | 4E-47 |
| Desulfovibrio africanus | Transcriptional regulator, LuxR family | 31 | 98 | 7E-21 | |
| Desulfovibrio africanus | 2 components transcriptional regulator | 32 | 97 | 2E-34 | |
| Desulfovibrio magneticus RS-1 | LuxR family transcriptional regulator | 48 | 34 | 1E-17 | |
| Desulfotomaculum nigrificans | 2 components transcriptional regulator, LuxR family | 35 | 99 | 5E-42 | |
| Desulfotomaculum acetoxidans | 2 components transcriptional regulator, LuxR family | 24 | 94 | 3E-08 | |
| Desulfotomaculum acetoxidans | Transcriptional regulator, LuxR family | 27 | 94 | 1E-15 | |
| Desulfotomaculum kuznetsovii | 2 components transcriptional regulator, LuxR family | 38 | 99 | 3E-49 | |
| Desulfotomaculum reducens | 2 components transcriptional regulator, LuxR family | 38 | 99 | 3E-41 | |
| Desulfotomaculum ruminis | Regulatory protein, LuxR | 52 | 16 | 2E-11 | |
| Desulfosarcina cetonica | LuxR family transcriptional regulator | 26 | 90 | 6E-16 | |
| Desulfobacterium autotrophicum | 2 components transcriptional regulator, LuxR family | 39 | 98 | 2E-43 | |
| Desulfobacterium autotrophicum | LuxR family transcriptional regulator | 47 | 17 | 2E-05 | |
| Desulfobulbus propionicus DSM 2032 | 2 components transcriptional regulator, LuxR family | 34 | 98 | 5E-41 | |
| Desulfovibrio vulgaris (str. Hildenborough) | LuxR family transcriptional regulator | 32 | 93 | 2E-29 | |
| Syntrophobacter fumaroxidans | 2 components transcriptional regulator, LuxR family | 40 | 96 | 2e-51 | |
| Thermodesulfobium narugense | 2 components transcriptional regulator, LuxR family | 33 | 90 | 4e-32 | |
| Thermodesulfovibrio aggregans | LuxR family transcriptional regulator | 35 | 99 | 3e-36 | |
| LuxP | Desulfovibrio piezophilus | AI-2 binding perisplatic protein, LuxP | 42 | 95 | 7E-95 |
| Desulfovibrio hydrothermalis | AI-2 binding perisplatic protein, LuxP | 43 | 94 | 6E-103 | |
| Desulfovibrio alaskensis | AI-2 binding perisplatic protein, LuxP precursor | 44 | 89 | 2E-105 | |
| LuxQ | Desulfovibrio salexigens | PAS/PAC sensor signal transduction histidine kinase | 29 | 31 | 2E-22 |
| Desulfovibrio hydrothermalis | Signal transduction histidine kinase | 31 | 31 | 5E-23 | |
| LuxO | Desulfotignum phosphitoxidans | Luminescence regulatory protein, LuxO | 45 | 67 | 7E-85 |
| Desulfovibrio salexigens | PAS modulated sigma54 specific transcriptional | 53 | 55 | 2E-83 | |
| Desulfovibrio magneticus RS-1 | Fis family transcriptional regulator | 43 | 68 | 5E-75 | |
| Desulfovibrio vulgaris | Sigma54 specific transcriptional regulator | 46 | 53 | 4E-82 | |
| Desulfovibrio hydrothermalis | PAS modulated sigma54 specific transcriptional | 54 | 55 | 1E-84 | |
| Desulfovibrio africanus | PAS modulated sigma54 specific transcriptional | 39 | 69 | 4E-81 | |
| CqsS | Desulfovibrio salexigens | PAS/ signal transduction histidine kinase | 34 | 52 | 1E-83 |
| Desulfovibrio magneticus | Multi-sensor hybrid histidine kinase | 39 | 28 | 8E-77 |
| Name | Origin | Structure | Action Mechanism | Treatment Condition | QSI Effect | Ref. |
|---|---|---|---|---|---|---|
| N (2′-phenylethyl)-Isobutyramide 3-methyl-N (2′-phenylethyl)-butyramide | Halobacillus salinus C42 (sea grass) | 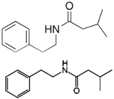 | Analog: Competes with N-acyl homoserine lactones for receptor binding | 30 °C in marine broth | QS system inhibition of AHL in biosensor Chromobacterium violaceum | [94] |
| Unknown compound; AHL QSI | Favia sp. coral isolate Fav 2-50-7 (>98% similarity to Vibrio harveyi) | Possible AHL analog | Maintains anti-QS activity at high temperature (from 26 °C to 95 °C) | Antibiofilm of P. aeruginosa by QS inhibition | [95] | |
| Penicillic acid | commercial |  | Autoinducer antagonist that may interfere with the stability and function of the autoinducer synthase or QS regulator protein | artificial seawater 30 °C | Inhibit AI-2 activity and biofilm formation of marine strain Halomonas pacific, QS inhibition dose is 25 µM concentration | [96] |
| Patulin | commercial | 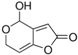 | ||||
| Vanillin | commercial | 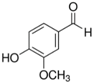 | Interfere or modify the structure of AHL to hinder the binding of AHL to receptor protein | Seawater environment | Reduce seawater desalination RO membrane biofouling. Vanillin suppresses EPS production for various marine bacterial communities on the RO membrane surface, QS inhibition dose is 1200 mg/L | [89] |
| Cinnamaldehyde |  | Reduce the DNA-binding ability of LuxR | ||||
| 4-nitropyridine-N-oxide | Synthetic Compound | QSI analogue | Seawater environment | Inhibits the formation of diatom-biofilm caused by two marine diatoms Cylinthrotheca sp. and Nitzschia closterium. QS inhibition dose is 10 mg/L | [97] | |
| Hexadecanoic acid | Marine cyanobac terium Synechococcus sp. Q-25 | antagonistic binding to the AHL receptor protein | Marine LB broth (pH 7.5 ± 0.2) at 30 °C | Reduces the biofilm and EPS formation of marine infectious pathogens Vibrio harveyi and Vibrio vulnificus | [98] | |
| Isonaamidine A | Marine sponge Leucetta chagosensis | AI-2 inhibitor | Artificial seawater | Inhibits strongly the AI-2 channel of Vibrio harveyi | [99] |
| Name | Origin | Property | Quenching Effect/Target | Ref. |
|---|---|---|---|---|
| AHL acylase | Bacillus pumilus S8-07 (Palk Bay) | Retains activity after incubation at 70 °C for 10 min. | Causes reduction of virulence factors and biofilm in Pseudomonas aeruginosa PAO1 and Serratia marcescens | [100] |
| AHL lactonase (AiiAB546) | Bacillus sp. B546 (mud of a fish pond) | Shows optimal activity at pH 8.0, 20 °C, stable at pH 8.0–12.0, however also remains thermostable at 70 °C and is highly resistant to proteases. | C10-HSL, C12-HSL, C6-HSL, 3-oxo-C6-HSL, 3-oxo-C8-HSL, C8-HSL Attenuates Aeromonas hydrophila infection in carp | [101] |
| AHL lactonase | Geobacillus caldoxylosilyticus YS-8, (volcano soil) | Exhibits activity over a wide temperature range of 30–70 °C, optimal temperature and pH: 50 °C and pH 7.5. | C6-HSL, 3-oxo-C12-HSL, 3-oxo-C6-AHL, C8-HSL | [102] |
| AHL lactonase (AiiA TSAWB) | Bacillus sp. TSAWB (salty soil) | Shows hydrolysis activity in presence of 0%–5% salinity. | C10-HSL | [103] |
| AHL lactonase (SisLac) | Bacillus sp. TSAWB (salty soil) | Optimal activity at pH 9.0, enzymatic half-life of 84 min at 85 °C. | C8-HSL, and C10-HSL | [104] |
| Phosphotriesterase-like lactonases (SsoPox) | Hyperthermophilic archaeon Sulfolobus solfataricus MT4 | Exhibits activity over a broad pH range of 5.0–9.5, thermostable at 70 °C to 85 °C. | 3-oxo-C8-HSL, 3-O-C6-HSL, C4-HSL | [105] |
| AHL lactonase (AiiAAI96) | Bacillus sp. AI96 (pond sediment) | Possesses high activity under broad conditions: ranging from pH 6.0 to 8.5 and 10 °C to 40 °C. Also stable at 70°C, pH 8.0 for at least 1 h. | C4-HSL, C6-HSL, C7-HSL, C8-HSL, C10-HSL, C12-HSL, C14-HSL, 3-oxo-C8-HSL, 3-oxo-C6-HSL, 3-oxo-C10-HSL, 3-oxo-C12-HSL, 3-oxo-C14-HSL, 3-hydroxy-C8-HSL, 3-hydroxy-C14-HSL. Attenuates Aeromonas hydrophila infection in zebrafish by oral feeding. | [106] |
| AHL lactonase (AiiA) | Bacillus licheniformis DAHB1 | Optimal activity at pH: 7.0–8.0 and temperature range: 30–50 °C. Maintains 90% activity after incubation at 60 °C–80 °C for 1 h. Resistant to acidic environment and proteases. | C4-HSL, C6-HSL, 3-oxo-C6-HSL, C8-HSL, 3-oxo-C8-HSL, C10-HSL, C12-HSL, C14-HSL, Inhibits biofilm formation and viable counts of Vibrio parahaemolyticus and attenuates infection and mortality of shrimps in aquaculture | [107] |
| AHL lactonase (Aii20J) | Marine bacteria Tenacibaculum sp. strain 20 J | Crude enzyme stays active under 100 °C for 10 min, resistant to proteinase K and α-chymotrypsin, unaffected by wide ranges of pH. | C4-HSL, C6-HSL, C8-HSL, C10-HSL, C12-HSL, C14-HSL, 3-oxo-C6-HSL, 3-oxo-C12-HSL, 3-oxo-C10-HSL, 3-OH-C10-HSL, 3-oxo-C12-HSL, 3-OH-C12-HSL, 3-oxo-C13-HSL, 3-oxo-C14-HSL, Quenches AHL-mediated acid resistance in Escherichia coli | [108] |
| AHL lactonase (AiiT) | Marine bacteria Thermaerobacter marianensis JCM 10246 | Shows AHL degradation activity at temperature ranging from 40 to 80°C. Maintains 80% of enzyme activity after incubation at 40, 60 and 70 °C for 10 min. | C6-HSL, C8-HSL, C10-HSL | [109] |
| AHL lactonase (QsdH) | Pseudoalteromonas byunsanensis strain 1A01261 | Exhibits activity over a temperature range of 20–60 °C. Stays active after 60 °C for 30 min. | 3-oxo-C8-HSL, 3-oxo-C6-HSL, C4-HSL, C6-HSL, C8-HSL, C10-HSL, C12-HSL, C14-HSL, Attenuates pathogenicity of plant pathogen Erwinia carotovora under 0.15 M NaCl | [110] |
| AHL lactonase (MomL) | Muricauda olearia Th120 | Exhibits high activity range from 20–50 °C. Retains 30% activity after incubation at 60 °C for 30 min. | C6-HSL, C12-HSL, 3-oxo-C6HSL, C8-HSL, 3-oxo-C8-HSL, C4-HSL, 3-oxo-C10-HSL, C14-HSL, 3-oxo-C14-HSL, C10-HSL. Attenuates virulence of Pseudomonas aeruginosa and Caenorhabditis elegans | [111] |
| AHL lactonase | Tenacibaculum soleae T173 | Maintains C6-HSL degrading activity after boiled for 30 min. | C6-HSL, 3-oxo-C6-HSL, C8-HSL,3-oxo-C8-HSL, C10-HSL, 3-oxo-C10-HSL, C12-HSL, 3-oxo-C12-HSL, C14-HSL and 3-oxo-C14-HSL | [112] |
| Phospshotriesterase-like Lactonase | Geobacillus kaustophilus HTA426 | Retains its catalytic activity at 60 °C for up to 72 h. | C4-HSL, C6-HSL, 3-oxo-C6-HSL, C8-HSL, 3-oxo- C8-HSL, C10-HSL | [113] |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Scarascia, G.; Wang, T.; Hong, P.-Y. Quorum Sensing and the Use of Quorum Quenchers as Natural Biocides to Inhibit Sulfate-Reducing Bacteria. Antibiotics 2016, 5, 39. https://doi.org/10.3390/antibiotics5040039
Scarascia G, Wang T, Hong P-Y. Quorum Sensing and the Use of Quorum Quenchers as Natural Biocides to Inhibit Sulfate-Reducing Bacteria. Antibiotics. 2016; 5(4):39. https://doi.org/10.3390/antibiotics5040039
Chicago/Turabian StyleScarascia, Giantommaso, Tiannyu Wang, and Pei-Ying Hong. 2016. "Quorum Sensing and the Use of Quorum Quenchers as Natural Biocides to Inhibit Sulfate-Reducing Bacteria" Antibiotics 5, no. 4: 39. https://doi.org/10.3390/antibiotics5040039
APA StyleScarascia, G., Wang, T., & Hong, P.-Y. (2016). Quorum Sensing and the Use of Quorum Quenchers as Natural Biocides to Inhibit Sulfate-Reducing Bacteria. Antibiotics, 5(4), 39. https://doi.org/10.3390/antibiotics5040039






