FTIR-Derived Feature Insights for Predicting Time-Dependent Antibiotic Resistance Progression
Abstract
1. Introduction
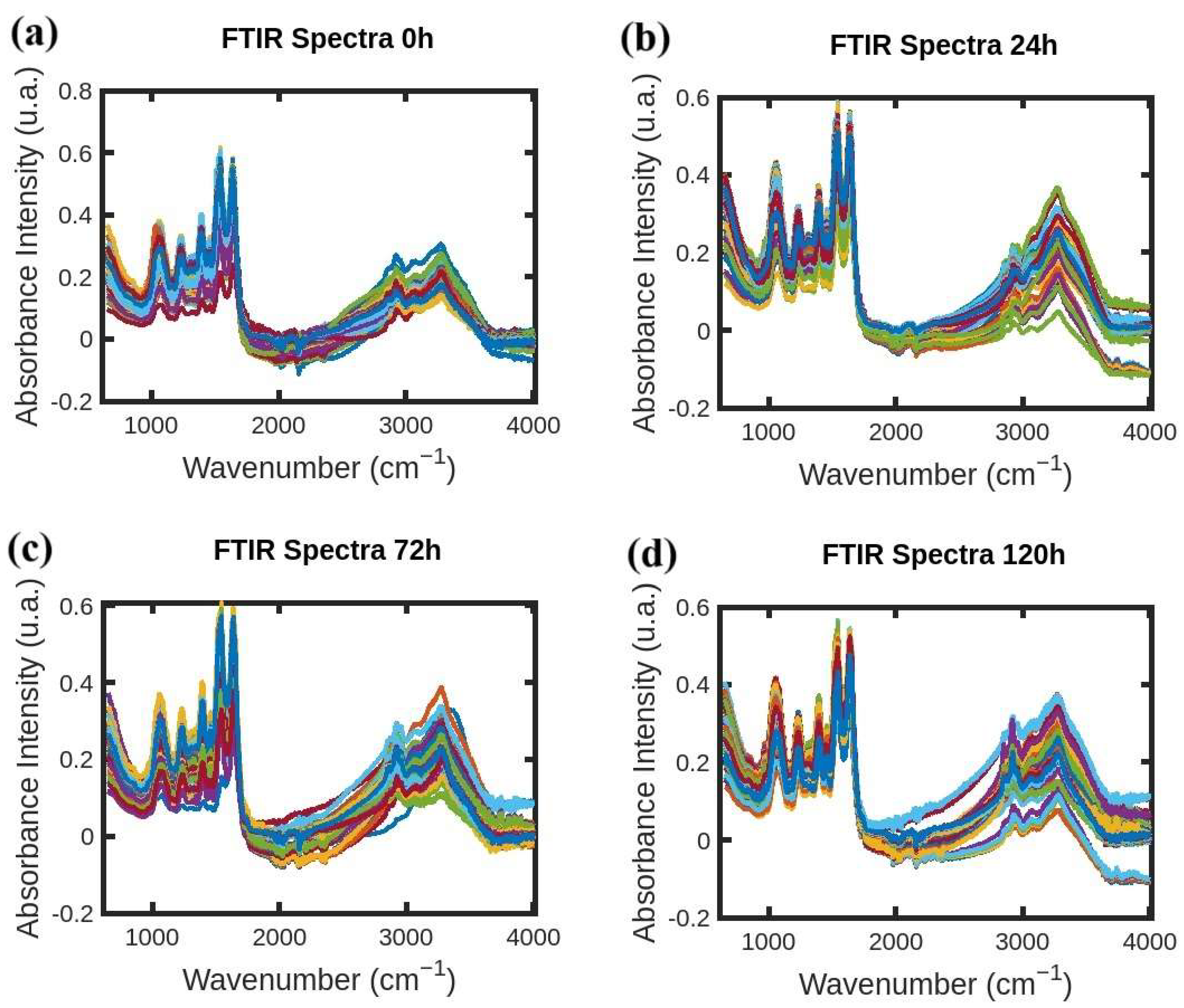
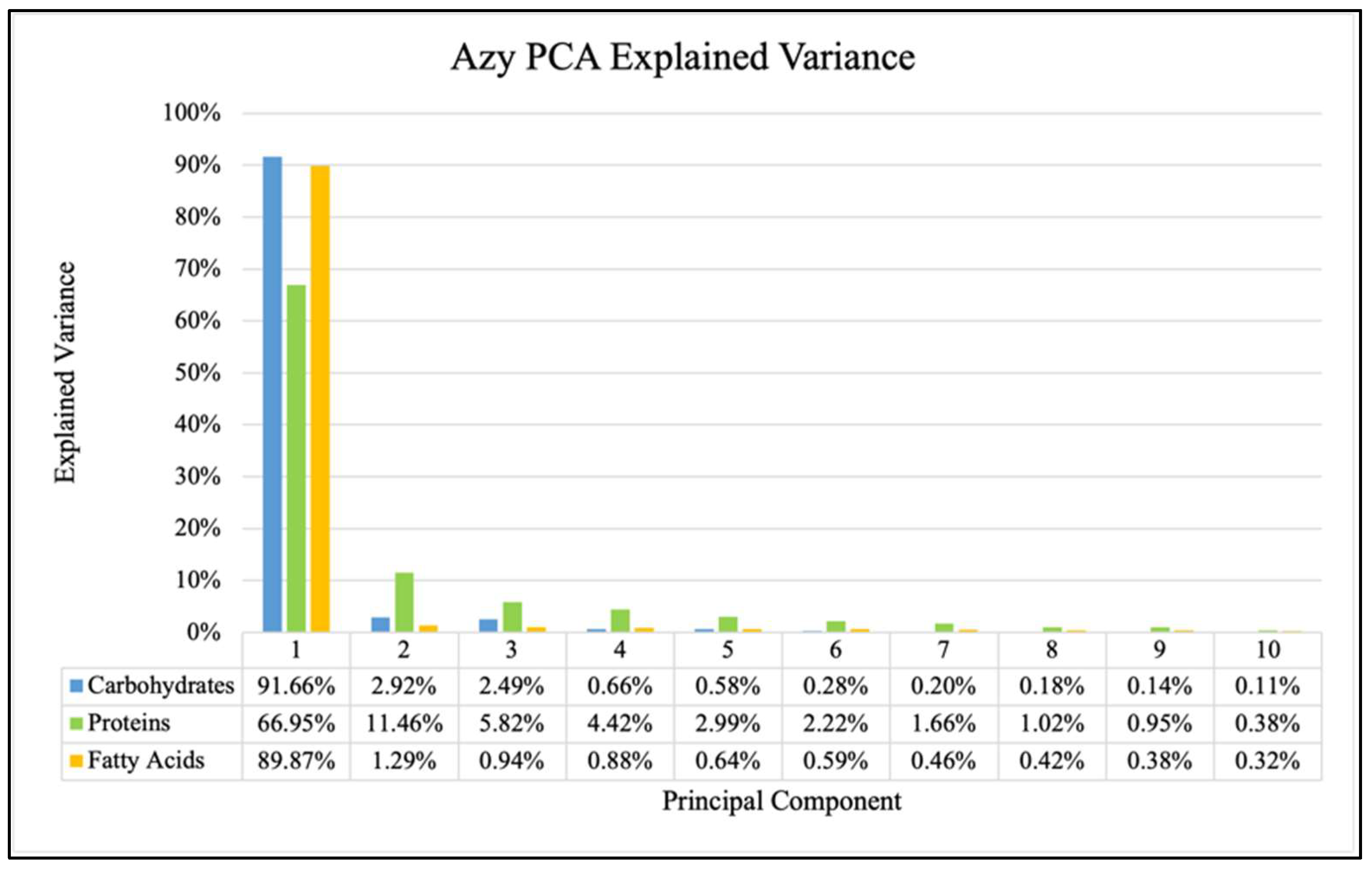
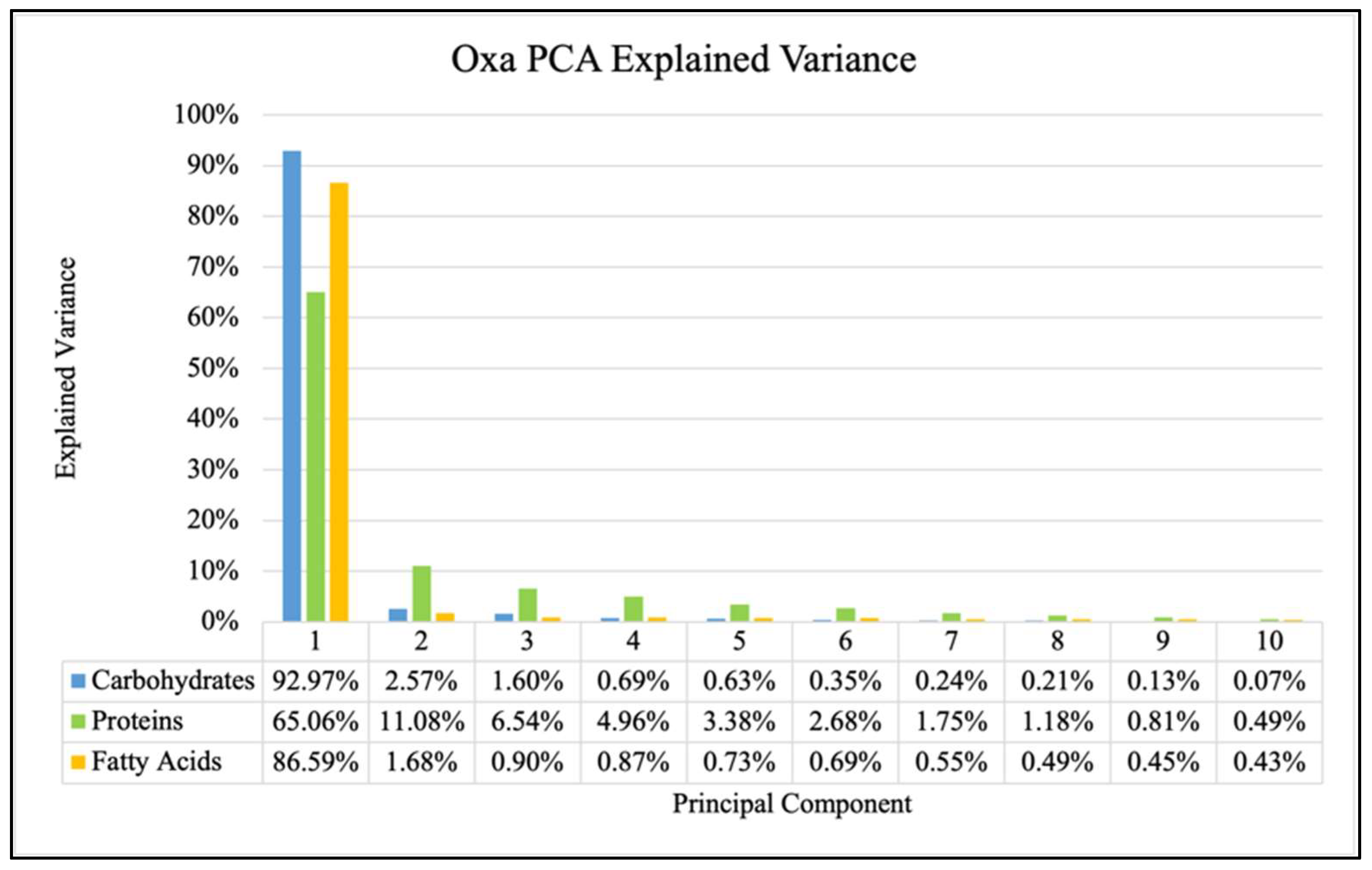
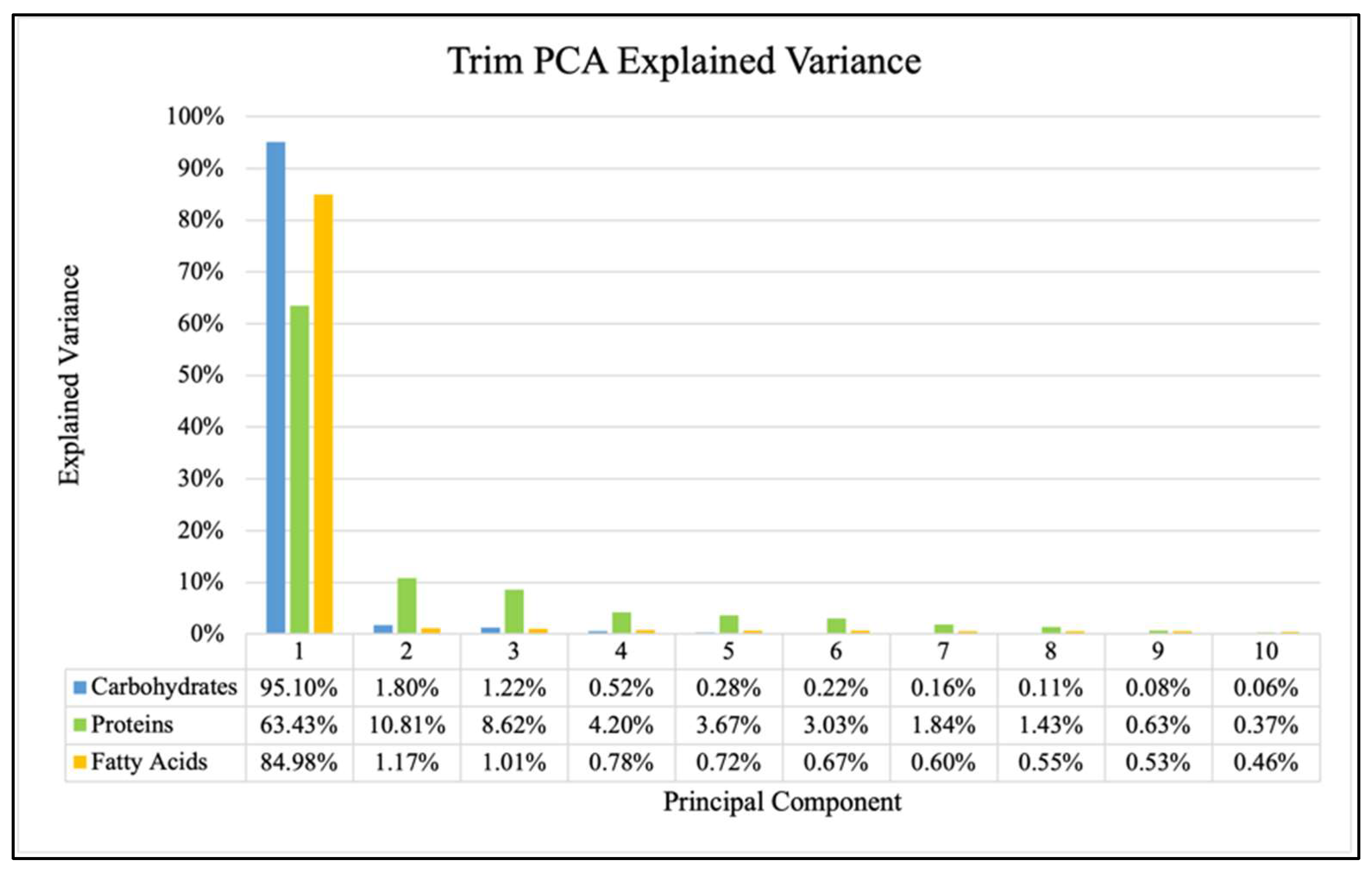
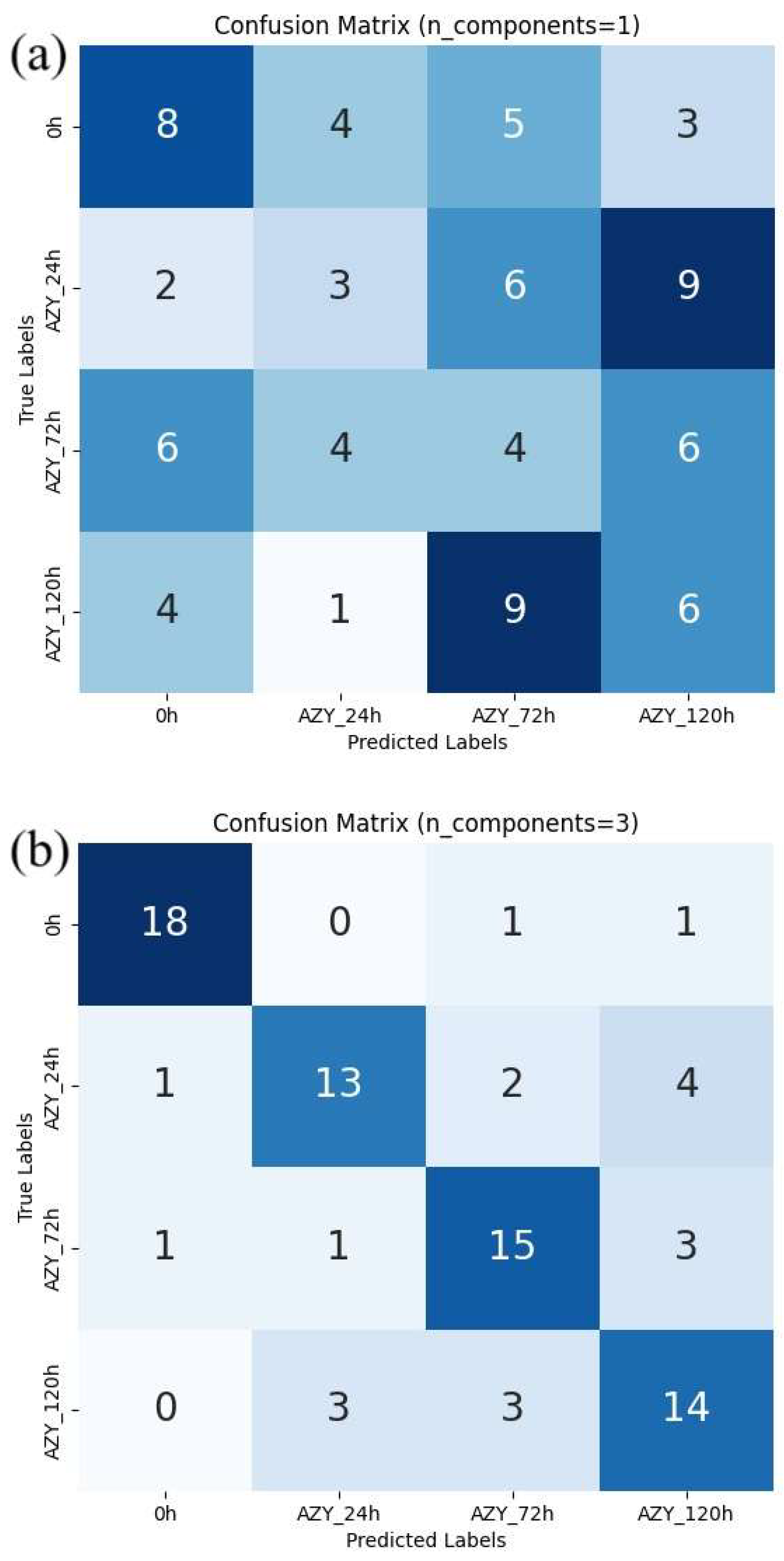
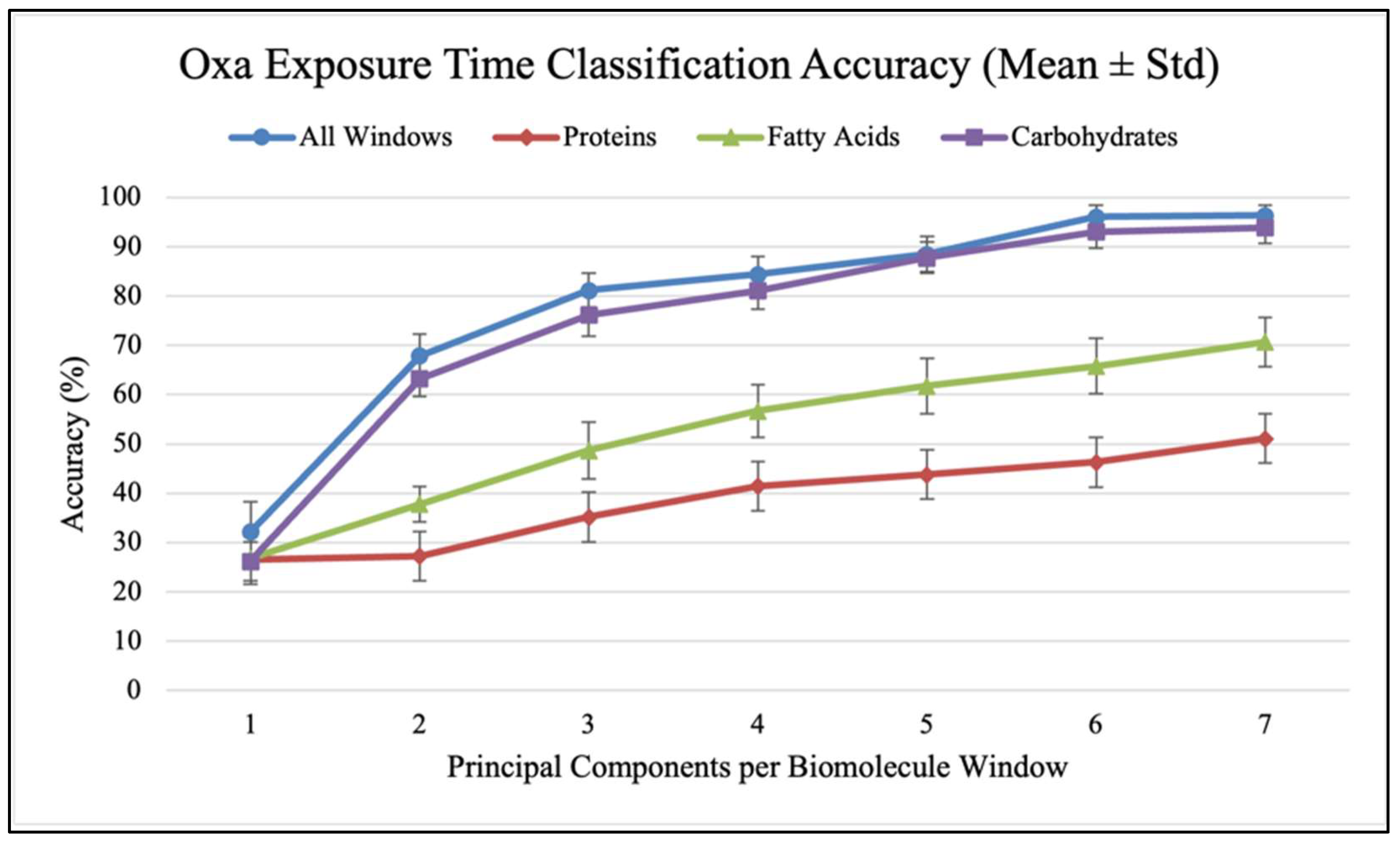
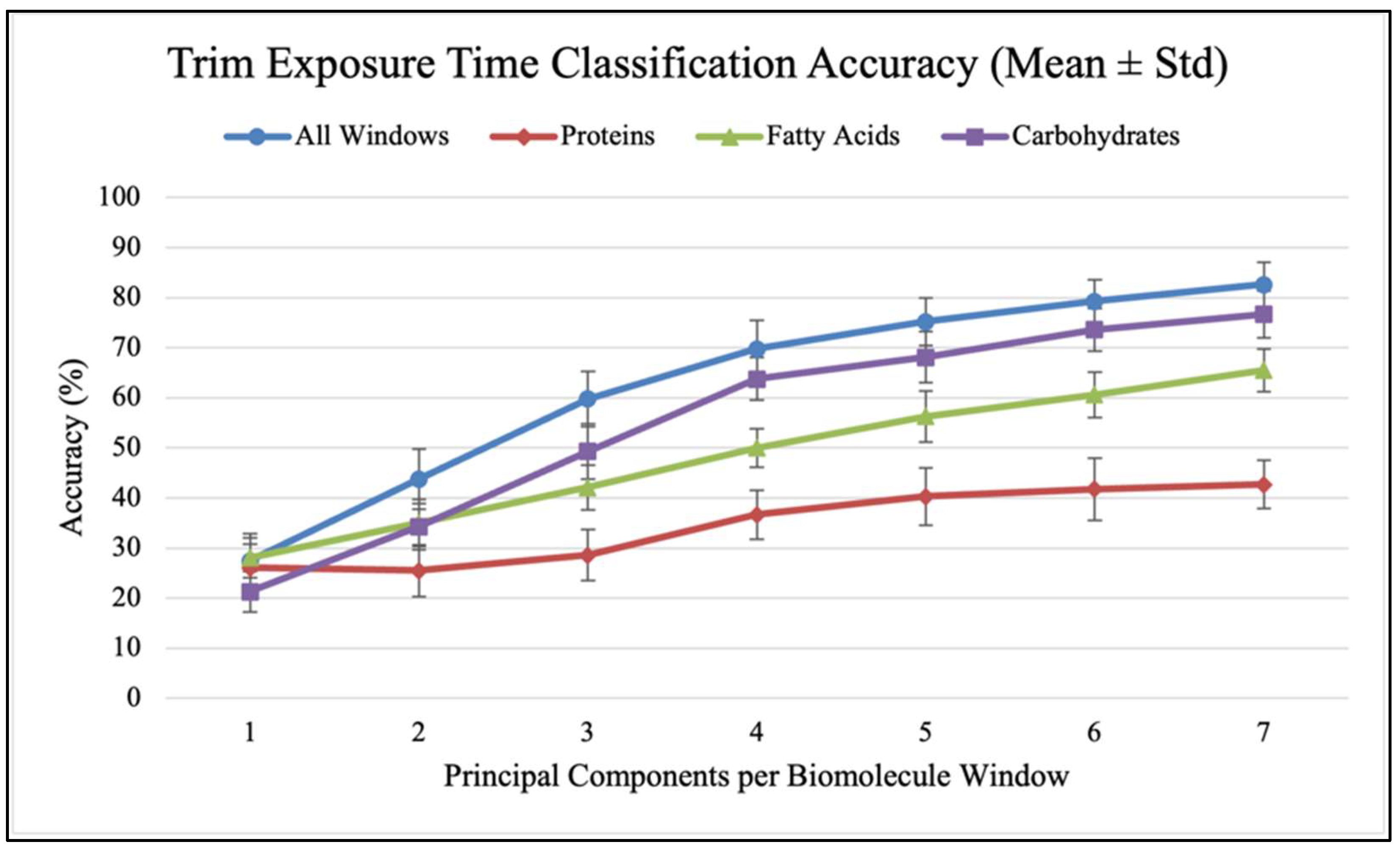
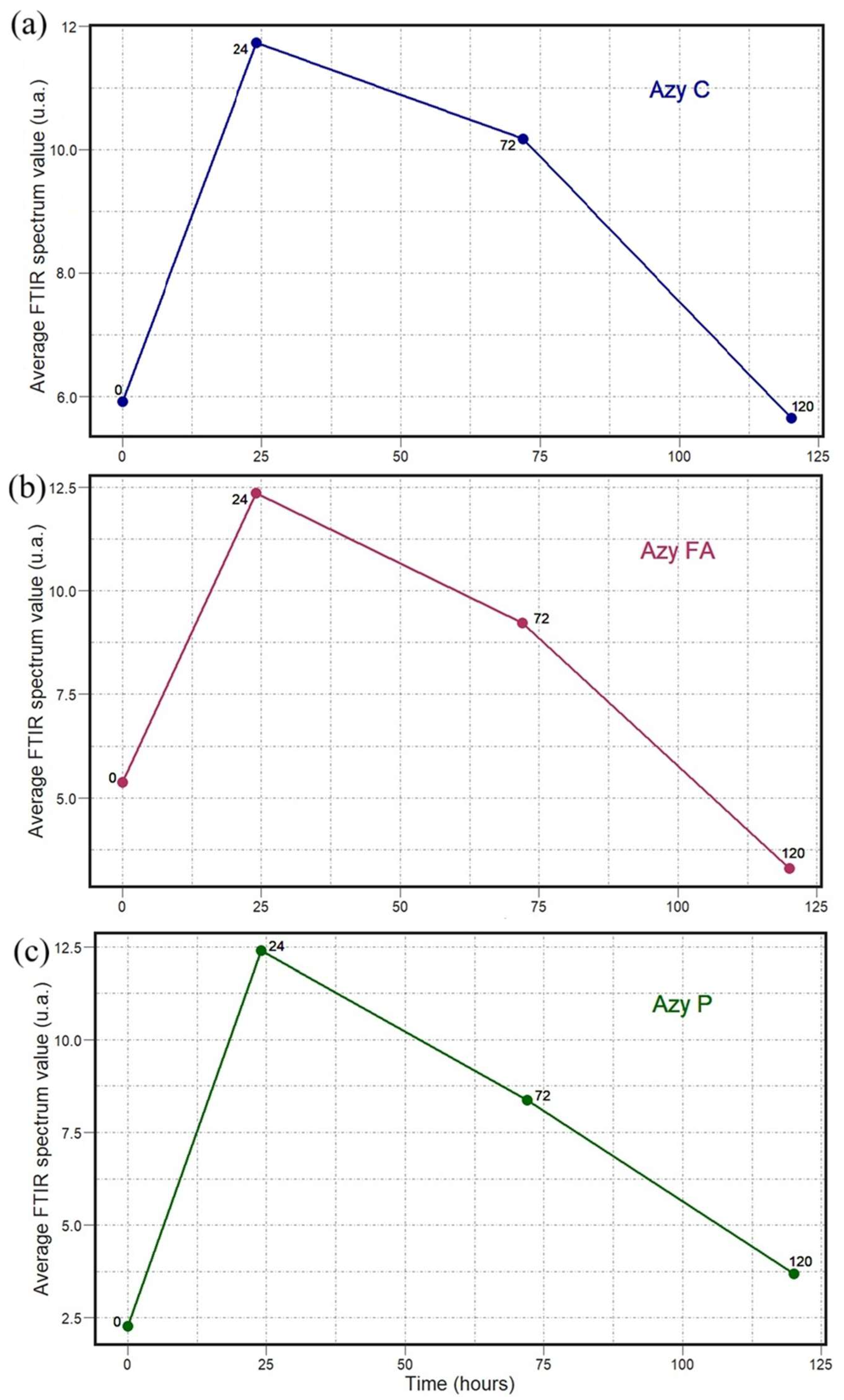
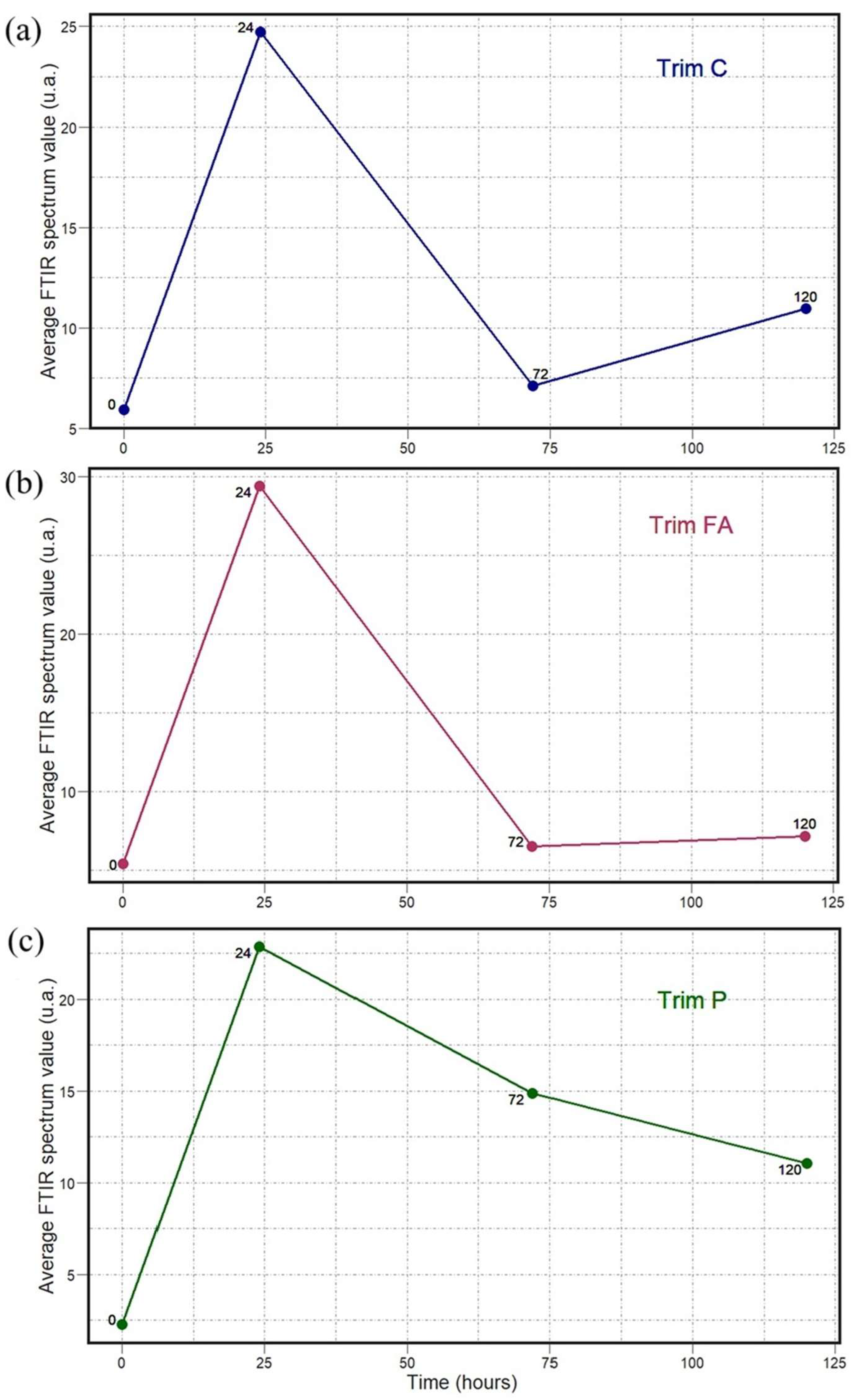
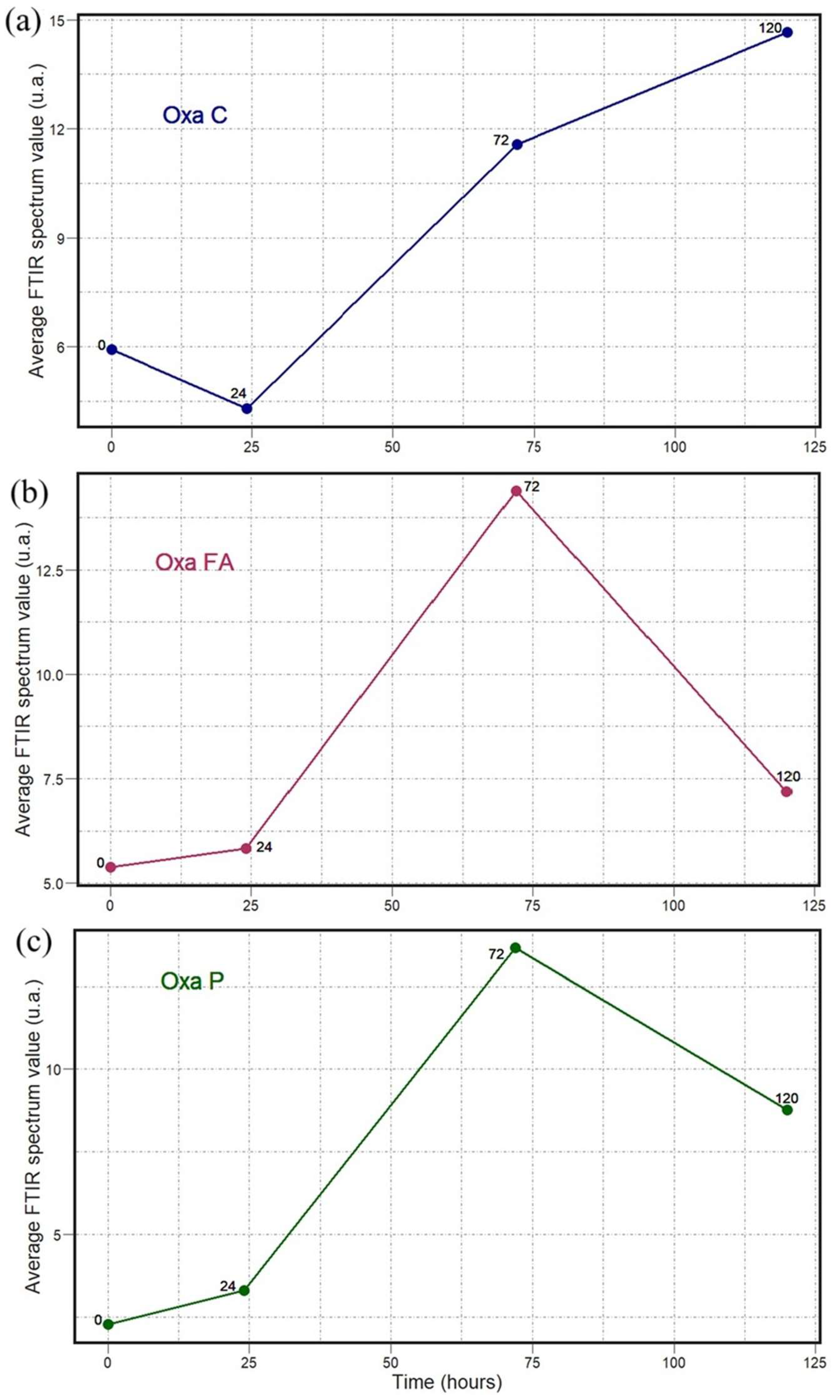
2. Results
3. Discussion
4. Materials and Methods
4.1. Sample Preparation and FTIR Spectra Acquisition
4.2. Data Preparation and Machine Learning Application
- (a)
- Data Processing and Principal Component Analysis (PCA)
- (b)
- Random Forest Model and Feature Evaluation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
| PCs Per Window | Overall Accuracy (Mean ± Std.) | |||
|---|---|---|---|---|
| All Windows | Proteins | Fatty Acids | Carbohydrates | |
| 1 | 30 ± 4.8% | 24.2 ± 4.2% | 26.3 ± 4.3% | 23 ± 3.1% |
| 2 | 50.4 ± 7.5% | 32.1 ± 5.8% | 38.2 ± 5.2% | 31 ± 4.3% |
| 3 | 68.3 ± 4.1% | 37.6 ± 4.0% | 54 ± 6.1% | 53.9 ± 4.2% |
| 4 | 77.8 ± 4.5% | 43.7 ± 4.0% | 65.2 ± 6.7% | 62.5 ± 4.2% |
| 5 | 82 ± 4.7% | 46.5 ± 4.8% | 69.2 ± 5.2% | 67 ± 4.5% |
| 6 | 83.8 ± 3.5% | 51.3 ± 4.5% | 72.4 ± 4.3% | 71.2 ± 4.2% |
| 7 | 84.9 ± 3.3% | 52.7 ± 4.3% | 75 ± 4.7% | 75.4 ± 3.5% |
| PCs Per Window | Overall Accuracy (Mean ± Std.) | |||
|---|---|---|---|---|
| All Windows | Proteins | Fatty Acids | Carbohydrates | |
| 1 | 32.2 ± 6.1% | 26.5 ± 5.0% | 26.8 ± 4.5% | 26.2 ± 3.9% |
| 2 | 67.9 ± 4.4% | 27.3 ± 3.8% | 37.8 ± 3.6% | 63.3 ± 3.7% |
| 3 | 81.2 ± 3.4% | 35.2 ± 3.5% | 48.7 ± 5.8% | 76.2 ± 4.3% |
| 4 | 84.4 ± 3.6% | 41.5 ± 4.8% | 56.7 ± 5.4% | 81.1 ± 3.7% |
| 5 | 88.5 ± 3.6% | 43.8 ± 4.7% | 61.8 ± 5.6% | 87.8 ± 3.2% |
| 6 | 96.1 ± 2.3% | 46.3 ± 5.0% | 65.8 ± 5.6% | 93.1 ± 3.4% |
| 7 | 96.4 ± 2.1% | 51.1 ± 6.0% | 70.7 ± 5.0% | 93.9 ± 3.2% |
| PCs Per Window | Overall Accuracy (Mean ± Std.) | |||
|---|---|---|---|---|
| All Windows | Proteins | Fatty Acids | Carbohydrates | |
| 1 | 27.5 ± 5.4% | 26.1 ± 4.7% | 28.1 ± 4.0% | 21.3 ± 4.0% |
| 2 | 43.8 ± 6.0% | 25.5 ± 5.2% | 35.0 ± 4.7% | 34.3 ± 4.6% |
| 3 | 59.8 ± 5.5% | 28.6 ± 5.1% | 42.1 ± 4.5% | 49.3 ± 5.5% |
| 4 | 69.8 ± 5.6% | 36.7 ± 4.9% | 50.0 ± 3.8% | 63.8 ± 4.2% |
| 5 | 75.2 ± 4.8% | 40.3 ± 5.7% | 56.3 ± 5.1% | 68.1 ± 5.1% |
| 6 | 79.3 ± 4.2% | 41.8 ± 6.2% | 60.6 ± 4.6% | 73.6 ± 4.3% |
| 7 | 82.6 ± 4.5% | 42.7 ± 4.8% | 65.5 ± 4.3% | 76.7 ± 4.7% |
| PCs Per Window | F1 Score (Mean ± Std.) | ||
|---|---|---|---|
| Positive: 120 h | Positive: 72, 120 h | Positive: 24, 72, 120 h | |
| 1 | 0.313 ± 0.090 | 0.556 ± 0.051 | 0.789 ± 0.036 |
| 2 | 0.431 ± 0.102 | 0.666 ± 0.073 | 0.892 ± 0.032 |
| 3 | 0.543 ± 0.074 | 0.776 ± 0.042 | 0.953 ± 0.022 |
| 4 | 0.655 ± 0.083 | 0.835 ± 0.035 | 0.987 ± 0.011 |
| 5 | 0.744 ± 0.079 | 0.859 ± 0.040 | 0.994 ± 0.007 |
| 6 | 0.781 ± 0.063 | 0.866 ± 0.044 | 0.994 ± 0.008 |
| 7 | 0.804 ± 0.048 | 0.873 ± 0.038 | 0.993 ± 0.008 |
| PCs Per Window | F1 Score (Mean ± Std.) | ||
|---|---|---|---|
| Positive: 120 h | Positive: 72, 120 h | Positive: 24, 72, 120 h | |
| 1 | 0.433 ± 0.100 | 0.534 ± 0.068 | 0.768 ± 0.042 |
| 2 | 0.904 ± 0.040 | 0.751 ± 0.044 | 0.860 ± 0.030 |
| 3 | 0.943 ± 0.028 | 0.874 ± 0.029 | 0.933 ± 0.018 |
| 4 | 0.947 ± 0.036 | 0.886 ± 0.036 | 0.939 ± 0.018 |
| 5 | 0.967 ± 0.028 | 0.905 ± 0.027 | 0.943 ± 0.017 |
| 6 | 0.989 ± 0.017 | 0.973 ± 0.020 | 0.981 ± 0.013 |
| 7 | 0.993 ± 0.015 | 0.976 ± 0.018 | 0.980 ± 0.014 |
| PCs Per Window | F1 Score (Mean ± Std.) | ||
|---|---|---|---|
| Positive: 120 h | Positive: 72, 120 h | Positive: 24, 72, 120 h | |
| 1 | 0.259 ± 0.073 | 0.485 ± 0.078 | 0.755 ± 0.046 |
| 2 | 0.412 ± 0.103 | 0.650 ± 0.058 | 0.788 ± 0.034 |
| 3 | 0.645 ± 0.081 | 0.746 ± 0.049 | 0.852 ± 0.030 |
| 4 | 0.703 ± 0.065 | 0.812 ± 0.053 | 0.888 ± 0.026 |
| 5 | 0.752 ± 0.052 | 0.836 ± 0.047 | 0.911 ± 0.026 |
| 6 | 0.780 ± 0.063 | 0.852 ± 0.040 | 0.929 ± 0.022 |
| 7 | 0.807 ± 0.059 | 0.874 ± 0.044 | 0.936 ± 0.022 |
References
- Ling, L.L.; Schneider, T.; Peoples, A.J.; Spoering, A.L.; Engels, I.; Conlon, B.P.; Mueller, A.; Schäberle, T.F.; Hughes, D.E.; Epstein, S.; et al. A New Antibiotic Kills Pathogens without Detectable Resistance. Nature 2015, 517, 455–459. [Google Scholar] [CrossRef]
- Yen, P.; Papin, J.A. History of Antibiotic Adaptation Influences Microbial Evolutionary Dynamics during Subsequent Treatment. PLoS Biol. 2017, 15, e2001586. [Google Scholar] [CrossRef]
- Barrera-Patiño, C.P.; Soares, J.M.; Branco, K.C.; Inada, N.M.; Bagnato, V.S. Spectroscopic Identification of Bacteria Resistance to Antibiotics by Means of Absorption of Specific Biochemical Groups and Special Machine Learning Algorithm. Antibiotics 2023, 12, 1502. [Google Scholar] [CrossRef] [PubMed]
- Barrera Patiño, C.P.; Soares, J.M.; Blanco, K.C.; Bagnato, V.S. Machine Learning in FTIR Spectrum for the Identification of Antibiotic Resistance: A Demonstration with Different Species of Microorganisms. Antibiotics 2024, 13, 821. [Google Scholar] [CrossRef] [PubMed]
- Smith, W.P.J.; Wucher, B.R.; Nadell, C.D.; Foster, K.R. Bacterial Defences: Mechanisms, Evolution and Antimicrobial Resistance. Nat. Rev. Microbiol. 2023, 21, 519–534. [Google Scholar] [CrossRef]
- Uddin, T.M.; Chakraborty, A.J.; Khusro, A.; Zidan, B.R.M.; Mitra, S.; Emran, T.B.; Dhama, K.; Ripon, M.K.H.; Gajdács, M.; Sahibzada, M.U.K.; et al. Antibiotic Resistance in Microbes: History, Mechanisms, Therapeutic Strategies and Future Prospects. J. Infect. Public Health 2021, 14, 1750–1766. [Google Scholar] [CrossRef] [PubMed]
- Imamovic, L.; Sommer, M.O.A. Use of Collateral Sensitivity Networks to Design Drug Cycling Protocols That Avoid Resistance Development. Sci. Transl. Med. 2013, 5, 204ra132. [Google Scholar] [CrossRef]
- Baym, M.; Stone, L.K.; Kishony, R. Multidrug Evolutionary Strategies to Reverse Antibiotic Resistance. Science 2016, 351, aad3292. [Google Scholar] [CrossRef]
- Barrera Patiño, C.P.; Bonner, M.; Borsatto, A.R.; Soares, J.M.; Blanco, K.C.; Bagnato, V.S. Time Evolution of Bacterial Resistance Observed with Principal Component Analysis. Antibiotics 2025, 14, 729. [Google Scholar] [CrossRef]
- Soares, J.M.; Guimarães, F.E.G.; Yakovlev, V.V.; Bagnato, V.S.; Blanco, K.C. Physicochemical Mechanisms of Bacterial Response in the Photodynamic Potentiation of Antibiotic Effects. Sci. Rep. 2022, 12, 21146. [Google Scholar] [CrossRef]
- Willis, J.A.; Cheburkanov, V.; Chen, S.; Soares, J.M.; Kassab, G.; Blanco, K.C.; Bagnato, V.S.; de Figueiredo, P.; Yakovlev, V.V. Breaking down Antibiotic Resistance in Methicillin-Resistant Staphylococcus aureus: Combining Antimicrobial Photodynamic and Antibiotic Treatments. Proc. Natl. Acad. Sci. USA 2022, 119, e2208378119. [Google Scholar] [CrossRef]
- Soares, J.M.; Yakovlev, V.V.; Blanco, K.C.; Bagnato, V.S. Recovering the Susceptibility of Antibiotic-Resistant Bacteria Using Photooxidative Damage. Proc. Natl. Acad. Sci. USA 2023, 120, e2311667120. [Google Scholar] [CrossRef]
- The MathWorks Inc. MATLAB, version R2021b; The MathWorks Inc.: Natick, MA, USA, 2021. Available online: https://www.mathworks.com (accessed on 3 February 2025).
- Naumann, D. Infrared Spectroscopy in Microbiology. Encycl. Anal. Chem. 2000, 102, 131. [Google Scholar]
- Morton, F.B.; Altschul, D. Data Reduction Analyses of Animal Behaviour: Avoiding Kaiser’s Criterion and Adopting More Robust Automated Methods. Anim. Behav. 2019, 149, 89–95. [Google Scholar] [CrossRef]
- Karlis, D.; Saporta, G.; Spinakis, A. A Simple Rule for the Selection of Principal Components. Commun. Stat. Theory Methods 2003, 32, 643–666. [Google Scholar] [CrossRef]
- Soares, J.M.; Inada, N.M.; Bagnato, V.S.; Blanco, K.C. Evolution of Surviving Streptoccocus Pyogenes from Pharyngotonsillitis Patients Submit to Multiple Cycles of Antimicrobial Photodynamic Therapy. J. Photochem. Photobiol. B Biol. 2020, 210, 111985. [Google Scholar] [CrossRef]
- National Center for Biotechnology Information. PubChem Compound Summary for CID 447043, Azithromycin. 2025. Available online: https://pubchem.ncbi.nlm.nih.gov/compound/447043?from=summary (accessed on 15 July 2025).
- National Center for Biotechnology Information. PubChem Compound Summary for CID 6196, Oxacillin. 2025. Available online: https://pubchem.ncbi.nlm.nih.gov/compound/Oxacillin (accessed on 15 July 2025).
- National Center for Biotechnology Information. PubChem Compound Summary for CID 5578, Trimethoprim. 2025. Available online: https://pubchem.ncbi.nlm.nih.gov/compound/Trimethoprim (accessed on 15 July 2025).
- Python Software Foundation. Python, version 3.12.3; Python Software Foundation: Wilmington, DE, USA, 2024. Available online: https://www.python.org/downloads/release/python-3123/ (accessed on 17 June 2025).
- Virtanen, P.; Gommers, R.; Oliphant, T.E.; Haberland, M.; Reddy, T.; Cournapeau, D.; Burovski, E.; Peterson, P.; Weckesser, W.; Bright, J.; et al. SciPy 1.0: Fundamental Algorithms for Scientific Computing in Python. Nat. Methods 2020, 17, 261–272. [Google Scholar] [CrossRef]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-Learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Nguyen, G.; Dlugolinsky, S.; Bobák, M.; Tran, V.; López García, Á.; Heredia, I.; Malík, P.; Hluchý, L. Machine Learning and Deep Learning Frameworks and Libraries for Large-Scale Data Mining: A Survey. Artif. Intell. Rev. 2019, 52, 77–124. [Google Scholar] [CrossRef]
- Claude Sammut, G.I.W. Encyclopedia of Machine Learning and Data Mining; Sammut, C., Webb, G.I., Eds.; Springer: Boston, MA, USA, 2017; ISBN 978-1-4899-7685-7. [Google Scholar]
- Rizzo, M.L. Statistical Computing with R; CRC Press: Boca Raton, FL, USA, 2019. [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2021. [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2013. [Google Scholar]
- Mair, P.; Hofmann, E.; Gruber, K.; Hatzinger, R.; Zeileis, A.; Hornik, K. Motivation, Values, and Work Design as Drivers of Participation in the R Open Source Project for Statistical Computing. Proc. Natl. Acad. Sci. USA 2015, 112, 14788–14792. [Google Scholar] [CrossRef]
- Chambers, J.M. Software for Data Analysis: Programming with R; Springer: Berlin/Heidelberg, Germany, 2008; Volume 2. [Google Scholar]
- Ripley, B.D. The R Project in Statistical Computing. MSOR Connect. Newsl. LTSN Maths Stats OR Netw. 2001, 1, 23–25. [Google Scholar] [CrossRef]







Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bonner, M.; Barrera Patiño, C.P.; Borsatto, A.R.; Soares, J.M.; Blanco, K.C.; Bagnato, V.S. FTIR-Derived Feature Insights for Predicting Time-Dependent Antibiotic Resistance Progression. Antibiotics 2025, 14, 831. https://doi.org/10.3390/antibiotics14080831
Bonner M, Barrera Patiño CP, Borsatto AR, Soares JM, Blanco KC, Bagnato VS. FTIR-Derived Feature Insights for Predicting Time-Dependent Antibiotic Resistance Progression. Antibiotics. 2025; 14(8):831. https://doi.org/10.3390/antibiotics14080831
Chicago/Turabian StyleBonner, Mitchell, Claudia P. Barrera Patiño, Andrew Ramos Borsatto, Jennifer M. Soares, Kate C. Blanco, and Vanderlei S. Bagnato. 2025. "FTIR-Derived Feature Insights for Predicting Time-Dependent Antibiotic Resistance Progression" Antibiotics 14, no. 8: 831. https://doi.org/10.3390/antibiotics14080831
APA StyleBonner, M., Barrera Patiño, C. P., Borsatto, A. R., Soares, J. M., Blanco, K. C., & Bagnato, V. S. (2025). FTIR-Derived Feature Insights for Predicting Time-Dependent Antibiotic Resistance Progression. Antibiotics, 14(8), 831. https://doi.org/10.3390/antibiotics14080831






