What Are the Drivers Triggering Antimicrobial Resistance Emergence and Spread? Outlook from a One Health Perspective
Abstract
1. Introduction
2. Antimicrobial Consumption
| Major Antimicrobial-Resistant Bacteria | Antimicrobial Resistance | Resistance Rates (%) | Resistance Genes | Reference(s) | |
|---|---|---|---|---|---|
| Human Clinical Points of View | Veterinary Points of View | ||||
| Staphylococcus aureus | Methicillin Resistance | 29.6 (China); 15.8 (Europe); 38.1 (the United States) | 7.6 (retail meats in the United States); 7.3 (raw meat in China) | mecA, mecC, fem | [19,20,21,22] |
| Escherichia coli | Extended-Spectrum β-Lactamases (ESBL) | 13.3 (the United States); 53.5 (China); 18.5 (Europe) | 0.9 (broilers in Europe); 69.3 (pigeons in China); 3.1 (companion animals in the United States) | blaTEM, blaSHV, blaOXA, blaCTX-M | [23,24,25,26] |
| Carbapenem Resistance | 0.3 (Europe); 1.0 (the United States); 2.1 (China) | 4.9 (chickens in China); not detected (chickens in the United States) | blaTEM, blaSHV, blaOXA, blaCTX-M, blaKPC | [19,20,27,28,29,30] | |
| Colistin Resistance | 1.3 (China); 0.3 (Europe); 0.3 (the United States) | 17.0 (chickens in China); 0.1 (retail chickens in the United States) | mcr, pmrAB, mgrB, phoPQ | [19,20,27,28,29,31] | |
| Klebsiella pneumoniae | ESBL | 11.8 (the United States); 21.8 (China); 35.5 (Europe) | 76 (mink in the United States); 44.5 (overall animal in Africa); 33.7 (overall animal in Asia) | blaKPC, blaSHV, blaOXA, blaCTX-M, blaTEM | [23,32,33,34,35,36] |
| Carbapenem Resistance | 13.3 (Europe); 10.8 (China); 24.6% (the United States) | 3.8 (pets in China); not detected (poultry meat in Greece) | blaOXA, blaKPC, blaVIM, blaIMP, blaNDM | [19,20,37,38,39] | |
| Pseudomonas aeruginosa | Carbapenem Resistance | 18.6 (Europe); 14.2 (the United States); 18.2 (China) | 13.5 (pets in China); 23.1 (shellfish in Croatia); not detected (horses, cows, and dogs in France) | blaVIM, blaGES, blaNDM, blaKPC | [19,20,27,37,40,41,42] |
| Acinetobacter baumannii | Carbapenem Resistance | 69.5 (Europe); 71.5 (China); 45.7 (the United States) | not detected (bovine in Germany); 17.9 (raw meat in Iran) | blaVIM, blaIMP, blaNDM, blaKPC | [27,43,44,45,46] |
| Enterococcus faecium | Vancomycin Resistance | 5.1 (China); 19.8 (Europe); 67.3 (the United States) | not detected (livestock in the United States); 0.4 (cats and dogs in China) | VanA, VanB, VanC, VanD, VanE, VanM | [19,20,47,48,49] |
| Salmonella | Fluoroquinolone Resistance | 21.8 (Europe); 0.6 (the United States); 26.6 (Salmonella Enteritidis in China); 19.7 (Salmonella Typhi and Salmonella Paratyphi in China) | 74.0 (ducks and wild geese in China); 14.9 (pigs in the European Union); 4.1 (market swine in the United States) | gyrA, gyrB, parC, parE | [24,29,50,51] |
3. Drivers of AMR
3.1. The Intrinsic Determinants of AMR
3.2. Extrinsic Drivers of AMR
3.2.1. Environmental Stress Factors
3.2.2. Social Factors
3.2.3. Economic Factor
3.2.4. Medicinal Factors
3.2.5. Health Factors
3.2.6. Climatic Factors
3.2.7. Ageing Factors
4. Transmission and Evolution of Antimicrobial-Resistant Bacteria
4.1. Transmission of Antimicrobial-Resistant Bacteria
4.2. Evolution of Antimicrobial-Resistant Bacteria
5. Impact of AMR on Humans
5.1. Human Health
5.2. Food Security and Safety
5.3. Healthy Carriers
5.4. Social Economy
6. Limitations and Prospects
Author Contributions
Funding
Institutional Review Board Statement
Conflicts of Interest
References
- McCulloch, T.R.; Wells, T.J.; Souza-Fonseca-Guimaraes, F. Towards efficient immunotherapy for bacterial infection. Trends Microbiol. 2022, 30, 158–169. [Google Scholar] [CrossRef] [PubMed]
- Antimicrobial Resistance. Available online: https://www.who.int/news-room/fact-sheets/detail/antimicrobial-resistance (accessed on 18 May 2025).
- Chang, R.Y.K.; Nang, S.C.; Chan, H.K.; Li, J. Novel antimicrobial agents for combating antibiotic-resistant bacteria. Adv. Drug Deliv. Rev. 2022, 187, 114378. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, S.; Ahmed, M.Z.; Rafique, S.; Almasoudi, S.E.; Shah, M.; Jalil, N.A.C.; Ojha, S.C. Recent Approaches for Downplaying Antibiotic Resistance: Molecular Mechanisms. Biomed. Res. Int. 2023, 2023, 5250040. [Google Scholar] [CrossRef] [PubMed]
- Naghavi, M.; Vollset, S.E.; Ikuta, K.S.; Swetschinski, L.R.; Gray, A.P.; Wool, E.E.; Aguilar, G.R.; Mestrovic, T.; Smith, G.; Han, C.; et al. Global burden of bacterial antimicrobial resistance 1990–2021: A systematic analysis with forecasts to 2050. Lancet 2024, 404, 1199–1226. [Google Scholar] [CrossRef]
- Matthiessen, L.E.; Hald, T.; Vigre, H. System Mapping of Antimicrobial Resistance to Combat a Rising Global Health Crisis. Front. Public Health 2022, 10, 816943. [Google Scholar] [CrossRef]
- Shaheen, M.N.F. The concept of one health applied to the problem of zoonotic diseases. Rev. Med. Virol. 2022, 32, e2326. [Google Scholar] [CrossRef]
- Aslam, B.; Khurshid, M.; Arshad, M.I.; Muzammil, S.; Rasool, M.; Yasmeen, N.; Shah, T.; Chaudhry, T.H.; Ra-sool, M.H.; Shahid, A.; et al. Antibiotic Resistance: One Health One World Outlook. Front. Cell Infect. Microbiol. 2021, 11, 771510. [Google Scholar] [CrossRef]
- Ferri, M.; Elena, R.; Romagnoli, P.; Giaccone, V. Antimicrobial resistance: A global emerging threat to public health systems. Crit. Rev. Food Sci. Nutr. 2017, 57, 2857–2876. [Google Scholar] [CrossRef]
- Klein, E.Y.; Impalli, I.; Poleon, S.; Denoel, P.; Cipriano, M.; Van Boeckel, T.P.; Pecetta, S.; Bloom, D.E.; Nandi, A. Global trends in antibiotic consumption during 2016–2023 and future projections through 2030. Proc. Natl. Acad. Sci. USA 2024, 121, e2411919121. [Google Scholar] [CrossRef]
- Zhao, Q.; Jiang, Z.; Li, T.; Cheng, M.; Sun, H.; Cui, M.; Zhang, C.; Xu, S.; Wang, H.; Wu, C. Current status and trends in antimicrobial use in food animals in China, 2018–2020. One Health Adv. 2023, 1, 29. [Google Scholar] [CrossRef]
- Yang, Y.; Geng, X.; Liu, X.; Wen, X.; Wu, R.; Cui, D.; Mao, Z. Antibiotic Use in China’s Public Healthcare Institutions During the COVID-19 Pandemic: An Analysis of Nationwide Procurement Data, 2018–2020. Front. Pharmacol. 2022, 13, 813213. [Google Scholar] [CrossRef] [PubMed]
- Antimicrobial Consumption in the EU/EEA (ESAC-Net)—Annual Epidemiological Report for 2020. 18 November 2021. Available online: https://www.ecdc.europa.eu/en/publications-data/surveillance-antimicrobial-consumption-europe-2020 (accessed on 17 May 2025).
- The State of the World’s Antibiotics in 2021. One Health Trust. Available online: https://onehealthtrust.org/publications/reports/the-state-of-the-worlds-antibiotic-in-2021/ (accessed on 17 May 2025).
- Zhang, Q.Q.; Ying, G.G.; Pan, C.G.; Liu, Y.S.; Zhao, J.L. Comprehensive Evaluation of Antibiotics Emission and Fate in the River Basins of China: Source Analysis, Multimedia Modeling, and Linkage to Bacterial Resistance. Environ. Sci. Technol. 2015, 49, 6772–6782. [Google Scholar] [CrossRef] [PubMed]
- Shao, Y.; Wang, Y.; Yuan, Y.; Xie, Y. A systematic review on antibiotics misuse in livestock and aquaculture and regulation implications in China. Sci. Total Environ. 2021, 798, 149205. [Google Scholar] [CrossRef] [PubMed]
- European Sales and Use of Antimicrobials for Veterinary Medicine (ESUAvet) annual surveillance reports | European Medicines Agency (EMA). 31 March 2025. Available online: https://www.ema.europa.eu/en/veterinary-regulatory-overview/antimicrobial-resistance-veterinary-medicine/european-sales-use-antimicrobials-veterinary-medicine-esuavet-annual-surveillance-reports (accessed on 29 April 2025).
- Van Boeckel, T.P.; Glennon, E.E.; Chen, D.; Gilbert, M.; Robinson, T.P.; Grenfell, B.T.; Levin, S.A.; Bonhoeffer, S.; Laxminarayan, R. Reducing antimicrobial use in food animals. Science 2017, 357, 1350–1352. [Google Scholar] [CrossRef]
- Antimicrobial Resistance & Patient Safety Portal. Available online: https://arpsp.cdc.gov/profile/antibiotic-resistance?tab=antibiotic-resistance (accessed on 18 May 2025).
- Antimicrobial resistance in the EU/EEA (EARS-Net)—Annual Epidemiological Report 2023. 18 November 2024. Available online: https://www.ecdc.europa.eu/en/publications-data/antimicrobial-resistance-eueea-ears-net-annual-epidemiological-report-2023 (accessed on 18 May 2025).
- Ge, B.; Mukherjee, S.; Hsu, C.-H.; Davis, J.A.; Tran, T.T.T.; Yang, Q.; Abbott, J.W.; Ayers, S.L.; Young, S.R.; Crarey, E.T.; et al. MRSA and multidrug-resistant Staphylococcus aureus in U.S. retail meats, 2010–2011. Food Microbiol. 2017, 62, 289–297. [Google Scholar] [CrossRef]
- Wu, S.; Huang, J.; Zhang, F.; Wu, Q.; Zhang, J.; Pang, R.; Zeng, H.; Yang, X.; Chen, M.; Wang, J.; et al. Prevalence and Characterization of Food-Related Methicillin-Resistant Staphylococcus aureus (MRSA) in China. Front. Microbiol. 2019, 10, 304. [Google Scholar] [CrossRef]
- Karlowsky, J.A.; Lob, S.H.; DeRyke, C.A.; Siddiqui, F.; Young, K.; Motyl, M.R.; Sahm, D.F. Prevalence of ESBL non-CRE Escherichia coli and Klebsiella pneumoniae among clinical isolates collected by the SMART global surveillance programme from 2015 to 2019. Int. J. Antimicrob. Agents 2022, 59, 106535. [Google Scholar] [CrossRef]
- Authority, E.F.S.; European Centre for Disease Prevention and Control. The European Union summary report on antimicrobial resistance in zoonotic and indicator bacteria from humans, animals and food in 2022–2023. EFSA J. 2025, 23, e9237. [Google Scholar] [CrossRef]
- Li, S.; Liu, X.; Zhao, H.; Zhang, Y.; Lu, Z.; Wang, J.; Li, R.; Xie, P.; Hu, Y.; Zhou, C.; et al. Genome-Based Molecular Diversity of Extended-Spectrum β-Lactamase-Producing Escherichia coli From Pigeons in China. Transbound. Emerg. Dis. 2024, 2024, 1828830. [Google Scholar] [CrossRef]
- Shaheen, B.W.; Nayak, R.; Foley, S.L.; Kweon, O.; Deck, J.; Park, M.; Rafii, F.; Boothe, D.M. Molecular Characterization of Resistance to Extended-Spectrum Cephalosporins in Clinical Escherichia coli Isolates from Companion Animals in the United States. Antimicrob. Agents Chemother. 2011, 55, 5666–5675. [Google Scholar] [CrossRef]
- Reports | ATLAS. Available online: https://atlas-surveillance.com/r/antibacterials/database/susceptibility (accessed on 18 May 2025).
- Zou, M.; Ma, P.-P.; Liu, W.-S.; Liang, X.; Li, X.-Y.; Li, Y.-Z.; Liu, B.-T. Prevalence and Antibiotic Resistance Characteristics of Extraintestinal Pathogenic Escherichia coli among Healthy Chickens from Farms and Live Poultry Markets in China. Animals 2021, 11, 1112. [Google Scholar] [CrossRef] [PubMed]
- Medicine C for V. NARMS Now: Integrated Data. FDA. 21 April 2025. Available online: https://www.fda.gov/animal-veterinary/national-antimicrobial-resistance-monitoring-system/narms-now-integrated-data (accessed on 16 May 2025).
- Seyedjavadi, S.S.; Goudarzi, M.; Sabzehali, F. Relation between blaTEM, blaSHV and blaCTX-M genes and acute urinary tract infections. J. Acute Dis. 2016, 5, 71–76. [Google Scholar] [CrossRef]
- Wang, C.H.; Siu, L.K.; Chang, F.Y.; Tsai, Y.K.; Huang, L.Y.; Lin, J.C. Influence of PhoPQ and PmrAB two component system alternations on colistin resistance from non-mcr colistin resistant clinical E. Coli strains. BMC Microbiol. 2024, 24, 109. [Google Scholar] [CrossRef] [PubMed]
- Ramatla, T.; Mafokwane, T.; Lekota, K.; Monyama, M.; Khasapane, G.; Serage, N.; Nkhebenyane, J.; Be-zuidenhout, C.; Thekisoe, O. “One Health” perspective on prevalence of co-existing extended-spectrum β-lactamase (ESBL)-producing Escherichia coli and Klebsiella pneumoniae: A comprehensive systematic review and meta-analysis. Ann. Clin. Microbiol. Antimicrob. 2023, 22, 88. [Google Scholar] [CrossRef]
- Castanheira, M.; Simner, P.J.; Bradford, P.A. Extended-spectrum β-lactamases: An update on their characteristics, epidemiology and detection. JAC Antimicrob. Resist. 2021, 3, dlab092. [Google Scholar] [CrossRef]
- Ballén, V.; Gabasa, Y.; Ratia, C.; Ortega, R.; Tejero, M.; Soto, S. Antibiotic Resistance and Virulence Profiles of Klebsiella pneumoniae Strains Isolated From Different Clinical Sources. Front. Cell Infect. Microbiol. 2021, 11, 738223. [Google Scholar] [CrossRef]
- Agga, G.E.; Silva, P.J.; Martin, R.S. Detection of Extended-Spectrum Beta-Lactamase-Producing and Carbapenem-Resistant Bacteria from Mink Feces and Feed in the United States. Foodborne Pathog. Dis. 2021, 18, 497–505. [Google Scholar] [CrossRef]
- Geleta, D.; Abebe, G.; Tilahun, T.; Abdissa, A.; Mihret, A.; Cataldo, R.J.; Workneh, N.; Negash, A.A.; Beyene, G. Molecular and clinical insights into extended-spectrum β-lactamase genes of Klebsiella pneumoniae isolated from neonatal sepsis in Ethiopia. BMC Infect. Dis. 2024, 24, 1442. [Google Scholar] [CrossRef]
- Song, Y.; An, Q.; Chen, S.; Dai, H.; Ma, S.; Wu, C.; Lyu, Y.; Shen, J.; Krüger-Haker, H.; Schwarz, S.; et al. Antimicrobial resistance of pet-derived bacteria in China, 2000–2020. Antimicrob. Agents Chemother. 2025, 69, e01657-24. [Google Scholar] [CrossRef]
- Tsitsos, A.; Damianos, A.; Tsiouris, V.; Papapanagiotou, E.; Soultos, N.; Papa, A.; Tyrodimos, I.; Economou, V. Prevalence, seasonal variation, and proteomic relationship of β-lactamase-producing Escherichia coli, Klebsiella pneumoniae, and Acinetobacter spp. in poultry meat at the abattoir level in Greece. Food Microbiol. 2025, 128, 104709. [Google Scholar] [CrossRef]
- Alkan Bilik, Ö.; Bayraktar, M.; Özcan, N.; Gül, K.; Akpolat, N. Dissemination of blaOXA-48 like, blaNDM, blaKPC, blaIMP-1, blaVIM genes among carbapenem-resistant Escherichia coli and Klebsiella pneumoniae strains in Southeastern Turkey: First report of Klebsiella pneumoniae co-producing blaOXA-48-like, blaVIM and blaIMP-1 genes. Rev. Res. Med. Microbiol. 2021, 32, 205. [Google Scholar] [CrossRef]
- Maravić, A.; Šamanić, I.; Šprung, M.; Fredotović, Ž.; Ilić, N.; Dragičević, J.; Puizina, J. Broad-spectrum resistance of Pseudomonas aeruginosa from shellfish: Infrequent acquisition of novel resistance mechanisms. Environ. Monit. Assess. 2018, 190, 81. [Google Scholar] [CrossRef] [PubMed]
- Haenni, M.; Hocquet, D.; Ponsin, C.; Cholley, P.; Guyeux, C.; Madec, J.-Y.; Bertrand, X. Population structure and antimicrobial susceptibility of Pseudomonas aeruginosa from animal infections in France. BMC Vet. Res. 2015, 11, 9. [Google Scholar] [CrossRef]
- Martínez-Zavaleta, M.G.; Fernández-Rodríguez, D.; Hernández-Durán, M.; Colín-Castro, C.A.; de Lourdes García-Hernández, M.; Becerra-Lobato, N.; Franco-Cendejas, R.; López-Jácome, L.E. Acquired blaVIM and blaGES Carbapenemase-Encoding Genes in Pseudomonas aeruginosa: A Seven-Year Survey Highlighting an Increasing Epidemiological Threat. Pathogens 2023, 12, 1256. [Google Scholar] [CrossRef] [PubMed]
- Klotz, P.; Higgins, P.G.; Schaubmar, A.R.; Failing, K.; Leidner, U.; Seifert, H.; Scheufen, S.; Semmler, T.; Ewers, C. Seasonal Occurrence and Carbapenem Susceptibility of Bovine Acinetobacter baumannii in Germany. Front. Microbiol. 2019, 10, 272. [Google Scholar] [CrossRef]
- Ghaffoori Kanaan, M.H.; Al-Shadeedi, S.M.J.; Al-Massody, A.J.; Ghasemian, A. Drug resistance and virulence traits of Acinetobacter baumannii from Turkey and chicken raw meat. Comp. Immunol. Microbiol. Infect. Dis. 2020, 70, 101451. [Google Scholar] [CrossRef]
- Askari, N.; Momtaz, H.; Tajbakhsh, E. Prevalence and phenotypic pattern of antibiotic resistance of Acinetobacter baumannii isolated from different types of raw meat samples in Isfahan, Iran. Vet. Med. Sci. 2020, 6, 147–153. [Google Scholar] [CrossRef]
- Khalid, H.M. Molecular study of blaVIM and blaIMP genes in Acinetobacter baumannii strains isolated from burn patients in Duhok City, Iraq. J. Infect. Dev. Ctries. 2024, 18, 101–105. [Google Scholar] [CrossRef]
- Kahn, L.H. Antimicrobial resistance: A One Health perspective. Trans. R. Soc. Trop. Med. Hyg. 2017, 111, 255–260. [Google Scholar] [CrossRef]
- Reinseth, I.S.; Ovchinnikov, K.V.; Tønnesen, H.H.; Carlsen, H.; Diep, D.B. The Increasing Issue of Vancomycin-Resistant Enterococci and the Bacteriocin Solution. Probiotics Antimicro. Prot. 2020, 12, 1203–1217. [Google Scholar] [CrossRef]
- He, Y.-H.; Ruan, G.-J.; Hao, H.; Xue, F.; Ma, Y.-K.; Zhu, S.-N.; Zheng, B. Real-time PCR for the rapid detection of vanA, vanB and vanM genes. J. Microbiol. Immunol. Infect. 2020, 53, 746–750. [Google Scholar] [CrossRef] [PubMed]
- Chu, Y.; Wang, D.; Hao, W.; Sun, R.; Sun, J.; Liu, Y.; Liao, X. Prevalence, antibiotic resistance, virulence genes and molecular characteristics of Salmonella isolated from ducks and wild geese in China. Food Microbiol. 2024, 118, 104423. [Google Scholar] [CrossRef]
- Nathania, I.; Nainggolan, I.M.; Yasmon, A.; Nusatia, A.C.M.; Tjoa, E.; Gunardi, W.D.; Moehario, L.H. Hotspots sequences of gyrA, gyrB, parC, and parE genes encoded for fluoroquinolones resistance from local Salmonella Typhi strains in Jakarta. BMC Microbiol. 2022, 22, 250. [Google Scholar] [CrossRef] [PubMed]
- Rousham, E.K.; Unicomb, L.; Islam, M.A. Human, animal and environmental contributors to antibiotic resistance in low-resource settings: Integrating behavioural, epidemiological and One Health approaches. Proc. Biol. Sci. 2018, 285, 20180332. [Google Scholar] [CrossRef] [PubMed]
- Allen, H.K.; Donato, J.; Wang, H.H.; Cloud-Hansen, K.A.; Davies, J.; Handelsman, J. Call of the wild: Antibiotic resistance genes in natural environments. Nat. Rev. Microbiol. 2010, 8, 251–259. [Google Scholar] [CrossRef]
- Yu, P.; Dong, P.; Wang, H. Deciphering changes in the abundance of intracellular and extracellular antibiotic resistance genes and mobile genetic elements under anaerobic fermentation: Driven by bacterial community. Bioresour. Technol. 2022, 355, 127264. [Google Scholar] [CrossRef]
- Irfan, M.; Almotiri, A.; AlZeyadi, Z.A. Antimicrobial Resistance and Its Drivers—A Review. Antibiotics 2022, 11, 1362. [Google Scholar] [CrossRef]
- Darby, E.M.; Trampari, E.; Siasat, P.; Gaya, M.S.; Alav, I.; Webber, M.A.; Blair, J.M.A. Molecular mechanisms of antibiotic resistance revisited. Nat. Rev. Microbiol. 2023, 21, 280–295. [Google Scholar] [CrossRef]
- AlMatar, M.; Albarri, O.; Makky, E.A.; Köksal, F. Efflux pump inhibitors: New updates. Pharmacol. Rep. 2021, 73, 1–16. [Google Scholar] [CrossRef]
- Nadeem, S.F.; Gohar, U.F.; Tahir, S.F.; Mukhtar, H.; Pornpukdeewattana, S.; Nukthamna, P.; Moula Ali, A.M.; Bavisetty, S.C.B.; Massa, S. Antimicrobial resistance: More than 70 years of war between humans and bacteria. Crit. Rev. Microbiol. 2020, 46, 578–599. [Google Scholar] [CrossRef]
- Jacques, M.; Malouin, F. One Health-One Biofilm. Vet. Res. 2022, 53, 51. [Google Scholar] [CrossRef] [PubMed]
- Tooke, C.L.; Hinchliffe, P.; Bragginton, E.C.; Colenso, C.K.; Hirvonen, V.H.A.; Takebayashi, Y.; Spencer, J. β-Lactamases and β-Lactamase Inhibitors in the 21st Century. J. Mol. Biol. 2019, 431, 3472–3500. [Google Scholar] [CrossRef] [PubMed]
- Pang, L.; Weeks, S.D.; Van Aerschot, A. Aminoacyl-tRNA Synthetases as Valuable Targets for Antimicrobial Drug Discovery. Int. J. Mol. Sci. 2021, 22, 1750. [Google Scholar] [CrossRef] [PubMed]
- Moura de Sousa, J.; Lourenço, M.; Gordo, I. Horizontal gene transfer among host-associated microbes. Cell Host Microbe. 2023, 31, 513–527. [Google Scholar] [CrossRef]
- Magnusson, U. Antimicrobial use and resistance in food-producing animals-How can we protect the efficacy of antibiotics for reproductive diseases? Reprod. Domest. Anim. 2022, 57, 13–20. [Google Scholar] [CrossRef]
- Sweileh, W.M. Global research publications on irrational use of antimicrobials: Call for more research to contain antimicrobial resistance. Glob. Health 2021, 17, 94. [Google Scholar] [CrossRef]
- Rocha, D.C.; da Silva Rocha, C.; Tavares, D.S.; de Morais Calado, S.L.; Gomes, M.P. Veterinary antibiotics and plant physiology: An overview. Sci. Total Environ. 2021, 767, 144902. [Google Scholar] [CrossRef]
- Anedda, E.; Farrell, M.L.; Morris, D.; Burgess, C.M. Evaluating the impact of heavy metals on antimicrobial resistance in the primary food production environment: A scoping review. Environ. Pollut. 2023, 320, 121035. [Google Scholar] [CrossRef]
- Morones-Ramirez, J.R.; Winkler, J.A.; Spina, C.S.; Collins, J.J. Silver enhances antibiotic activity against gram-negative bacteria. Sci. Transl. Med. 2013, 5, 190ra81. [Google Scholar] [CrossRef]
- Rilstone, V.; Vignale, L.; Craddock, J.; Cushing, A.; Filion, Y.; Champagne, P. The role of antibiotics and heavy metals on the development, promotion, and dissemination of antimicrobial resistance in drinking water biofilms. Chemosphere 2021, 282, 131048. [Google Scholar] [CrossRef]
- Pu, Q.; Fan, X.T.; Li, H.; An, X.L.; Lassen, S.B.; Su, J.Q. Cadmium enhances conjugative plasmid transfer to a fresh water microbial community. Environ. Pollut. 2021, 268, 115903. [Google Scholar] [CrossRef] [PubMed]
- Maillard, J.Y.; Pascoe, M. Disinfectants and antiseptics: Mechanisms of action and resistance. Nat. Rev. Microbiol. 2024, 22, 4–17. [Google Scholar] [CrossRef] [PubMed]
- Razzaque, M.S. Exacerbation of antimicrobial resistance: Another casualty of the COVID-19 pandemic? Expert Rev. Anti. Infect. Ther. 2021, 19, 967–971. [Google Scholar] [CrossRef] [PubMed]
- Wushouer, H.; Luo, Z.; Guan, X.; Shi, L. Influence of Government Price Regulation on the Price, Volume and Spending of Antibiotics in China: A Controlled Interrupted Time Series Study. Int. J. Health Policy Manag. 2022, 11, 218–223. [Google Scholar] [CrossRef]
- Okubo, Y.; Nishi, A.; Michels, K.B.; Nariai, H.; Kim-Farley, R.J.; Arah, O.A.; Uda, K.; Kinoshita, N.; Miyairi, I. The consequence of financial incentives for not prescribing antibiotics: A Japan’s nationwide quasi-experiment. Int. J. Epidemiol. 2022, 51, 1645–1655. [Google Scholar] [CrossRef]
- Li, Z.; Li, M.; Zhang, Z.; Li, P.; Zang, Y.; Liu, X. Antibiotics in aquatic environments of China: A review and meta-analysis. Ecotoxicol. Environ. Saf. 2020, 199, 110668. [Google Scholar] [CrossRef]
- Xu, L.; Zang, J.; Cong, W.; Holton, E.; Jiang, L.; Sheppard, S.K.; Wang, Y.; Wang, N.; Weeks, J.; Fu, C.; et al. Assessment of community-wide antimicrobials usage in Eastern China using wastewater-based epidemiology. Water Res. 2022, 222, 118942. [Google Scholar] [CrossRef]
- Nguyen, A.Q.; Vu, H.P.; Nguyen, L.N.; Wang, Q.; Djordjevic, S.P.; Donner, E.; Yin, H.; Nghiem, L.D. Monitoring antibiotic resistance genes in wastewater treatment: Current strategies and future challenges. Sci. Total Environ. 2021, 783, 146964. [Google Scholar] [CrossRef]
- Guenin, M.J.; Studnitz, M.; Molia, S. Interventions to change antimicrobial use in livestock: A scoping review and an impact pathway analysis of what works, how, for whom and why. Prev. Vet. Med. 2023, 220, 106025. [Google Scholar] [CrossRef]
- Gao, J.; Yang, Z.; Zhao, C.; Tang, X.; Jiang, Q.; Yin, Y. A comprehensive review on natural phenolic compounds as alternatives to in-feed antibiotics. Sci. China Life Sci. 2023, 66, 1518–1534. [Google Scholar] [CrossRef]
- Huang, J.; Wang, Z.; Chen, Z.; Liang, H.; Li, X.; Li, B. Occurrence and Removal of Antibiotic Resistance in Nationwide Hospital Wastewater Deciphered by Metagenomics Approach—China, 2018–2022. China CDC Wkly. 2023, 5, 1023–1028. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.J.; Kim, D.W.; Kim, C.; Ryu, H.D.; Chung, E.G.; Kim, K. Concentrations and Risk Assessments of Antibiotics in an Urban-Rural Complex Watershed with Intensive Livestock Farming. Int. J. Environ. Res. Public Health 2021, 18, 10797. [Google Scholar] [CrossRef] [PubMed]
- Gharbi, M.; Moore, L.S.P.; Castro-Sánchez, E.; Spanoudaki, E.; Grady, C.; Holmes, A.H.; Drumright, L.N. A needs assessment study for optimising prescribing practice in secondary care junior doctors: The Antibiotic Prescribing Education among Doctors (APED). BMC Infect. Dis. 2016, 16, 456. [Google Scholar] [CrossRef] [PubMed]
- Pandolfo, A.M.; Horne, R.; Jani, Y.; Reader, T.W.; Bidad, N.; Brealey, D.; Enne, V.I.; Livermore, D.M.; Gant, V.; Brett, S.J. Understanding decisions about antibiotic prescribing in ICU: An application of the Necessity Concerns Framework. BMJ Qual. Saf. 2022, 31, 199–210. [Google Scholar] [CrossRef]
- De Waele, J.J.; Akova, M.; Antonelli, M.; Canton, R.; Carlet, J.; De Backer, D.; Dimopoulos, G.; Gar-nacho-Montero, J.; Kesecioglu, J.; Lipman, J.; et al. Antimicrobial resistance and antibiotic stewardship programs in the ICU: Insistence and persistence in the fight against resistance. A position statement from ESICM/ESCMID/WAAAR round table on multi-drug resistance. Intensive Care Med. 2018, 44, 189–196. [Google Scholar] [CrossRef]
- Mathew, P.; Chandy, S.J.; Ranjalkar, J. Community engagement to mitigate antimicrobial resistance in low-and middle-income countries—An essential strategy for implementation of national action plans on AMR. Lancet Reg. Health—Southeast Asia 2024, 24, 100379. [Google Scholar] [CrossRef]
- Ravinetto, R.M.; Dujardin, C. The threat of poor quality antibiotics in increasing antimicrobial resistance. BMJ 2016, 354, i3618. [Google Scholar] [CrossRef]
- Wu, Y.; Zeng, Z. Antibiotic Residues, Antimicrobial Resistance and Intervention Strategies of Foodborne Pathogens. Antibiotics 2024, 13, 321. [Google Scholar] [CrossRef]
- Khan, M.Z.H. Recent Biosensors for Detection of Antibiotics in Animal Derived Food. Crit. Rev. Anal. Chem. 2022, 52, 780–790. [Google Scholar] [CrossRef]
- Nguyen, B.A.T.; Chen, Q.L.; He, J.Z.; Hu, H.W. Livestock manure spiked with the antibiotic tylosin significantly altered soil protist functional groups. J. Hazard. Mater. 2022, 427, 127867. [Google Scholar] [CrossRef]
- Magnano San Lio, R.; Favara, G.; Maugeri, A.; Barchitta, M.; Agodi, A. How Antimicrobial Resistance Is Linked to Climate Change: An Overview of Two Intertwined Global Challenges. Int. J. Environ. Res. Public Health 2023, 20, 1681. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Verdugo, A.; Lozano-Huntelman, N.; Cruz-Loya, M.; Savage, V.; Yeh, P. Compounding Effects of Climate Warming and Antibiotic Resistance. iScience 2020, 23, 101024. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Li, Z.; Hu, J.; Yan, L.; He, Y.; Li, X.; Wang, M.; Sun, X.; Xu, H. The biological and chemical contents of atmospheric particulate matter and implication of its role in the transmission of bacterial pathogenesis. Environ. Microbiol. 2021, 23, 5481–5486. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Lim, J.; Koh, J.; Beard, J.; Rowe, J.W. Research Network on an Aging Society. A global analysis of adaptation to societal aging across low-, middle- and high-income countries using the Global Aging Society Index. Nat. Aging 2025, 5, 113–121. [Google Scholar] [CrossRef]
- Cattaneo, D.; Falcone, M.; Gervasoni, C.; Marriott, D.J.E. Therapeutic Drug Monitoring of Antibiotics in the Elderly: A Narrative Review. Ther. Drug Monit. 2022, 44, 75. [Google Scholar] [CrossRef]
- Wang, Y.; Lu, J.; Engelstädter, J.; Zhang, S.; Ding, P.; Mao, L.; Yuan, Z.; Bond, P.L.; Guo, J. Non-antibiotic pharmaceuticals enhance the transmission of exogenous antibiotic resistance genes through bacterial transformation. ISME J. 2020, 14, 2179–2196. [Google Scholar] [CrossRef]
- Su, G.; Xu, H.; Riggi, E.; He, Z.; Lu, L.; Lindholm, B.; Marrone, G.; Wen, Z.; Liu, X.; Johnson, D.W.; et al. Association of Kidney Function with Infections by Multidrug-Resistant Organisms: An Electronic Medical Record Analysis. Sci. Rep. 2018, 8, 13372. [Google Scholar] [CrossRef]
- Fisher, M.C.; Alastruey-Izquierdo, A.; Berman, J.; Bicanic, T.; Bignell, E.M.; Bowyer, P.; Bromley, M.; Brügge-mann, R.; Garber, G.; Cornely, O.A.; et al. Tackling the emerging threat of antifungal resistance to human health. Nat. Rev. Microbiol. 2022, 20, 557–571. [Google Scholar] [CrossRef]
- Komp Lindgren, P.; Higgins, P.G.; Seifert, H.; Cars, O. Prevalence of hypermutators among clinical Acinetobacter baumannii isolates. J. Antimicrob. Chemother. 2016, 71, 661–665. [Google Scholar] [CrossRef]
- Huijbers, P.M.C.; Blaak, H.; de Jong, M.C.M.; Graat, E.A.M.; Vandenbroucke-Grauls, C.M.J.E.; de Roda Husman, A.M. Role of the Environment in the Transmission of Antimicrobial Resistance to Humans: A Review. Environ. Sci. Technol. 2015, 49, 11993–12004. [Google Scholar] [CrossRef]
- Sakeena, M.H.F.; Bennett, A.A.; McLachlan, A.J. Non-prescription sales of antimicrobial agents at community pharmacies in developing countries: A systematic review. Int. J. Antimicrob. Agents 2018, 52, 771–782. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, J.; Paul, C.; Collins, A.; Abriti, A.; Sushil, B.; Laura, C.; Rajib, D.; Fariza, F.; Mariana, F.-B.; Huque, R.; et al. Community engagement: The key to tackling Antimicrobial Resistance (AMR) across a One Health context? Glob. Public Health 2022, 17, 2647–2664. [Google Scholar] [CrossRef] [PubMed]
- Holmes, A.H.; Moore, L.S.P.; Sundsfjord, A.; Steinbakk, M.; Regmi, S.; Karkey, A.; Guerin, P.J.; Piddock, L.J.V. Understanding the mechanisms and drivers of antimicrobial resistance. Lancet 2016, 387, 176–187. [Google Scholar] [CrossRef] [PubMed]
- World Population Prospects 2022: Summary of Results | Population Division. Available online: https://www.un.org/development/desa/pd/content/World-Population-Prospects-2022 (accessed on 12 May 2025).
- Ramirez, J.; Guarner, F.; Bustos Fernandez, L.; Maruy, A.; Sdepanian, V.L.; Cohen, H. Antibiotics as Major Disruptors of Gut Microbiota. Front. Cell Infect. Microbiol. 2020, 10, 572912. [Google Scholar] [CrossRef]
- Theophilus, R.J.; Taft, D.H. Antimicrobial Resistance Genes (ARGs), the Gut Microbiome, and Infant Nutrition. Nutrients 2023, 15, 3177. [Google Scholar] [CrossRef]
- Liu, C.; Chen, J.; Yang, Y.; Teng, Y.; Chen, H. Biogeography and diversity patterns of antibiotic resistome in the sediments of global lakes. J. Environ. Sci. 2023, 127, 421–430. [Google Scholar] [CrossRef]
- Yang, L.; Shen, Y.; Jiang, J.; Wang, X.; Shao, D.; Lam, M.M.C.; Holt, K.E.; Shao, B.; Wu, C.; Shen, J.; et al. Distinct increase in antimicrobial resistance genes among Escherichia coli during 50 years of antimicrobial use in livestock production in China. Nat. Food 2022, 3, 197–205. [Google Scholar] [CrossRef]
- Bai, H.; He, L.-Y.; Wu, D.-L.; Gao, F.-Z.; Zhang, M.; Zou, H.-Y.; Yao, M.-S.; Ying, G.-G. Spread of airborne antibiotic resistance from animal farms to the environment: Dispersal pattern and exposure risk. Environ. Int. 2022, 158, 106927. [Google Scholar] [CrossRef]
- Peng, Z.; Maciel-Guerra, A.; Baker, M.; Zhang, X.; Hu, Y.; Wang, W.; Rong, J.; Zhang, J.; Xue, N.; Barrow, P.; et al. Whole-genome sequencing and gene sharing network analysis powered by machine learning identifies antibiotic resistance sharing between animals, humans and environment in livestock farming. PLoS Comput. Biol. 2022, 18, e1010018. [Google Scholar] [CrossRef]
- Yang, Y.; Hu, X.; Cai, S.; Hu, N.; Yuan, Y.; Wu, Y.; Wang, Y.; Mi, J.; Liao, X. Pet cats may shape the antibiotic resistome of their owner’s gut and living environment. Microbiome 2023, 11, 235. [Google Scholar] [CrossRef]
- Finisterra, L.; Duarte, B.; Peixe, L.; Novais, C.; Freitas, A.R. Industrial dog food is a vehicle of multidrug-resistant enterococci carrying virulence genes often linked to human infections. Int. J. Food Microbiol. 2021, 358, 109284. [Google Scholar] [CrossRef] [PubMed]
- Hamame, A.; Davoust, B.; Cherak, Z.; Rolain, J.M.; Diene, S.M. Mobile Colistin Resistance (mcr) Genes in Cats and Dogs and Their Zoonotic Transmission Risks. Pathogens 2022, 11, 698. [Google Scholar] [CrossRef] [PubMed]
- Desai, A.N.; Mohareb, A.M.; Hauser, N.; Abbara, A. Antimicrobial Resistance and Human Mobility. Infect. Drug Resist. 2022, 15, 127–133. [Google Scholar] [CrossRef] [PubMed]
- Allel, K.; Day, L.; Hamilton, A.; Lin, L.; Furuya-Kanamori, L.; Moore, C.E.; Van Boeckel, T.; Laxminarayan, R.; Yakob, L. Global antimicrobial-resistance drivers: An ecological country-level study at the human-animal interface. Lancet Planet Health 2023, 7, e291–e303. [Google Scholar] [CrossRef]
- Andersson, D.I.; Hughes, D. Selection and Transmission of Antibiotic-Resistant Bacteria. Microbiol. Spectr. 2017, 5, 10–1128. [Google Scholar] [CrossRef]
- Baquero, F.; Martínez, J.L.; F Lanza, V.; Rodríguez-Beltrán, J.; Galán, J.C.; San Millán, A.; Cantón, R.; Coque, T.M. Evolutionary Pathways and Trajectories in Antibiotic Resistance. Clin. Microbiol. Rev. 2021, 34, e0005019. [Google Scholar] [CrossRef]
- Lebreton, F.; van Schaik, W.; McGuire, A.M.; Godfrey, P.; Griggs, A.; Mazumdar, V.; Corander, J.; Cheng, L.; Saif, S.; Young, S.; et al. Emergence of epidemic multidrug-resistant Enterococcus faecium from animal and commensal strains. mBio 2013, 4, e00534-13. [Google Scholar] [CrossRef]
- Ho, C.S.; Wong, C.T.H.; Aung, T.T.; Lakshminarayanan, R.; Mehta, J.S.; Rauz, S.; McNally, A.; Kintses, B.; Peacock, S.J.; de la Fuente-Nunez, C.; et al. Antimicrobial resistance: A concise update. Lancet Microbe 2025, 6, 100947. [Google Scholar] [CrossRef]
- Wu, X.; Yang, L.; Wu, Y.; Li, H.; Shao, B. Spread of multidrug-resistant Pseudomonas aeruginosa in animal-derived foods in Beijing, China. Int. J. Food Microbiol. 2023, 403, 110296. [Google Scholar] [CrossRef]
- Baker, M.; Zhang, X.; Maciel-Guerra, A.; Dong, Y.; Wang, W.; Hu, Y.; Renney, D.; Hu, Y.; Liu, L.; Li, H.; et al. Machine learning and metagenomics reveal shared antimicrobial resistance profiles across multiple chicken farms and abattoirs in China. Nat. Food 2023, 4, 707–720. [Google Scholar] [CrossRef]
- Wu, H.; Wang, Y.; Wu, Y.; Qiao, J.; Li, H.; Zheng, S.; Xia, X.; Cui, S.; Wang, X.; Xi, M.; et al. Emergence of β-lactamases and extended-spectrum β-lactamases (ESBLs) producing Salmonella in retail raw chicken in China. Foodborne Pathog. Dis. 2015, 12, 228–234. [Google Scholar] [CrossRef] [PubMed]
- Wei, Z.; Xu, X.; Yan, M.; Chang, H.; Li, Y.; Kan, B.; Zeng, M. Salmonella Typhimurium and Salmonella Enteritidis Infections in Sporadic Diarrhea in Children: Source Tracing and Resistance to Third-Generation Cephalosporins and Ciprofloxacin. Foodborne Pathog. Dis. 2019, 16, 244–255. [Google Scholar] [CrossRef] [PubMed]
- Grudlewska-Buda, K.; Bauza-Kaszewska, J.; Wiktorczyk-Kapischke, N.; Budzyńska, A.; Gospodarek-Komkowska, E.; Skowron, K. Antibiotic Resistance in Selected Emerging Bacterial Foodborne Pathogens-An Issue of Concern? Antibiotics 2023, 12, 880. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Li, G.; Hong, Z.; Zhang, C.; Zhu, N.; Tan, Y.; Gao, T. Prevalence of Multidrug-Resistant Organisms in Healthy Adults in Shenzhen, China. Health Secur. 2023, 21, 122–129. [Google Scholar] [CrossRef] [PubMed]
- Lozano, C.; Gómez-Sanz, E.; Benito, D.; Aspiroz, C.; Zarazaga, M.; Torres, C. Staphylococcus aureus nasal carriage, virulence traits, antibiotic resistance mechanisms, and genetic lineages in healthy humans in Spain, with detection of CC398 and CC97 strains. Int. J. Med. Microbiol. 2011, 301, 500–505. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Zhang, E.; Abdulai, M.K.; Tia, A.B.; Ngegba, E.D.; Yin, J.; Xu, S.; Wang, L.; Dong, X.; Harding, D.; et al. Dissemination of Antibiotic Resistance Genes Among Patients with Diarrhea—Freetown, Sierra Leone, 2018. CCDCW 2022, 4, 1093–1096. [Google Scholar] [CrossRef]
- Ju, X.; Wang, S.; Yang, X.; Han, W.; Cai, C.; Wu, Y.; Liu, C.; Qian, J.; Zhao, X.; Qian, X.; et al. Epidemiology and Molecular Characteristics of mcr-9 in Citrobacter spp. from Healthy Individuals and Patients in China. Microbiol. Spectr. 2022, 10, e0134622. [Google Scholar] [CrossRef]
- Rojas García, P.; Antoñanzas Villar, F. Effects of economic and health policies on the consumption of antibiotics in a Spanish region. Expert Rev. Pharmacoecon. Outcomes Res. 2020, 20, 379–386. [Google Scholar] [CrossRef]
- Pulingam, T.; Parumasivam, T.; Gazzali, A.M.; Sulaiman, A.M.; Chee, J.Y.; Lakshmanan, M.; Chin, C.F.; Su-desh, K. Antimicrobial resistance: Prevalence, economic burden, mechanisms of resistance and strategies to overcome. Eur. J. Pharm. Sci. 2022, 170, 106103. [Google Scholar] [CrossRef]
- Shen, L.; Wei, X.; Yin, J.; Haley, D.R.; Sun, Q.; Lundborg, C.S. Interventions to optimize the use of antibiotics in China: A scoping review of evidence from humans, animals, and the environment from a One Health perspective. One Health 2022, 14, 100388. [Google Scholar] [CrossRef]
- AWaRe Classification of Antibiotics for Evaluation and Monitoring of Use. 2023. Available online: https://www.who.int/publications/i/item/WHO-MHP-HPS-EML-2023.04 (accessed on 27 February 2025).
- Khursheed, N.; Adnan, F.; Khan, M.A.; Hatif, R. Regional insights on the prevalence and antimicrobial susceptibility pattern of carbapenem and colistin-resistant gram-negative bacteria: An observational cross-sectional study from Karachi, Pakistan. BMC Infect. Dis. 2025, 25, 186. [Google Scholar] [CrossRef] [PubMed]
- Gobezie, M.Y.; Hassen, M.; Tesfaye, N.A.; Solomon, T.; Demessie, M.B.; Kassa, T.D.; Wendie, T.F.; Andualem, A.; Alemayehu, E.; Belayneh, Y.M. Prevalence of meropenem-resistant Pseudomonas Aeruginosa in Ethiopia: A systematic review and meta-analysis. Antimicrob. Resist. Infect. Control 2024, 13, 37. [Google Scholar] [CrossRef] [PubMed]
- Vikesland, P.J.; Pruden, A.; Alvarez, P.J.J.; Aga, D.; Bürgmann, H.; Li, X.-D.; Manaia, C.M.; Nambi, I.; Wig-ginton, K.; Zhang, T.; et al. Toward a Comprehensive Strategy to Mitigate Dissemination of Environmental Sources of Antibiotic Resistance. Environ. Sci. Technol. 2017, 51, 13061–13069. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zhang, J.; Wu, Y. National Action Plan in Antimicrobial Resistance Using Framework Analysis for China. China CDC Wkly. 2023, 5, 492–498. [Google Scholar] [CrossRef]
- Muloi, D.M.; Jauneikaite, E.; Anjum, M.F.; Essack, S.Y.; Singleton, D.A.; Kasudi, M.R.; Wade, M.J.; Egyir, B.; Nunn, J.G.; Midega, J.T.; et al. Exploiting genomics for antimicrobial resistance surveillance at One Health interfaces. Lancet Microbe 2023, 4, e1056–e1062. [Google Scholar] [CrossRef]
- Report Signals Increasing Resistance to Antibiotics in Bacterial Infections in Humans and Need for Better Data. Available online: https://www.who.int/news/item/09-12-2022-report-signals-increasing-resistance-to-antibiotics-in-bacterial-infections-in-humans-and-need-for-better-data (accessed on 5 March 2025).
- Temkin, E.; Fallach, N.; Almagor, J.; Gladstone, B.P.; Tacconelli, E.; Carmeli, Y. Estimating the number of infections caused by antibiotic-resistant Escherichia coli and Klebsiella pneumoniae in 2014: A modelling study. Lancet Glob. Health 2018, 6, e969–e979. [Google Scholar] [CrossRef]
- Shah, A.S.; Karunaratne, K.; Shakya, G.; Barreto, I.; Khare, S.; Paveenkittiporn, W.; Wangchuk, S.; Tin, H.H.; Muhsin, M.A.; Aung, L.; et al. Strengthening laboratory surveillance of antimicrobial resistance in South East Asia. BMJ 2017, 358, j3474. [Google Scholar] [CrossRef][Green Version]
- Gandra, S.; Alvarez-Uria, G.; Turner, P.; Joshi, J.; Limmathurotsakul, D.; van Doorn, H.R. Antimicrobial Resistance Surveillance in Low- and Middle-Income Countries: Progress and Challenges in Eight South Asian and Southeast Asian Countries. Clin. Microbiol. Rev. 2020, 33, 10.1128/cmr.00048-19. [Google Scholar] [CrossRef]
- Kim, J.I.; Maguire, F.; Tsang, K.K.; Gouliouris, T.; Peacock, S.J.; McAllister, T.A.; McArthur, A.G.; Beiko, R.G. Machine Learning for Antimicrobial Resistance Prediction: Current Practice, Limitations, and Clinical Perspective. Clin. Microbiol. Rev. 2022, 35, e0017921. [Google Scholar] [CrossRef]
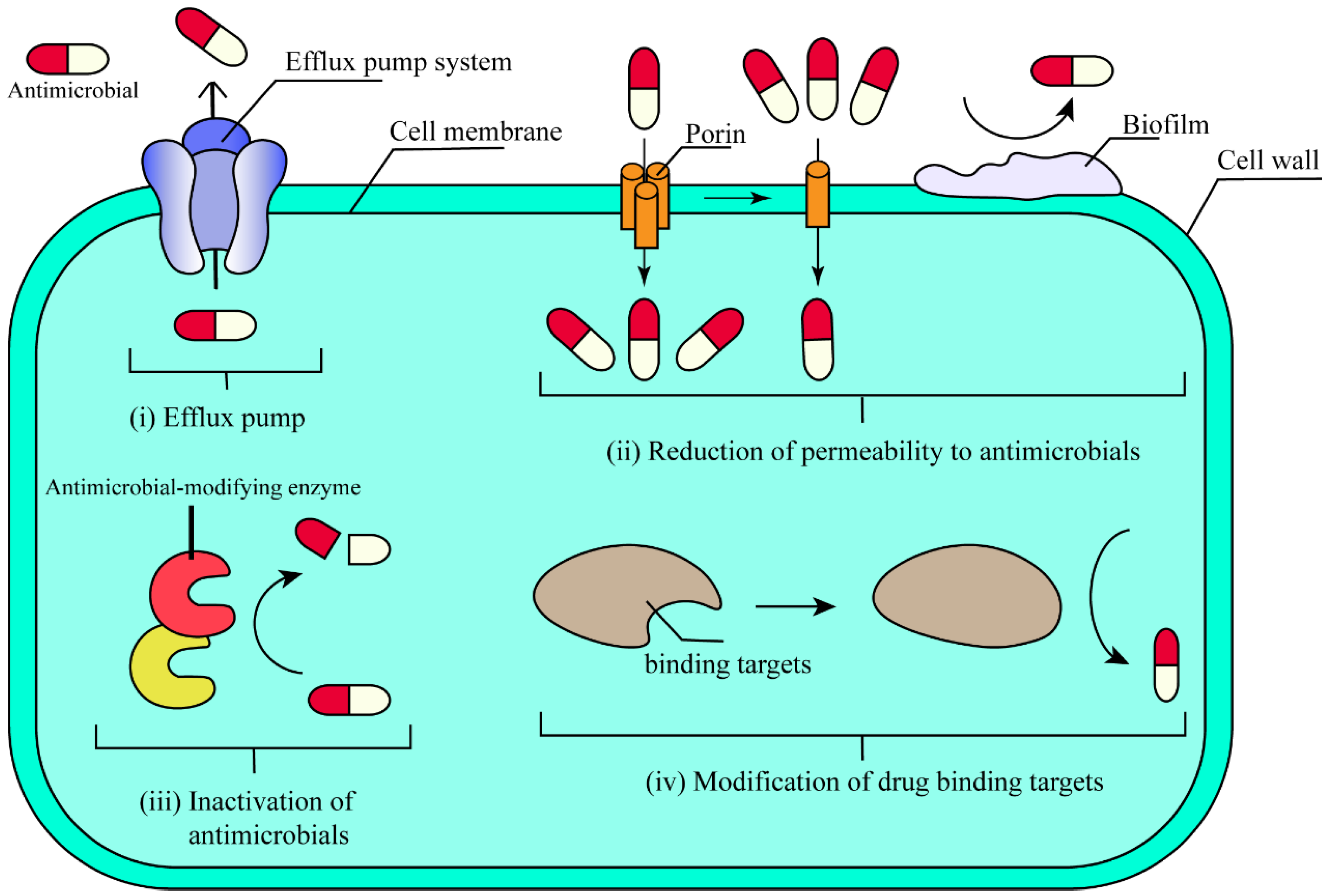
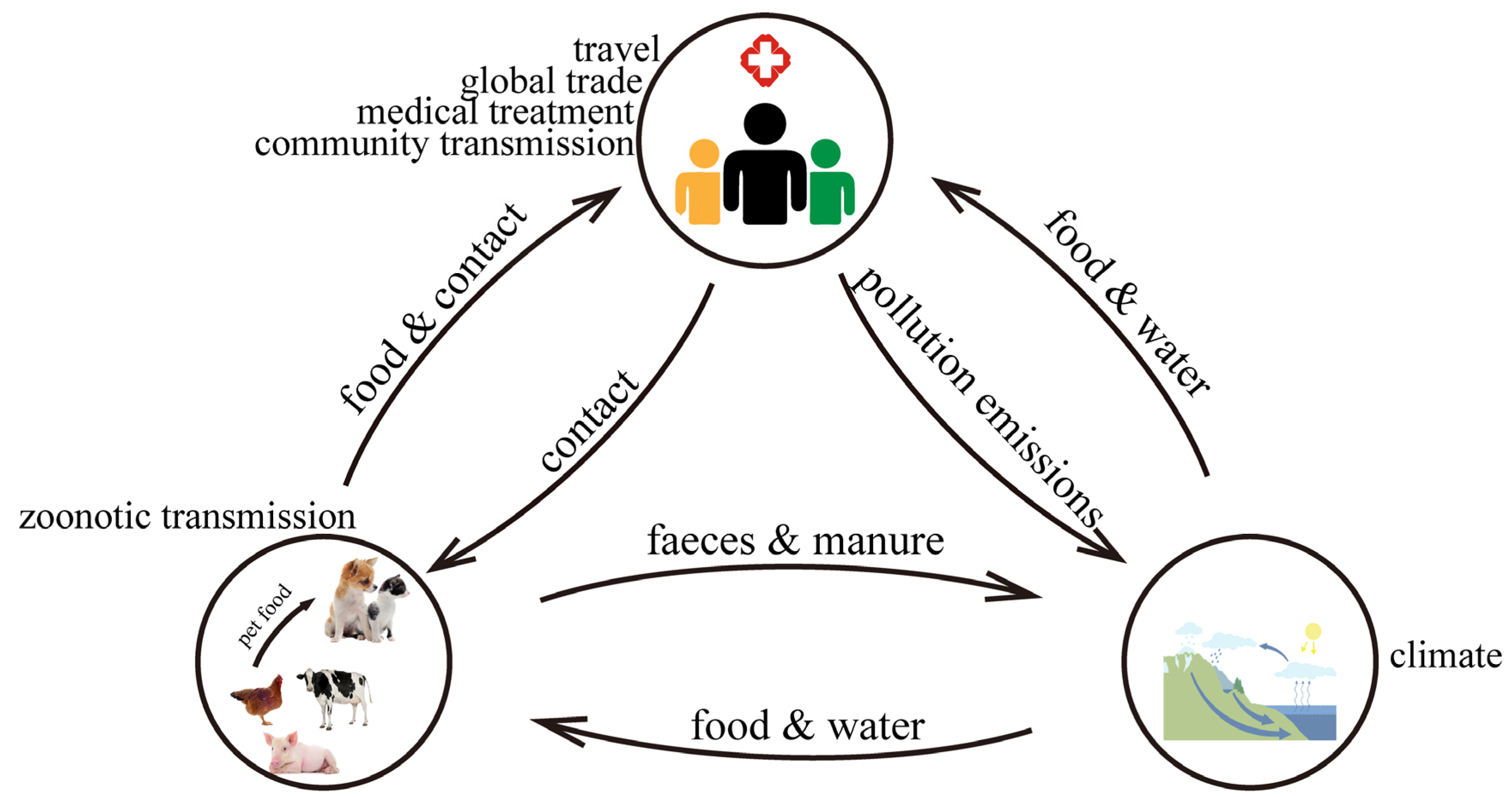
| Driving Factor | Current Status | Causes | Reference(s) | |
|---|---|---|---|---|
| Intrinsic determinants | Antimicrobial Resistance Genes | Antimicrobial resistance genes are widely distributed in bacterial populations, enabling resistance through enzymatic or structural adaptations. | Mutations and vertical gene transfer during bacterial replication. | [56,57,58,59,60,61] |
| Mobile Gene Elements | Mobile gene elements act as carriers for antimicrobial resistance genes, facilitating rapid horizontal spread. | Conjugation, transformation, and transduction mechanisms. | [62] | |
| Extrinsic drivers | Antimicrobial Misuse | Accumulation of antibiotic residues in the environment enhances bacterial resistance. | Overuse in healthcare and livestock, incomplete metabolism, and environmental discharge of antimicrobials. | [63,64,65] |
| Heavy Metal Effects | Synergistic effects between heavy metals and antimicrobials promote co-resistance. | Industrial/agricultural pollution; bacterial efflux pumps resisting both heavy metals and antimicrobials. | [66,67,68,69] | |
| Disinfectant Misuse | Increased cross-resistance to disinfectants and antimicrobials. | Excessive disinfectant use during pandemics; bacterial adaptation via target modification or efflux pumps. | [70,71] | |
| Social Factors | Widespread antimicrobial misuse and weak regulatory policies. | Inadequate national antimicrobial policies; poor enforcement of environmental regulations (e.g., wastewater discharge). | [72,73,74,75,76] | |
| Economic Factors | Intensive farming practices amplify antimicrobial residues and resistant bacteria. | Declining meat prices drive antimicrobial-dependent farming; surging antimicrobial use in livestock. | [77,78,79,80] | |
| Medicinal factors | (1) Hospitals and communities promote the development of AMR. (2) Substandard antimicrobials create subtherapeutic selection pressure. | Inappropriate prescribing and cross-infection in hospitals drive AMR transmission, while ICU-specific interventions exacerbate AMR risks. Community antimicrobial misuse drives AMR, compounded by weak healthcare systems and human–animal–environment interactions. Production of low-quality antimicrobials; irrational prescribing practices (e.g., underdosing). | [81,82,83,84,85] | |
| Health Factors | Contaminated food/environment increases human exposure to resistant bacteria. | Antimicrobial residues in animal-derived products; manure fertilization spreading AMR to soil and crops. | [86,87,88] | |
| Climate Factors | Climate change accelerates AMR spread via airborne particles and extreme weather. | Rising temperatures enhance bacterial growth/gene transfer; storms disperse pollutants and ARGs. | [89,90,91] | |
| Aging Factors | The aging population has accelerated the development of AMR. | Older adults’ susceptibility to infections and high prevalence of chronic diseases lead to increased medical visits in AMR-prone settings and antimicrobial usage. | [92,93,94,95] | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ye, Z.; Li, M.; Jing, Y.; Liu, K.; Wu, Y.; Peng, Z. What Are the Drivers Triggering Antimicrobial Resistance Emergence and Spread? Outlook from a One Health Perspective. Antibiotics 2025, 14, 543. https://doi.org/10.3390/antibiotics14060543
Ye Z, Li M, Jing Y, Liu K, Wu Y, Peng Z. What Are the Drivers Triggering Antimicrobial Resistance Emergence and Spread? Outlook from a One Health Perspective. Antibiotics. 2025; 14(6):543. https://doi.org/10.3390/antibiotics14060543
Chicago/Turabian StyleYe, Zehong, Menghan Li, Yiwen Jing, Kejun Liu, Yongning Wu, and Zixin Peng. 2025. "What Are the Drivers Triggering Antimicrobial Resistance Emergence and Spread? Outlook from a One Health Perspective" Antibiotics 14, no. 6: 543. https://doi.org/10.3390/antibiotics14060543
APA StyleYe, Z., Li, M., Jing, Y., Liu, K., Wu, Y., & Peng, Z. (2025). What Are the Drivers Triggering Antimicrobial Resistance Emergence and Spread? Outlook from a One Health Perspective. Antibiotics, 14(6), 543. https://doi.org/10.3390/antibiotics14060543







