Helicobacter pylori Antibiotic Resistance in Russia: A Systematic Review and Meta-Analysis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Search Strategy
2.2. Eligibility Criteria and Quality Assessment
2.3. Data Extraction
2.4. Statistical Analysis
3. Results
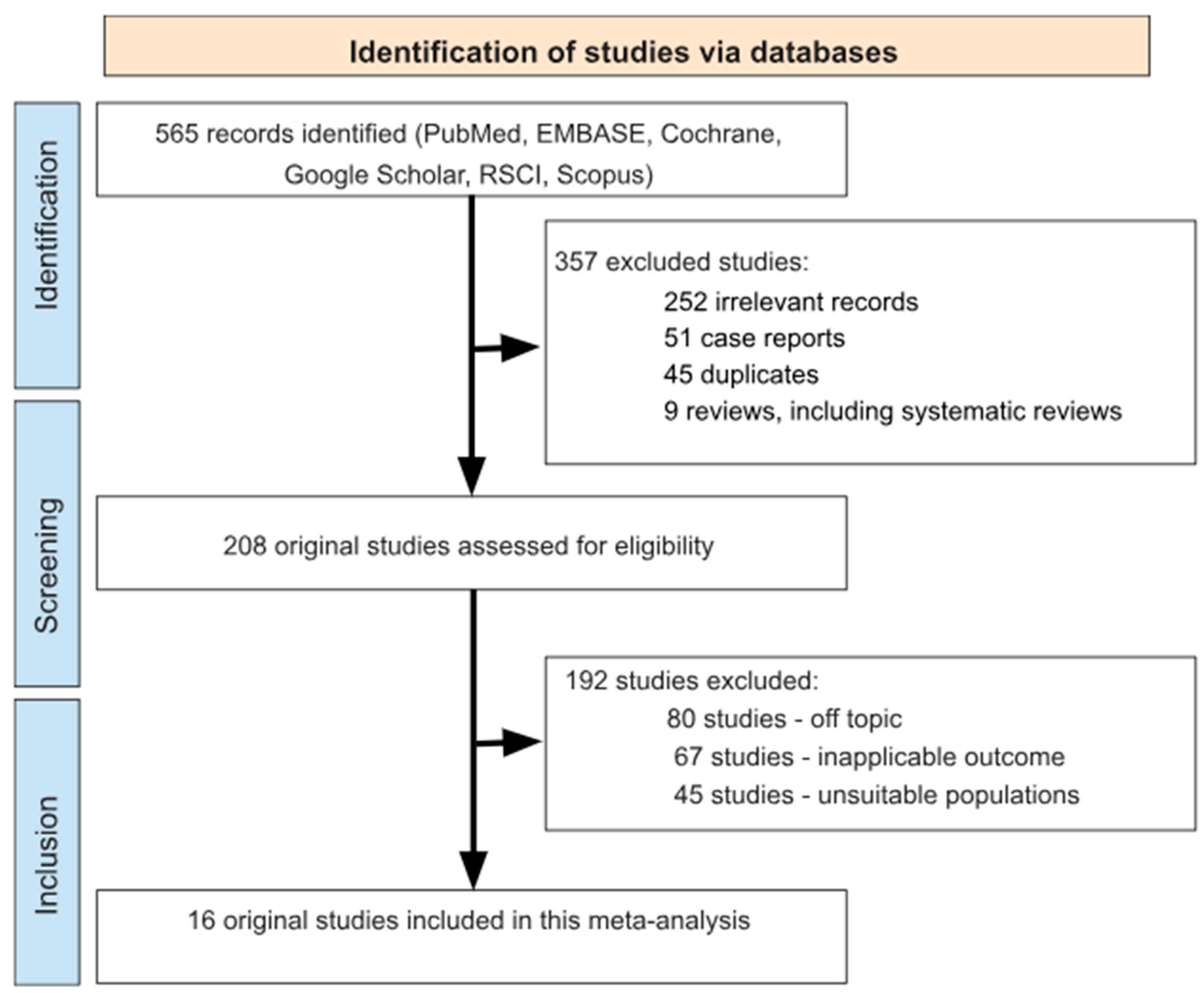
3.1. Search Results
3.2. Characteristics of Included Studies
3.3. Study Quality Evaluation, Reliability Analysis and Publication Bias
3.4. Pooled Resistance Rates
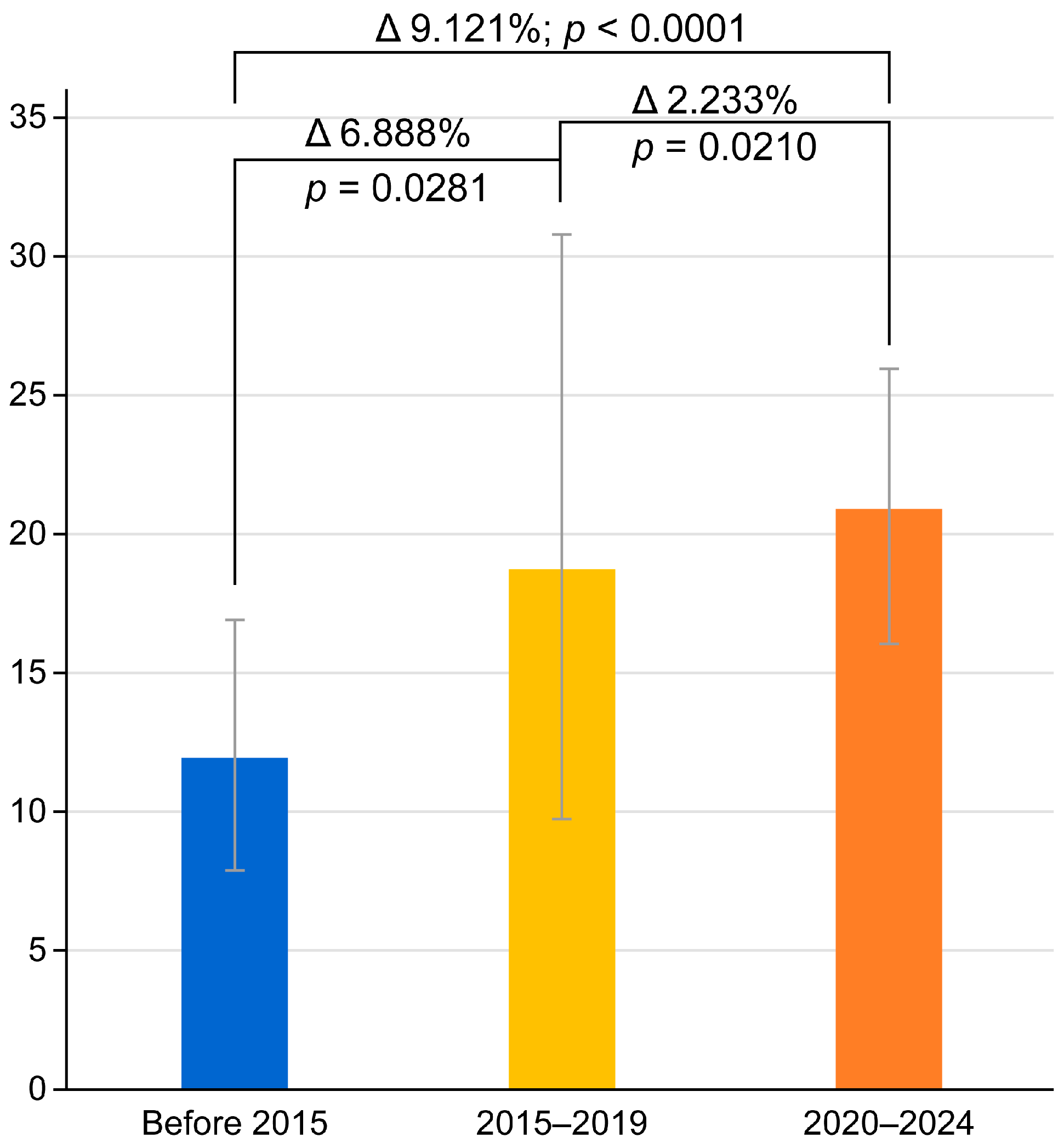
3.5. Chronological Dynamics of Antibiotic Resistance in H. pylori
3.6. Subanalyses
3.7. Sensitivity Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Malfertheiner, P.; Megraud, F.; Rokkas, T.; Gisbert, J.P.; Liou, J.M.; Schulz, C.; Gasbarrini, A.; Hunt, R.H.; Leja, M.; O’Morain, C.; et al. Management of Helicobacter pylori infection: The Maastricht VI/Florence consensus report. Gut 2022, 71, 1724–1762. [Google Scholar] [CrossRef] [PubMed]
- Chey, W.D.; Howden, C.W.; Moss, S.F.; Morgan, D.R.; Greer, K.B.; Grover, S.; Shah, S.C. ACG Clinical Guideline: Treatment of Helicobacter pylori Infection. Am. J. Gastroenterol. 2024, 119, 1730–1753. [Google Scholar] [CrossRef]
- Katelaris, P.; Hunt, R.; Bazzoli, F.; Cohen, H.; Fock, K.M.; Gemilyan, M.; Malfertheiner, P.; Mégraud, F.; Piscoya, A.; Quach, D.; et al. Helicobacter pylori World Gastroenterology Organization Global Guideline. J. Clin. Gastroenterol. 2023, 57, 111–126. [Google Scholar] [CrossRef]
- Iarc, L. Schistosomes, liver flukes and Helicobacter pylori. IARC Monogr. Evalutaion Carcinog. Risks Hum. 1994, 61, 1–241. [Google Scholar] [PubMed] [PubMed Central]
- Plummer, M.; Franceschi, S.; Vignat, J.; Forman, D.; de Martel, C. Global burden of gastric cancer attributable to Helicobacter pylori. Int. J. Cancer 2015, 136, 487–490. [Google Scholar] [CrossRef] [PubMed]
- Gu, J.; He, F.; Clifford, G.M.; Li, M.; Fan, Z.; Li, X.; Wang, S.; Wei, W. A systematic review and meta-analysis on the relative and attributable risk of Helicobacter pylori infection and cardia and non-cardia gastric cancer. Expert Rev. Mol. Diagn. 2023, 23, 1251–1261. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.C.; Malfertheiner, P.; Yu, H.T.; Kuo, C.L.; Chang, Y.Y.; Meng, F.T.; Wu, Y.X.; Hsiao, J.L.; Chen, M.J.; Lin, K.P.; et al. Global Prevalence of Helicobacter pylori Infection and Incidence of Gastric Cancer Between 1980 and 2022. Gastroenterology 2024, 166, 605–619, Erratum in Gastroenterology 2025, 168, 850. [Google Scholar] [CrossRef]
- Bordin, D.; Morozov, S.; Plavnik, R.; Bakulina, N.; Voynovan, I.; Skibo, I.; Isakov, V.; Bakulin, I.; Andreev, D.; Maev, I. Helicobacter pylori infection prevalence in ambulatory settings in 2017–2019 in RUSSIA: The data of real-world national multicenter trial. Helicobacter 2022, 27, e12924. [Google Scholar] [CrossRef] [PubMed]
- Mladenova, I. Epidemiology of Helicobacter pylori Resistance to Antibiotics (A Narrative Review). Antibiotics 2023, 12, 1184. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Salahi-Niri, A.; Nabavi-Rad, A.; Monaghan, T.M.; Rokkas, T.; Doulberis, M.; Sadeghi, A.; Zali, M.R.; Yamaoka, Y.; Tacconelli, E.; Yadegar, A. Global prevalence of Helicobacter pylori antibiotic resistance among children in the world health organization regions between 2000 and 2023: A systematic review and meta-analysis. BMC Med. 2024, 22, 598. [Google Scholar] [CrossRef]
- Gisbert, J.P.; McNicholl, A.G. Optimization strategies aimed to increase the efficacy of H. pylori eradication therapies. Helicobacter 2017, 22, e12392. [Google Scholar] [CrossRef]
- Graham, D.Y.; Rokkas, T. Overcoming the effects of increasing antimicrobial resistance on Helicobacter pylori therapy. Expert Rev. Gastroenterol. Hepatol. 2024, 18, 705–711. [Google Scholar] [CrossRef]
- Zou, Y.; Qian, X.; Liu, X.; Song, Y.; Song, C.; Wu, S.; An, Y.; Yuan, R.; Wang, Y.; Xie, Y. The effect of antibiotic resistance on Helicobacter pylori eradication efficacy: A systematic review and meta-analysis. Helicobacter 2020, 25, e12714. [Google Scholar] [CrossRef]
- Maev, I.V.; Andreev, D.N.; Fomenko, A.K.; Podporin, M.S.; Lyamina, S.V.; Zaborovsky, A.V.; Khimina, I.N.; Cheremushkin, S.V.; Bagdasarian, A.S.; Cheremushkina, N.V.; et al. Trends of antibiotic resistance of Helicobacter pylori in Moscow. Ter. Arkhiv 2025, 97, 163–168. (In Russian) [Google Scholar] [CrossRef] [PubMed]
- Jukic, I.; Vukovic, J. The Challenges of Treating a Helicobacter pylori Infection following the COVID-19 Pandemic in Croatia: A Review. J. Clin. Med. 2024, 13, 5762. [Google Scholar] [CrossRef] [PubMed]
- Andreev, D.N.; Maev, I.V.; Kucheryavyy, Y.A. Helicobacter pylori resistance in the Russian Federation: A meta-analysis of studies over the past 10 years. Ter. Arkhiv 2020, 92, 24–30. (In Russian) [Google Scholar] [PubMed]
- Nyssen, O.P.; Vaira, D.; Tepes, B.; Kupcinskas, L.; Bordin, D.; Pérez-Aisa, Á.; Gasbarrini, A.; Castro-Fernández, M.; Bujanda, L.; Garre, A.; et al. Room for Improvement in the Treatment of Helicobacter pylori Infection: Lessons from the European Registry on H. pylori Management (Hp-EuReg). J. Clin. Gastroenterol. 2022, 56, e98–e108. [Google Scholar] [CrossRef]
- Bordin, D.S.; Abdulkhakov, S.R.; Andreev, D.N.; Voynovan, I.; Bakulin, I.G.; Bakulina, N.V.; Baryshnikova, N.V.; Ilchishina, T.A.; Starostin, B.D.; Vologzhanina, L.G.; et al. Effectiveness of the first-line eradication therapy in 14 cities in Russia: Results for the period 2013-2022 of the European registry on Helicobacter pylori management (HpEuReg). United Eur. Gastroenterol. J. 2024, 12, 812–813. [Google Scholar]
- Bodunova, N.; Tsapkova, L.; Polyakova, V.; Baratova, I.; Rumyantsev, K.; Dekhnich, N.; Nikolskaya, K.; Chebotareva, M.; Voynovan, I.; Parfenchikova, E.; et al. Genetic Markers of Helicobacter pylori Resistance to Clarithromycin and Levofloxacin in Moscow, Russia. Curr. Issues Mol. Biol. 2024, 46, 6665–6674. [Google Scholar] [CrossRef]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 statement: An updated guideline for reporting systematic reviews. BMJ 2021, 372, 7. [Google Scholar] [CrossRef]
- Dekhnich, N.N.; Kostyakova, E.A.; Punin, A.A.; Alimov, A.V.; Ivanchik, N.V.; Kozlov, R.S. Antibiotic-resistance of H. pylori: Results of microbiologic regional investigation. Russ. J. Gastroenterol. Hepatol. Coloproctol. 2011, 21, 37–42. (In Russian) [Google Scholar]
- Lazebnik, L.B.; Belousova, N.L.; Bordin, D.S.; Mikheeva, O.M.; Dubtsova, E.A.; Vorobyeva, N.N.; Zelenikin, S.A. Helicobacter pylori resistance to clarithromycin in Moscow and propolis as a means of increasing eradication effectiveness. Eksp. Klin. Gastroenterol. 2012, 8, 10–14. (In Russian) [Google Scholar]
- Osipenko, M.F.; Bikbulatova, E.A.; Shakalite, Y.D.; Chernova, L.N.; Ustinov, S.N.; Kulikov, I.V.; Maksimov, V.N. Clarithromycin resistance of Helicobacter pylori in Novosibirsk. Exp. Clin. Gastroenterol. 2012, 8, 15–17. (In Russian) [Google Scholar]
- Abdulkhakov, R.A.; Abuzarova, E.R.; Abdulkhakov, S.R.; Safin, A.G.; Sayfutdinov, I.M.; Chernov, V.M.; Chernova, O.A. Clarithromycin resistance of Helicobacter pylori in Kazan. Exp. Clin. Gastroenterol. 2012, 8, 24–29. (In Russian) [Google Scholar]
- Sablin, O.A.; Mikhailov, N.V.; Iurin, M.V.; Ilchishina, T.A.; Kondrashin, A.S.; Kobiashvili, M.G.; Mikhailova, I.A.; Svarval, A.V.; Zhibrun, A.B. Primary antibiotic resistance of Helicobacter pylori in Saint Petersburg. Exp. Clin. Gastroenterol. 2012, 8, 18–23. (In Russian) [Google Scholar]
- Reva, I.; Takano, T.; Higuchi, W.; Iwao, Y.; Taneike, I.; Nakagawa, S.; Ike, M.; Pererva, O.; Tarankov, A.; Agapov, M.; et al. Virulence genotypes and drug resistance of Helicobacter pylori from Vladivostok, Russia: Another feature in the Far East. Microbiol. Immunol. 2012, 56, 198–202. [Google Scholar] [CrossRef] [PubMed]
- Simanenkov, V.I.; Zakharova, N.V.; Zhebrun, A.B.; Svarval, A.V.; Ferman, R.S.; Savilova, I.V. Resistance of Helicobacter pylori to antimicrobial agents. Lechashchiy Vrach 2015, 4, 91–95. (In Russian) [Google Scholar]
- Dekhnich, N.N.; Ivanchik, N.V.; Kozlov, R.S.; Alimov, A.V.; Lukyanova, A.V.; Nagaeva, O.A.; Steshits, A.S.; Bruk, P.G. Antimicrobial susceptibility of Helicobacter pylori isolates in Smolensk in 2015-2016. Russ. J. Gastroenterol. Hepatol. Coloproctol. 2016, 26, 24–31. (In Russian) [Google Scholar] [CrossRef]
- Kalugin, A.A.; Stepchenko, A.A.; Voropaev, E.V.; Osipkina, O.V.; Zyatkov, A.A. Frequency of detection of Helicobacter pylori gene polymorphisms associated with clarithromycin resistance. Chelovek I Ego Zdorov’e 2016, 3, 17–21. (In Russian) [Google Scholar]
- Dekhnich, N.; Ivanchik, N.; Kozlov, R.; Alimov, A.; Steshits, A.; Kirsov, P.; Pandav, K. Dynamics of antimicrobial resistance of Helicobacter pylori isolates in the Smolensk region of Russian Federation. Helicobacter 2018, 23, e12545. [Google Scholar] [CrossRef] [PubMed]
- Maev, I.V.; Andreev, D.N.; Govorun, V.M.; Ilina, E.N.; Kucheryavyi, Y.A.; Oganesyan, T.S.; Melnikova, E.V.; Zayratyants, O.V.; Parfenova, T.V.; Dzhedzheiya, L.V.; et al. Antibiotic resistance of Helicobacter pylori in the European part of the Russian Federation: Preliminary results. Ter. Arkhiv 2020, 92, 24–28. (In Russian) [Google Scholar] [CrossRef]
- Luzina, E.V.; Lazebnik, L.B.; Lareva, N.V.; Chartorizhskaya, N.N.; Dutova, A.A.; Melnikov, V.V.; Mutsolgova, T.B. Chita experience of the “Doctors Against Helicobacteriosis” program by the Russian Gastroenterological Society and Russian Scientific Medical Society of Therapists. Eksperimental’naya I Klin. Gastroenterol. 2020, 175, 34–46. (In Russian) [Google Scholar] [CrossRef]
- Starkova, D.; Gladyshev, N.; Polev, D.; Saitova, A.; Egorova, S.; Svarval, A. First insight into the whole genome sequence variations in clarithromycin resistant Helicobacter pylori clinical isolates in Russia. Sci. Rep. 2024, 14, 20108. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Kuprivanova, E.A.; Abdulkhakov, S.R.; Ismagilova, R.K.; Safina, D.D.; Akhtereva, A.R.; Galimova, R.R.; Safin, A.G.; Grigoreva, T.V.; Abdulkhakov, R.A. Prevalence of mutations associated with Helicobacter pylori antibiotic resistance in Kazan. Ter. Arkhiv 2024, 96, 739–743. (In Russian) [Google Scholar] [CrossRef]
- Okubo, M.; Tahara, T.; Shibata, T.; Nakamura, M.; Yoshioka, D.; Maeda, Y.; Yonemura, J.; Ishizuka, T.; Arisawa, T.; Hirata, I. Changes in gastric mucosal patterns seen by magnifying NBI during H. pylori eradication. J. Gastroenterol. 2011, 46, 175–182. [Google Scholar] [CrossRef]
- Lee, Y.C.; Chen, T.H.; Chiu, H.M.; Shun, C.T.; Chiang, H.; Liu, T.Y.; Wu, M.S.; Lin, J.T. The benefit of mass eradication of Helicobacter pylori infection: A community-based study of gastric cancer prevention. Gut 2013, 62, 676–682. [Google Scholar] [CrossRef]
- Bujanda, L.; Nyssen, O.P.; Ramos, J.; Bordin, D.S.; Tepes, B.; Perez-Aisa, A.; Pavoni, M.; Castro-Fernandez, M.; Lerang, F.; Leja, M.; et al. Effectiveness of Helicobacter pylori Treatments According to Antibiotic Resistance. Am. J. Gastroenterol. 2024, 119, 646–654. [Google Scholar] [CrossRef]
- Ng, H.Y.; Leung, W.K.; Cheung, K.S. Antibiotic Resistance, Susceptibility Testing and Stewardship in Helicobacter pylori Infection. Int. J. Mol. Sci. 2023, 24, 11708. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Grau, S.; Echeverria-Esnal, D.; Gómez-Zorrilla, S.; Navarrete-Rouco, M.E.; Masclans, J.R.; Espona, M.; Gracia-Arnillas, M.P.; Duran, X.; Comas, M.; Horcajada, J.P.; et al. Evolution of Antimicrobial Consumption During the First Wave of COVID-19 Pandemic. Antibiotics 2021, 10, 132. [Google Scholar] [CrossRef]
- Lucien, M.A.B.; Canarie, M.F.; Kilgore, P.E.; Jean-Denis, G.; Fénélon, N.; Pierre, M.; Cerpa, M.; Joseph, G.A.; Maki, G.; Zervos, M.J.; et al. Antibiotics and antimicrobial resistance in the COVID-19 era: Perspective from resource-limited settings. Int. J. Infect. Dis. 2021, 104, 250–254. [Google Scholar] [CrossRef]
- Zakharenkov, I.A.; Rachina, S.A.; Kozlov, R.S.; Belkova, Y.A. Consumption of systemic antibiotics in the Russian Federation in 2017–2021. Clin. Microbiol. Antimicrob. Chemother. 2022, 24, 220–225. (In Russian) [Google Scholar] [CrossRef]
- Yu, Y.; Xue, J.; Lin, F.; Liu, D.; Zhang, W.; Ru, S.; Jiang, F. Global Primary Antibiotic Resistance Rate of Helicobacter pylori in Recent 10 years: A Systematic Review and Meta-Analysis. Helicobacter 2024, 29, e13103. [Google Scholar] [CrossRef]
- Maev, I.V.; Andreev, D.N.; Bordin, D.S.; Kicheryavyy, Y.A. Resistance of Helicobacter pylori to clarithromycin in the Russian Federation. Eff. Farmakoter. 2020, 16, 16–22. (In Russian) [Google Scholar] [CrossRef]
- Kurkova, A.A.; Rachina, S.A.; Kozlov, R.S.; Portnyagina, U.S.; Palyutin, S.K.; Reshetko, O.V.; Zhuravleva, M.V.; Karpova, O.Y.; Myagkova, O.G.; Kuznetsova, E.V.; et al. Patterns of antimicrobial dispensing in community pharmacies in Russia during the COVID-19 pandemic. Clin. Microbiol. Antimicrob. Chemother. 2023, 25, 84–92. (In Russian) [Google Scholar] [CrossRef]
- Ivashkin, V.T.; Lapina, T.L.; Maev, I.V.; Drapkina, O.M.; Kozlov, R.S.; Sheptulin, A.A.; Trukhmanov, A.S.; Abdulkhakov, S.R.; Alekseeva, O.P.; Alekseenko, S.A.; et al. Fice Guidelines of Russian Gastroenterological Association, Scientific Society for the Clinical Study of Human Microbiome, Russian Society for the Prevention of Non-Communicable Diseases, Interregional Association for Clinical Microbiology and Antimicrobial Chemotherapy for H. pylori Diagnostics and Treatment in Adults. Russ. J. Gastroenterol. Hepatol. Coloproctol. 2022, 32, 72–93. (In Russian) [Google Scholar] [CrossRef]
- Ko, S.W.; Kim, Y.J.; Chung, W.C.; Lee, S.J. Bismuth supplements as the first-line regimen for Helicobacter pylori eradication therapy: Systemic review and meta-analysis. Helicobacter 2019, 24, e12565. [Google Scholar] [CrossRef]
- Zagari, R.M.; Dajti, E.; Cominardi, A.; Frazzoni, L.; Fuccio, L.; Eusebi, L.H.; Vestito, A.; Lisotti, A.; Galloro, G.; Romano, M.; et al. Standard Bismuth Quadruple Therapy versus Concomitant Therapy for the First-Line Treatment of Helicobacter pylori Infection: A Systematic Review and Meta-Analysis of Randomized Controlled Trials. J. Clin. Med. 2023, 12, 3258. [Google Scholar] [CrossRef]
- Tacconelli, E.; Carrara, E.; Savoldi, A.; Harbarth, S.; Mendelson, M.; Monnet, D.; Pulcini, C.; Kahlmeter, G.; Kluytmans, J.; Carmeli, Y.; et al. Discovery, research, and development of new antibiotics: The WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 2017, 18, 318–327. [Google Scholar] [CrossRef]
- Savoldi, A.; Carrara, E.; Graham, D.Y.; Conti, M.; Tacconelli, E. Prevalence of Antibiotic Resistance in Helicobacter pylori: A Systematic Review and Meta-analysis in World Health Organization Regions. Gastroenterology 2018, 155, 1372–1382.e17. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Du, J.; Zhang, D.; Jin, C.; Chen, J.; Wang, Z.; Mei, T.; Fu, K.; Qian, Q.; Pang, T. Primary antibiotic resistance in Helicobacter pylori in China: A systematic review and meta-analysis. J. Glob. Antimicrob. Resist. 2023, 34, 30–38, Erratum in J. Glob. Antimicrob. Resist. 2023, 35, 354. [Google Scholar] [CrossRef] [PubMed]
- Ho, J.J.C.; Navarro, M.; Sawyer, K.; Elfanagely, Y.; Moss, S.F. Helicobacter pylori Antibiotic Resistance in the United States Between 2011 and 2021: A Systematic Review and Meta-Analysis. Am. J. Gastroenterol. 2022, 117, 1221–1230. [Google Scholar] [CrossRef] [PubMed]
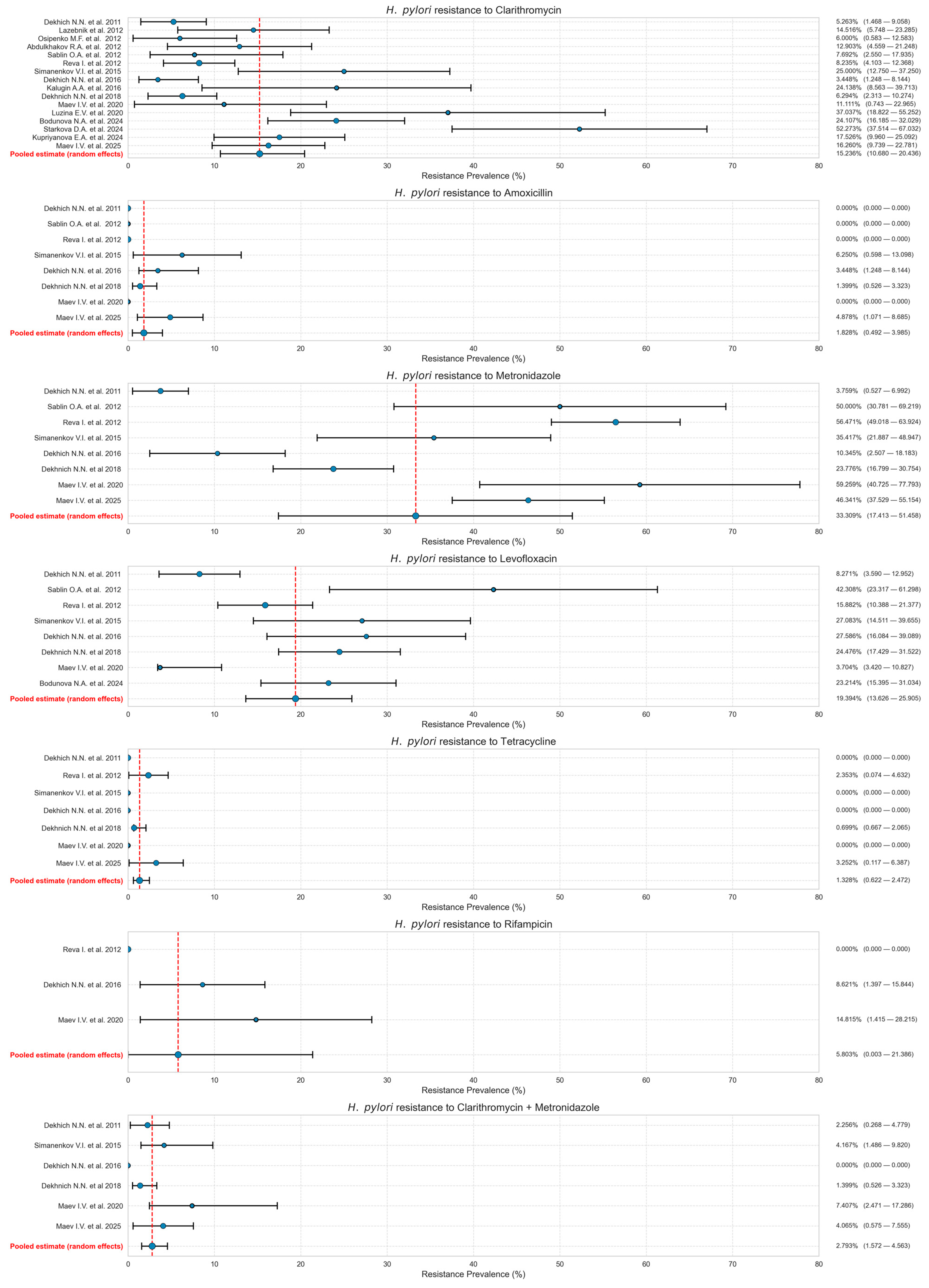

| Authorship and Year | City | Timeframe of Isolate Collection | Methodology | Number of Isolates | CLA-R | AMO-R | MET-R | LEV-R | TET-R | RIF-R | CLA-MET-R | NOS |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Dekhich N.N. et al., 2011 [21] | Smolensk | 2009–2010 | Serial Dilution Method | 133 | 7 | 0 | 5 | 11 | 0 | n/a | 3 | 7 |
| Lazebnik et al., 2012 [22] | Moscow | Before 2012 | Molecular Genetic Testing | 62 | 9 | n/a | n/a | n/a | n/a | n/a | n/a | 6 |
| Osipenko M.F. et al., 2012 [23] | Novosibirsk | Before 2012 | Molecular Genetic Testing | 50 | 3 | n/a | n/a | n/a | n/a | n/a | n/a | 6 |
| Abdulkhakov R.A. et al., 2012 [24] | Kazan | Before 2012 | Molecular Genetic Testing | 62 | 8 | n/a | n/a | n/a | n/a | n/a | n/a | 7 |
| Sablin O.A. et al., 2012 [25] | Saint-Petersburg | 2012 | Disk Diffusion Method | 26 | 2 | 0 | 18 | 11 | n/a | n/a | n/a | 8 |
| Reva I. et al., 2012 [26] | Vladivostok | 2004–2009 | Serial Dilution Method | 170 | 13 | 0 | 96 | 27 | 4 | 0 | n/a | 7 |
| Simanenkov V.I. et al., 2015 [27] | Saint-Petersburg | 2013–2014 | Serial Dilution Method | 48 | 12 | 3 | 17 | 13 | 0 | n/a | 2 | 7 |
| Dekhich N.N. et al., 2016 [28] | Smolensk | 2015–2016 | Serial Dilution Method | 58 | 2 | 2 | 6 | 16 | 0 | 5 | 0 | 5 |
| Kalugin A.A. et al., 2016 [29] | Kursk | 2016 | Molecular Genetic Testing | 29 | 7 | n/a | n/a | n/a | n/a | n/a | n/a | 4 |
| Dekhnich N. et al., 2018 [30] | Smolensk | 2015–2017 | Serial Dilution Method | 143 | 9 | 2 | 34 | 35 | 1 | n/a | 2 | 8 |
| Maev I.V. et al., 2020 [31] | Moscow, Yaroslavl | 2015–2018 | Disk Diffusion Method | 27 | 3 | 0 | 16 | 1 | 0 | 4 | 2 | 7 |
| Luzina E.V. et al., 2020 [32] | Chita | 2019 | Molecular Genetic Testing | 27 | 10 | n/a | n/a | n/a | n/a | n/a | n/a | 5 |
| Bodunova N et al., 2024 [19] | Moscow | 2022–2023 | Molecular Genetic Testing | 112 | 27 | n/a | n/a | 26 | n/a | n/a | n/a | 7 |
| Starkova D et al., 2024 [33] | Saint-Petersburg | 2014–2022 | Disk Diffusion Method | 44 | 23 | n/a | n/a | n/a | n/a | n/a | n/a | 6 |
| Kupriyanova E.A. et al., 2024 [34] | Kazan | 2019–2021 | Molecular Genetic Testing | 97 | 17 | n/a | n/a | 12 | n/a | n/a | n/a | 7 |
| Maev I.V. et al., 2025 [14] | Moscow | 2015–2024 | Disk Diffusion Method | 123 | 20 | 6 | 57 | n/a | 4 | n/a | 5 | 8 |
| Antibiotic | Before 2015 | 2015–2019 | 2020–2024 |
|---|---|---|---|
| Clarithromycin | 11.903% (95% CI: 7.602–17.013) | 18.791% (95% CI: 9.258–30.727) | 21.024% (95% CI: 16.086–26.680) |
| Metronidazole | 33.542% (95% CI: 6.645–68.477) | 33.412% (95% CI: 15.414–54.372) | 1 study, n/a for meta-analysis |
| Amoxicillin | 0.978% (95% CI: 0.000968–3.753) | 1.955% (95% CI: 0.645–4.475) | 1 study, n/a for meta-analysis |
| Levofloxacin | 20.831% (95% CI: 10.481–33.591) | 18.322% (95% CI: 10.144–28.262) | 17.122% (95% CI: 6.010–32.399) |
| Tetracycline | 0.958% (95% CI: 0.0224–3.244) | 0.831% (95% CI: 0.113–2.847) | 1 study, n/a for meta-analysis |
| Clarithromycin + Metronidazole | 3.180% (95% CI: 1.153–6.867) | 3.088% (95% CI: 0.530–7.660) | 1 study, n/a for meta-analysis |
| Antibiotic | Molecular Genetic Testing | Serial Dilution Method | Disk Diffusion Method |
|---|---|---|---|
| Clarithromycin | 18.494% (95% CI: 13.138–24.532) | 8.545% (95% CI: 4.491–13.742) | 21.122% (95% CI: 6.817–40.608) |
| Metronidazole | n/a | 23.752% (95% CI: 6.518–47.458) | 48.889% (95% CI: 41.604–56.198) |
| Amoxicillin | n/a | 1.526% (95% CI: 0.204–4.047) | 2.955% (95% CI: 0.573–7.102) |
| Levofloxacin | 17.876% (95% CI: 8.676–29.491) | 19.740% (95% CI: 12.539–28.111) | 20.537% (95% CI: 0.052–65.345) |
| Tetracycline | n/a | 0.951% (95% CI: 0.260–2.069) | 2.985% (95% CI: 0.884–6.272) |
| Clarithromycin + Metronidazole | n/a | 2.060% (95% CI: 0.891–4.026) | 5.134% (95% CI: 2.206–9.193) |
| Antibiotic | Moscow | Saint-Petersburg | Kazan |
|---|---|---|---|
| Clarithromycin | 18.763% (95% CI: 14.014–24.028) | 28.540% (95% CI: 11.376–49.776) | 16.279% (95% CI: 10.953–22.882) |
| Metronidazole | 46.398% (95% CI: 37.436–55.534) | 27.612% (95% CI: 7.795–53.813) | n/a |
| Amoxicillin | 3.791% (95% CI: 0.111–12.314) | 3.769% (95% CI: 0.075–12.630) | n/a |
| Tetracycline | 3.530% (95% CI: 1.052–8.448) | n/a | n/a |
| Levofloxacin | 13.736% (95% CI: 1.239–36.503) | n/a | 13.015% (95% CI: 7.093–21.271) |
| Dual Therapy (Clarithromycin + Metronidazole) | 4.593% (95% CI: 1.657–9.881) | n/a | n/a |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Andreev, D.N.; Khurmatullina, A.R.; Maev, I.V.; Bordin, D.S.; Zaborovskiy, A.V.; Abdulkhakov, S.R.; Kucheryavyy, Y.A.; Sokolov, F.S.; Beliy, P.A. Helicobacter pylori Antibiotic Resistance in Russia: A Systematic Review and Meta-Analysis. Antibiotics 2025, 14, 524. https://doi.org/10.3390/antibiotics14050524
Andreev DN, Khurmatullina AR, Maev IV, Bordin DS, Zaborovskiy AV, Abdulkhakov SR, Kucheryavyy YA, Sokolov FS, Beliy PA. Helicobacter pylori Antibiotic Resistance in Russia: A Systematic Review and Meta-Analysis. Antibiotics. 2025; 14(5):524. https://doi.org/10.3390/antibiotics14050524
Chicago/Turabian StyleAndreev, Dmitrii N., Alsu R. Khurmatullina, Igor V. Maev, Dmitry S. Bordin, Andrey V. Zaborovskiy, Sayar R. Abdulkhakov, Yury A. Kucheryavyy, Filipp S. Sokolov, and Petr A. Beliy. 2025. "Helicobacter pylori Antibiotic Resistance in Russia: A Systematic Review and Meta-Analysis" Antibiotics 14, no. 5: 524. https://doi.org/10.3390/antibiotics14050524
APA StyleAndreev, D. N., Khurmatullina, A. R., Maev, I. V., Bordin, D. S., Zaborovskiy, A. V., Abdulkhakov, S. R., Kucheryavyy, Y. A., Sokolov, F. S., & Beliy, P. A. (2025). Helicobacter pylori Antibiotic Resistance in Russia: A Systematic Review and Meta-Analysis. Antibiotics, 14(5), 524. https://doi.org/10.3390/antibiotics14050524






