Determination of Colistin Resistance in Clinical Isolates from Healthcare Facilities in Mthatha and Surrounding Areas
Abstract
1. Introduction
Resurgence of Colistin
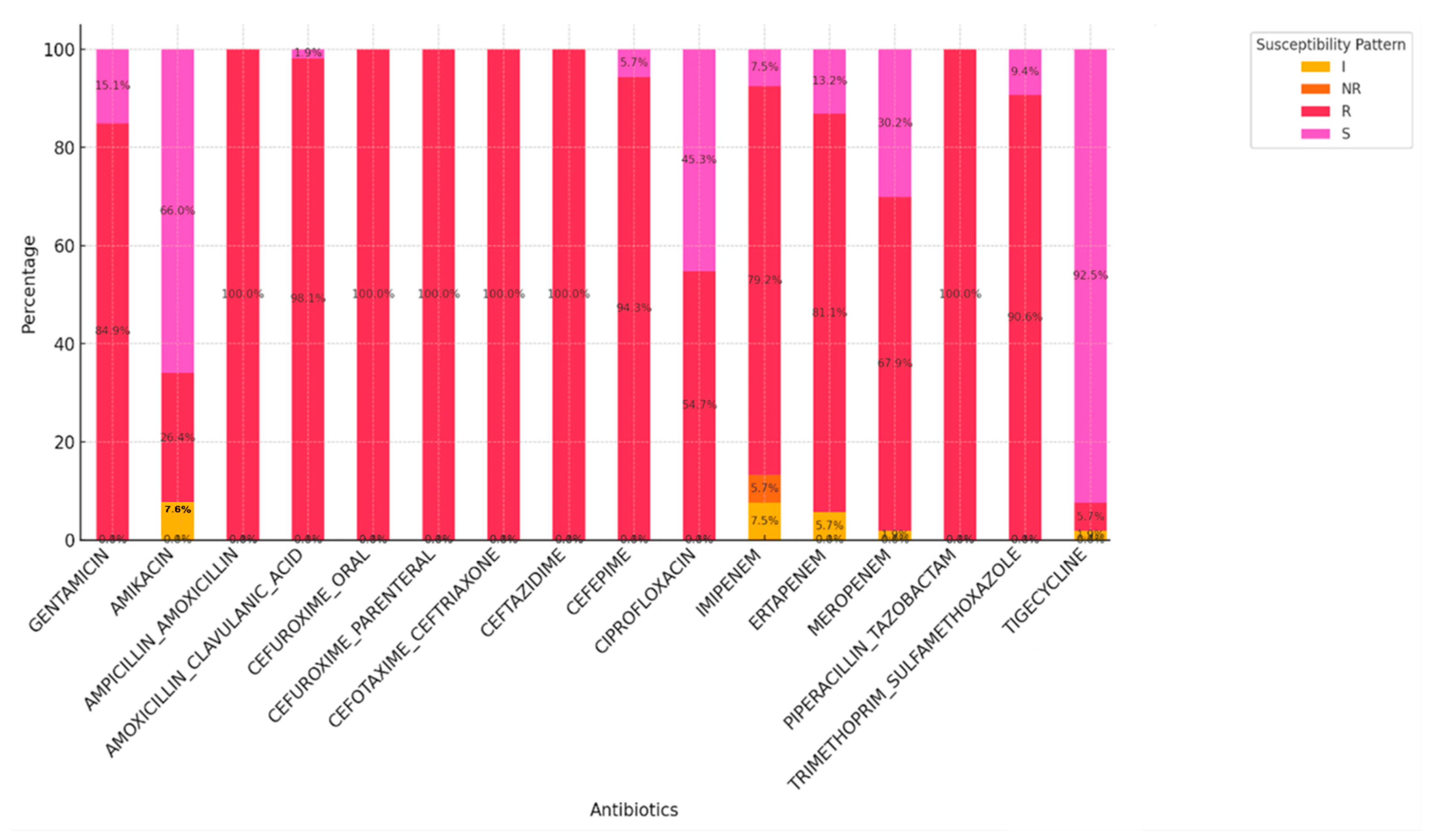
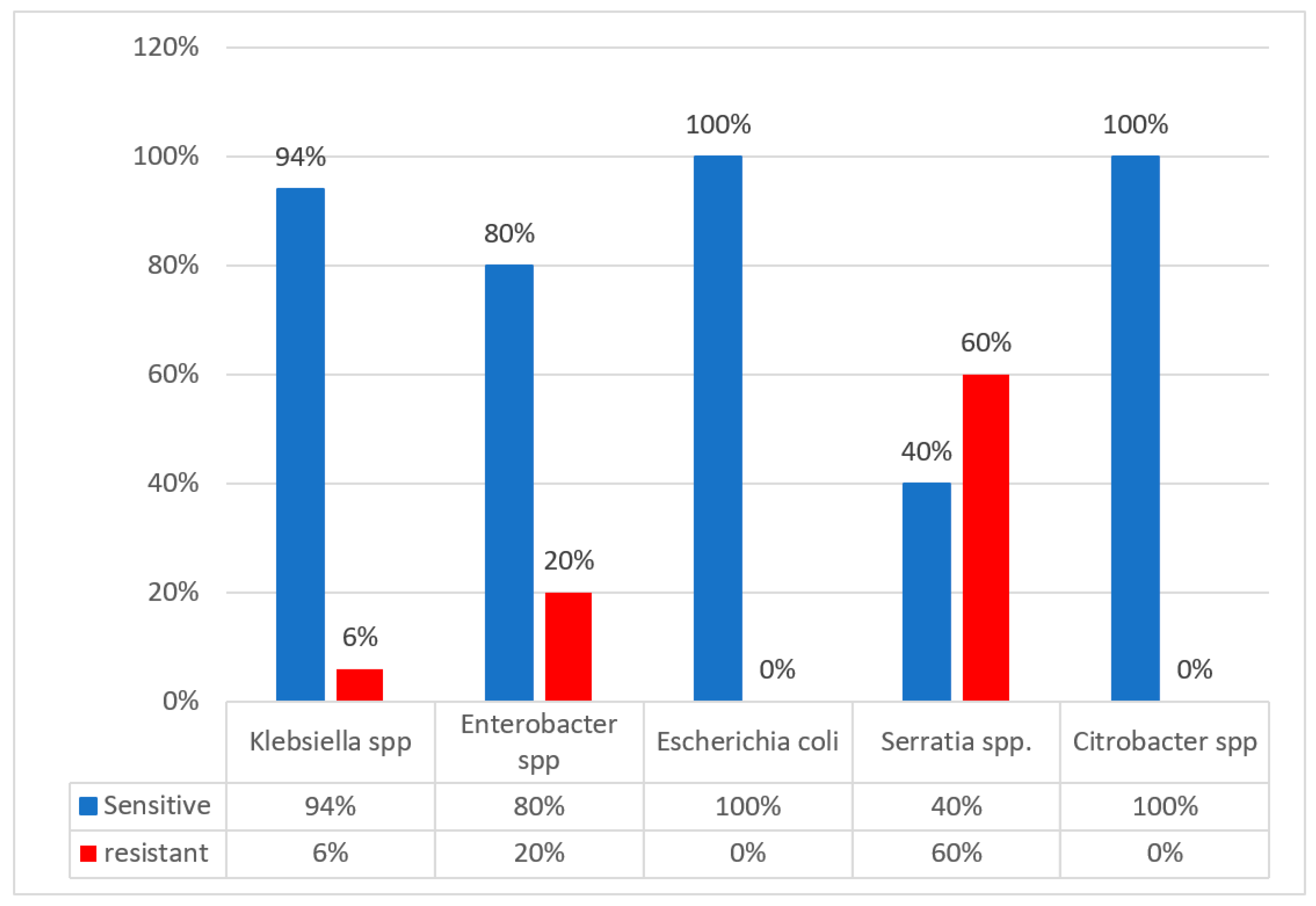
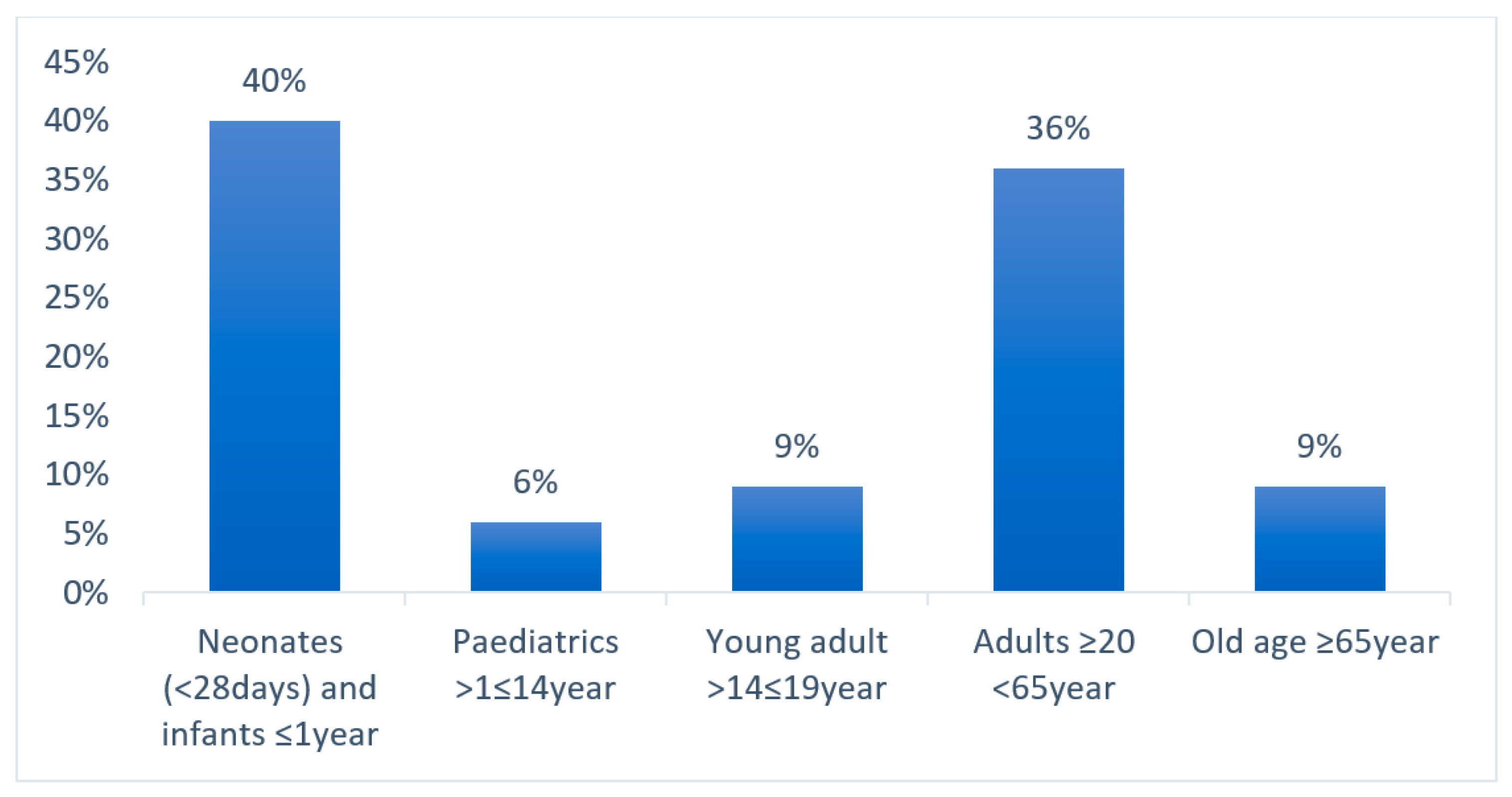
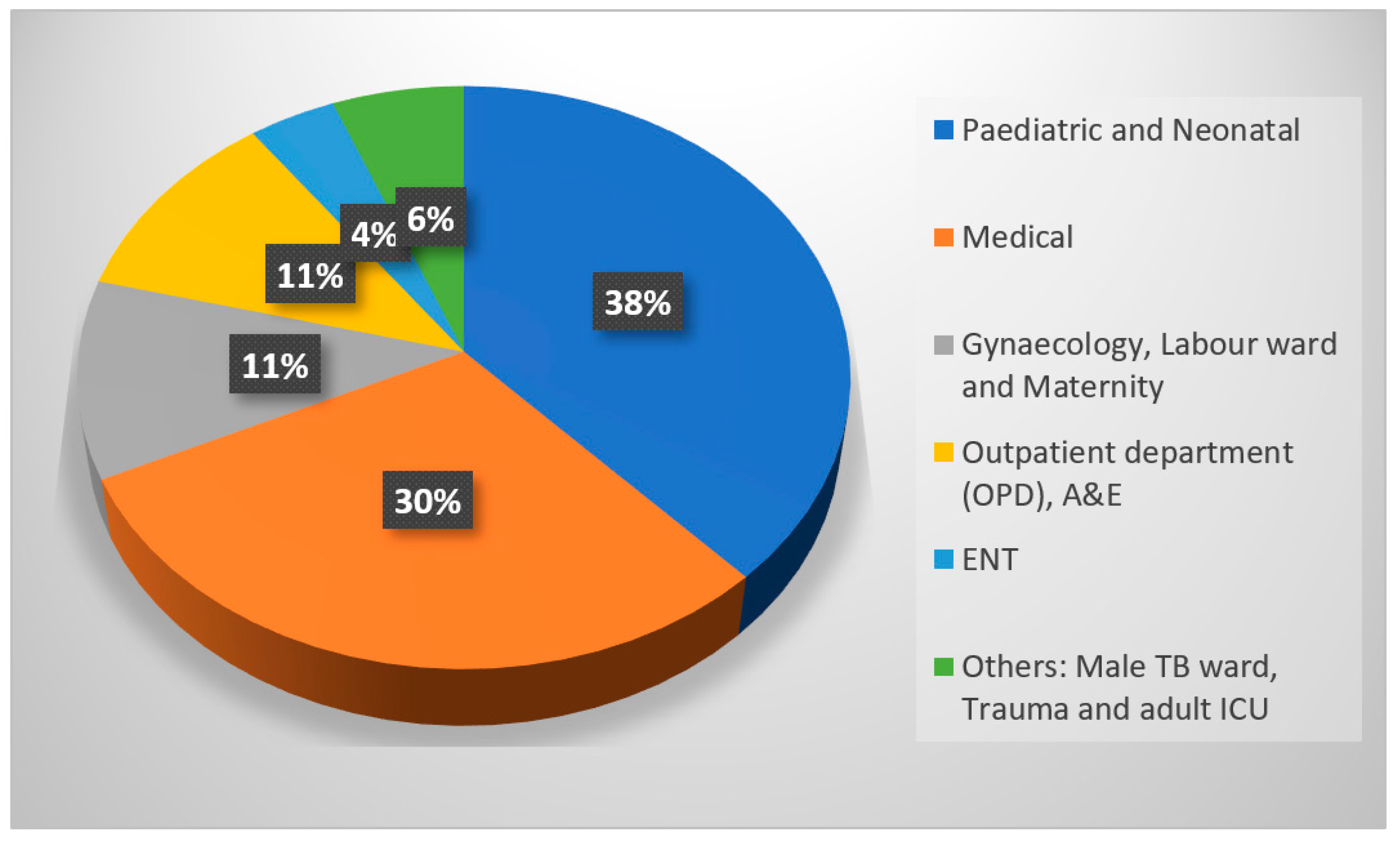
2. Results
Participant’s Eligibility Inclusion and Exclusion
3. Discussion
4. Materials and Methods
4.1. Research Design
4.2. Study Population and Sampling Method
4.3. Inclusion and Exclusion Criteria
4.4. Sampling Method and Size
4.5. Data Collection
4.6. Data Analysis
4.7. Microbiological Investigation
4.7.1. Bacterial Isolation and Identification
4.7.2. Carbapenemase Confirmatory Tests
4.7.3. Antimicrobial Susceptibility Testing of Colistin Using ComASP Colistin 0.25–16 mg/L (LIofilchem® srl, Roseto degli Abruzzi, Italy) Method
4.7.4. Detection of Mobile Colistin Resistance (mcr-1) Gene
DNA Extraction
DNA Library Preparation
Ethical Considerations
5. Conclusions
Limitations
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bartsch, S.M.; McKinnell, J.A.; Mueller, L.E.; Miller, L.G.; Gohil, S.K.; Huang, S.S.; Lee, B.Y. Potential economic burden of carbapenem-resistant Enterobacteriaceae (CRE) in the United States. Clin. Microbiol. Infect. 2017, 23, 48.e9–48.e16. [Google Scholar] [CrossRef] [PubMed]
- Cannatelli, A.; Giani, T.; Antonelli, A.; Principe, L.; Luzzaro, F.; Rossolini, G.M. First Detection of the mcr-1 Colistin Resistance Gene in Escherichia coli in Italy. Antimicrob. Agents Chemother. 2016, 60, 3257–3258. [Google Scholar] [CrossRef] [PubMed]
- Coetzee, J.; Corcoran, C.; Prentice, E.; Moodley, M.; Mendelson, M.; Poirel, L.; Nordmann, P.; Brink, A.J. Emergence of plasmid-mediated colistin resistance (MCR-1) among Escherichia coli isolated from South African patients. S. Afr. Med. J. 2016, 106, 449–450. [Google Scholar] [CrossRef] [PubMed]
- The Center for Disease Dynamics, Economics and Policy. The State of the World’s Antibiotics; The Center for Disease Dynamics, Economics and Policy: Washington, DC, USA, 2015; Available online: http://cddep.org/publications/state_worlds_antibiotics_2015#sthash.S3a3ME0o.dpbs (accessed on 1 February 2025).
- Prim, N.; Turbau, M.; Rivera, A.; Rodríguez-Navarro, J.; Coll, P.; Mirelis, B. Prevalence of colistin resistance in clinical isolates of Enterobacteriaceae: A four-year cross-sectional study. J. Infect. 2017, 75, 493–498. [Google Scholar] [CrossRef]
- Galani, I.; Adamou, P.; Karaiskos, I.; Giamarellou, H.; Souli, M. Evaluation of ComASP Colistin (formerly SensiTest Colistin), a commercial broth microdilution-based method to evaluate the colistin minimum inhibitor concentration for car-bapenem-resistant Klebsiella pneumoniae isolates. Glob. Antimicrob. Resist. 2018, 215, 123–126. [Google Scholar] [CrossRef]
- Lim, L.M.; Ly, N.; Anderson, D.; Yang, J.C.; Macander, L.; Jarkowski, A., III; Forrest, A.; Bulitta, J.B.; Tsuji, B.T. Resurgence of Colistin: A Review of Resistance, Toxicity, Pharmacodynamics, and Dosing. Pharmacotherapy 2010, 30, 1279–1291. [Google Scholar] [CrossRef]
- Tsuji, B.; Pogue, J.M.; Zavascki, A.P.; Paul, M.A.; Forrest, D.R.; Giacobbe, C.; Viscoli, I.; Karaiskos, D.; Kaye, J.; Mouton, V.H.; et al. International Consensus Guidelines for the Optimal Use of Polymyxins: Endorsed by the American College of Clinical Pharmacy (ACCP), European Society of Clinical Mi-crobiology and Infectious Diseases (ESCMID), Infectious Diseases Society of America (IDSA), International Society for Anti-infective Pharmacology (ISAP), Society of Critical Care Medicine (SCCM), and Society of Infectious Diseases Pharmacists (SIDP). Pharmacother. J. Hum. Pharmacol. Drug Ther. 2019, 39, 10–39. [Google Scholar]
- Labuschagne, Q.; Schellack, N.; Gous, A.; Bronkhorst, E.; Schellack, G.; van Tonder, L.; Truter, A.; Smith, C.; Lancaster, R.; Kolman, S. COLISTIN: Adult and paediatric guideline for South Africa, 2016. S. Afr. J. Infect. Dis. 2016, 31, 3–7. [Google Scholar] [CrossRef][Green Version]
- Bialvaei, A.Z.; Kafil, H.S. Colistin, mechanisms and prevalence of resistance. Curr. Med. Res. Opin. 2015, 31, 707–721. [Google Scholar] [CrossRef]
- Yap, P.S.-X.; Cheng, W.-H.; Chang, S.-K.; Lim, S.-H.E.; Lai, K.-S. MgrB Mutations and Altered Cell Permeability in Colistin Resistance in Klebsiella pneumoniae. Cells 2022, 11, 2995. [Google Scholar] [CrossRef]
- Okenwa, S.C.; Ugwuezea, J.C.; Ekweozor, C.A.; Anunwa, I.G.; Nwachukwu, J.C.; Nwankwo, F.M.; Njoku, C.C.; Mbakamma, C.E.; Onyeishi, E.E.; Obiekwe, I.J.; et al. Prevalence and Molecular Characterization of Colistin Resistance in Gram-negative Bacteria from Nigeria Hospitals: A Scoping Review. Asian J. Res. Med. Pharm. Sci. 2024, 13, 137–159. [Google Scholar] [CrossRef]
- Irrgang, A.; Roschanski, N.; Tenhagen, B.; Grobbel, M. Prevalence of mcr-1 in E. coli from Livestock and Food in Germany. PLoS ONE 2016, 11, e0159863. [Google Scholar] [CrossRef] [PubMed]
- Poirel, L.; Kieffer, N.; Brink, A.; Coetze, J.; Jayol, A.; Nordmann, P. Genetic Features of MCR-1-Producing Colistin-Resistant Escherichia coli Isolates in South Africa. Antimicrob. Agents Chemother. 2016, 60, 4394–4397. [Google Scholar] [CrossRef] [PubMed]
- Hamel, M.; Rolain, J.M.; Baron, S.A. The history of colistin resistance mechanisms in bacteria: Progress and challenges. Microorganisms 2021, 9, 442. [Google Scholar] [CrossRef]
- Puljko, A.; Barišić, I.; Rozman, S.D.; Križanović, S.; Babić, I.; Jelić, M.; Maravić, A.; Udiković-Kolić, N. Molecular epidemiology and mechanisms of carbapenem and colistin resistance in Klebsiella and other Enterobacterales from treated wastewater in Croatia. Environ. Int. 2024, 185, 108554. [Google Scholar] [CrossRef]
- Chiotos, K.; Hayes, M.; Gerber, J.S.; Tamma, P.D. Treatment of Carbapenem-Resistant Enterobacteriaceae Infections in Children. J. Pediatr. Infect. Dis. Soc. 2019, 9, 56–66. [Google Scholar] [CrossRef]
- Ibrahim, E.R.; Ahmed, Y.M.; Mohamed, A.K.; Ibrahim, A. Detection of colistin resistant Gram-negative bacilli intensive. Microbes Infect. Dis. 2021, 2, 92–99. [Google Scholar]
- Sarker, S.; Neeloy, R.M.; Habib, M.B.; Urmi, U.L.; Al Asad, M.; Mosaddek, A.S.M.; Khan, M.R.K.; Nahar, S.; Godman, B.; Islam, S. Mobile Colistin-Resistant Genes mcr-1, mcr-2, and mcr-3 Identified in Diarrheal Pathogens among Infants, Children, and Adults in Bangladesh: Implications for the Future. Antibiotics 2024, 13, 534. [Google Scholar] [CrossRef]
- Bir, R.; Gautam, H.; Arif, N.; Chakravarti, P.; Verma, J.; Banerjee, S.; Tyagi, S.; Mohapatra, S.; Sood, S.; Dhawan, B.; et al. Analysis of colistin resistance in carbapenem-resistant Enterobacterales and XDR Klebsiella pneumoniae. Ther. Adv. Infect. Dis. 2022, 9, 20499361221080650. [Google Scholar] [CrossRef]
- Moosavian, M.; Emam, N. The first report of emerging mobilized colistin-resistance (mcr) genes and ERIC-PCR typing in Escherichia coli and Klebsiella pneumoniae clinical isolates in southwest Iran. Infect. Drug Resist. 2019, 12, 1001–1010. [Google Scholar] [CrossRef]
- Liu, Y.-Y.; Wang, Y.; Walsh, T.R.; Yi, L.-X.; Zhang, R.; Spencer, J.; Doi, Y.; Tian, G.; Dong, B.; Huang, X.; et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: A microbiological and molecular biological study. Lancet Infect. Dis. 2016, 16, 161–168. [Google Scholar] [CrossRef] [PubMed]
- Vasaikar, S.D.; Hanise, P.; Abaver, D.T. Epidemiology, risk factors and molecular analysis of carbapenem-resistant Enterobacteriaceae (CRE) in Mthatha, Eastern Cape, South Africa. Int. J. Infect. Dis. 2020, 101, 54. [Google Scholar] [CrossRef]
- Xayimpi, O. A map showing the location of hospitals covered by NHLS Mthatha. In QGIS; Geography Department, Walter Sisulu University: Mthatha, South Africa, 2024. [Google Scholar]
- Clinical & Laboratory Standard and Institute. Performance Standard for Antimicrobial Susceptibility Testing Twenty-Sixth Informational Supplement CLSI Document M100-S26; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2016. [Google Scholar]
- Clinical Laboratory Standard Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing; Clinical Laboratory Standard Institute (CLSI): Wayne, PA, USA, 2022. [Google Scholar]
- Kolenda, C.; Benoit, R.; Carricajo, A.; Bonnet, R.; Dauwalder, O.; Laurent, F. Evaluation of the New Multiplex Immunochromatographic O.K.N.V. K.-SeT Assay for Rapid Detection of OXA-48-like, KPC, NDM, and VIM Carbapenemases. J. Clin. Microbiol. 2018, 56, e01247-18. [Google Scholar] [CrossRef] [PubMed]
- European Committee on Antimicrobial Susceptibility Testing. Breakpoint Tables for Interpretation of MICs and Zone Diameters. Version 12.0. Available online: http://www.eucast.org (accessed on 2 June 2024).
- Hosu, M.C.; Vasaikar, S.; Okuthe, G.E.; Apalata, T. Molecular Detection of Antibiotic-Resistant Genes in Pseudomonas aeruginosa from Nonclinical Environment: Public Health Implications in Mthatha, Eastern Cape Province, South Africa. Int. J. Microbiol. 2021, 2021, 8861074. [Google Scholar] [CrossRef]
- Hassan, I.Z.; Wandrag, B.; Gouws, J.J.; Qekwana, D.N.; Naidoo, V. Antimicrobial resistance and mcr-1 gene in Escherichia coli isolated from poultry samples submitted to a bacteriology laboratory in South Africa. Vet.-World 2021, 14, 2662–2669. [Google Scholar] [CrossRef]
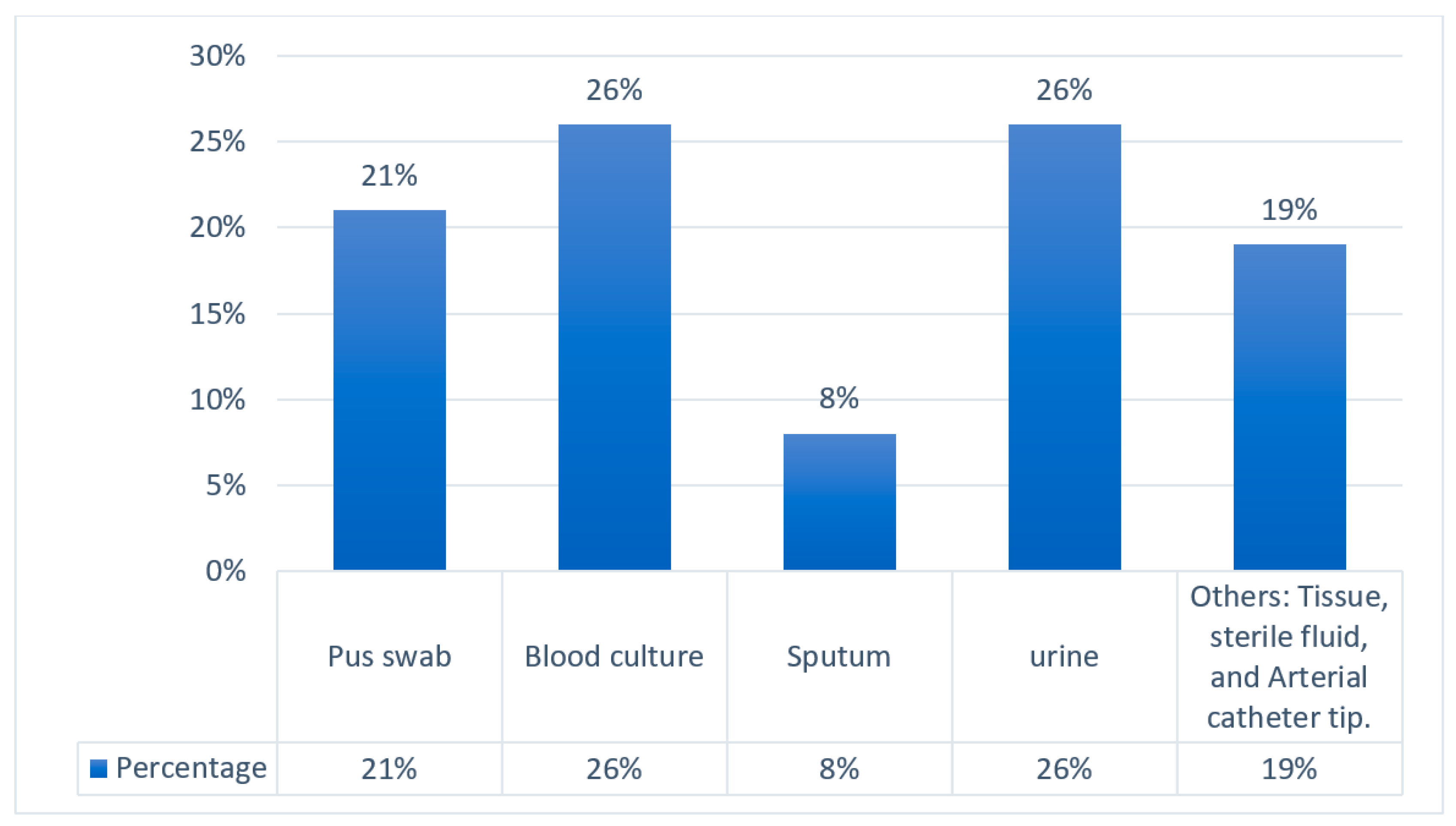
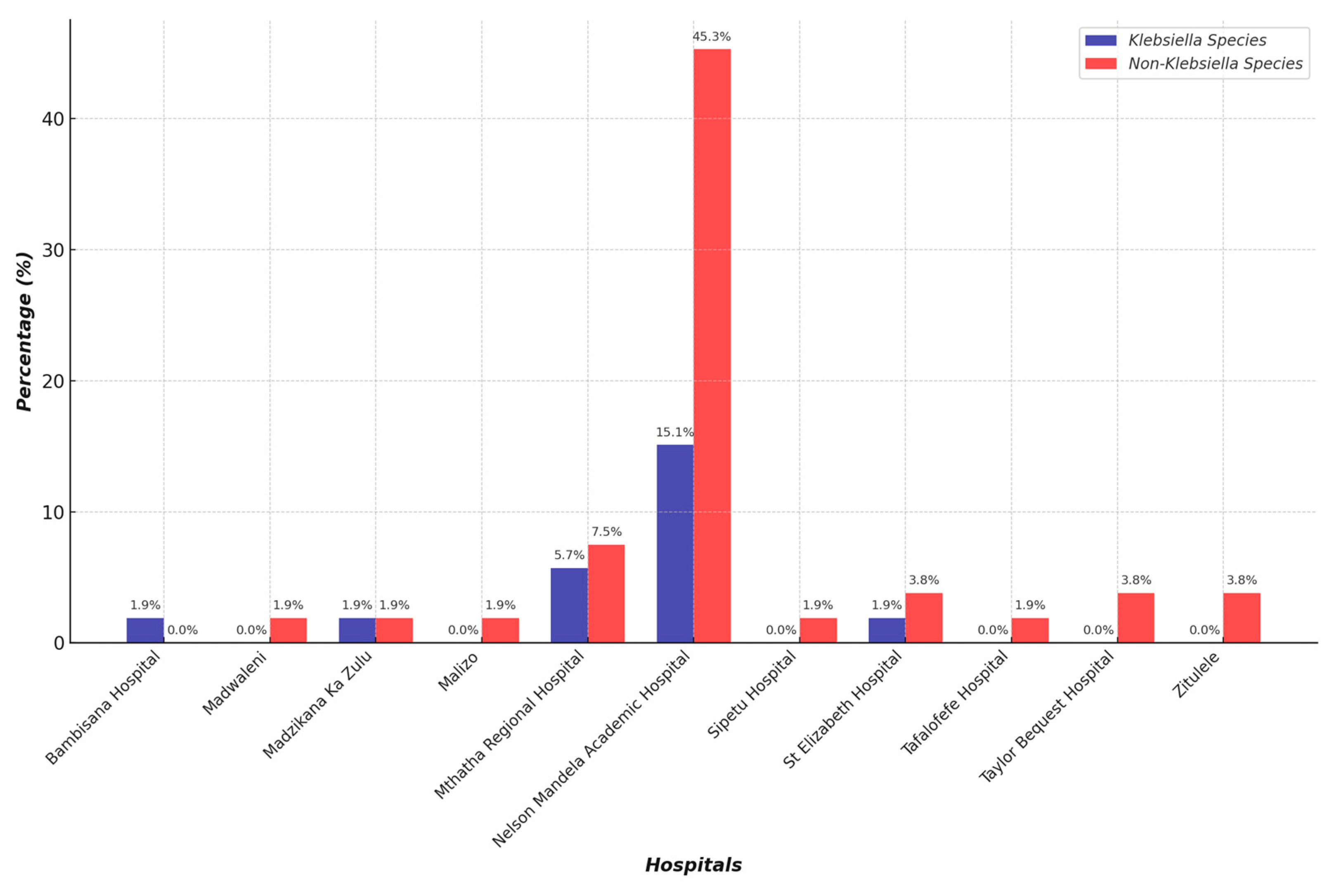
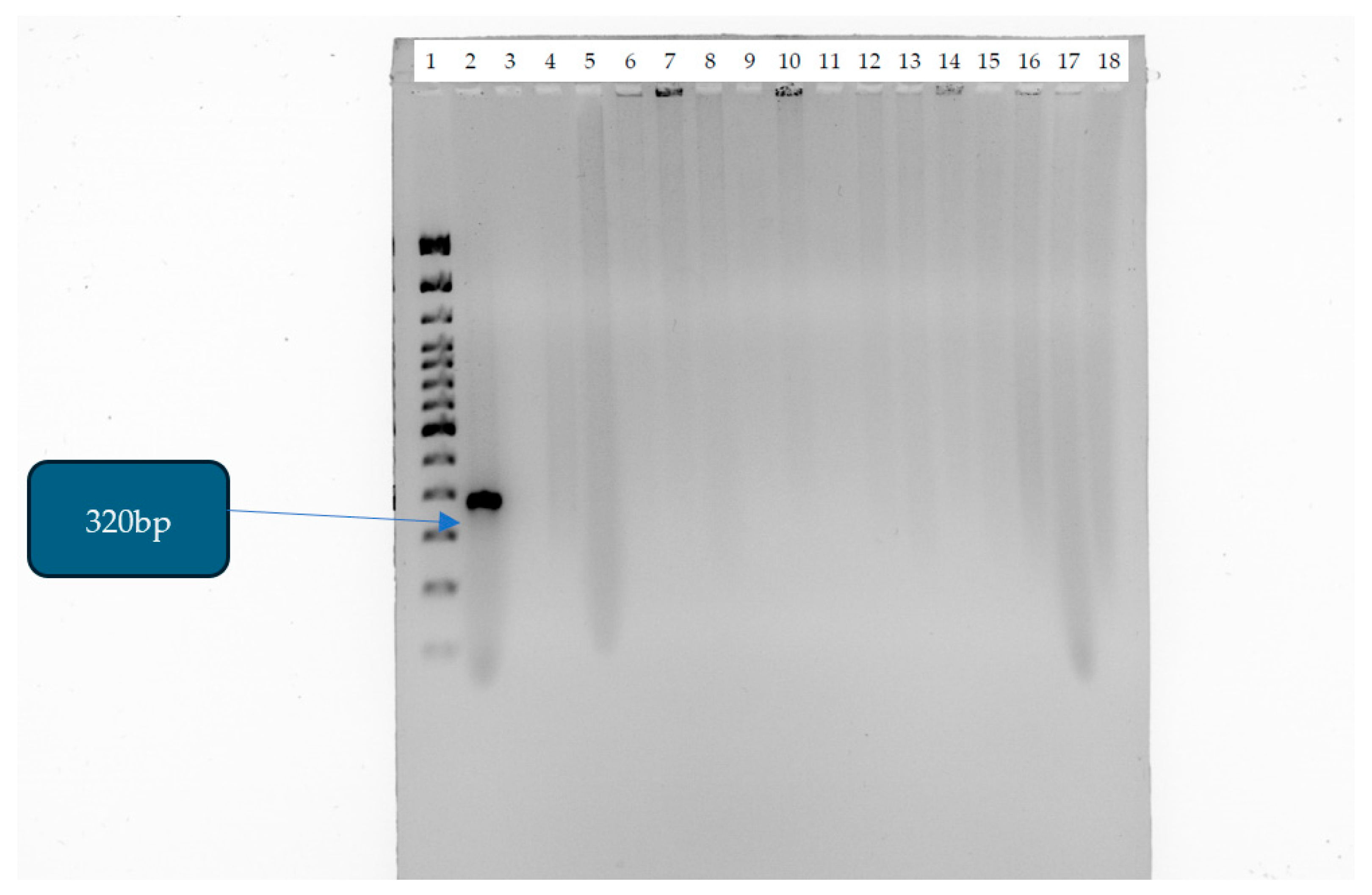
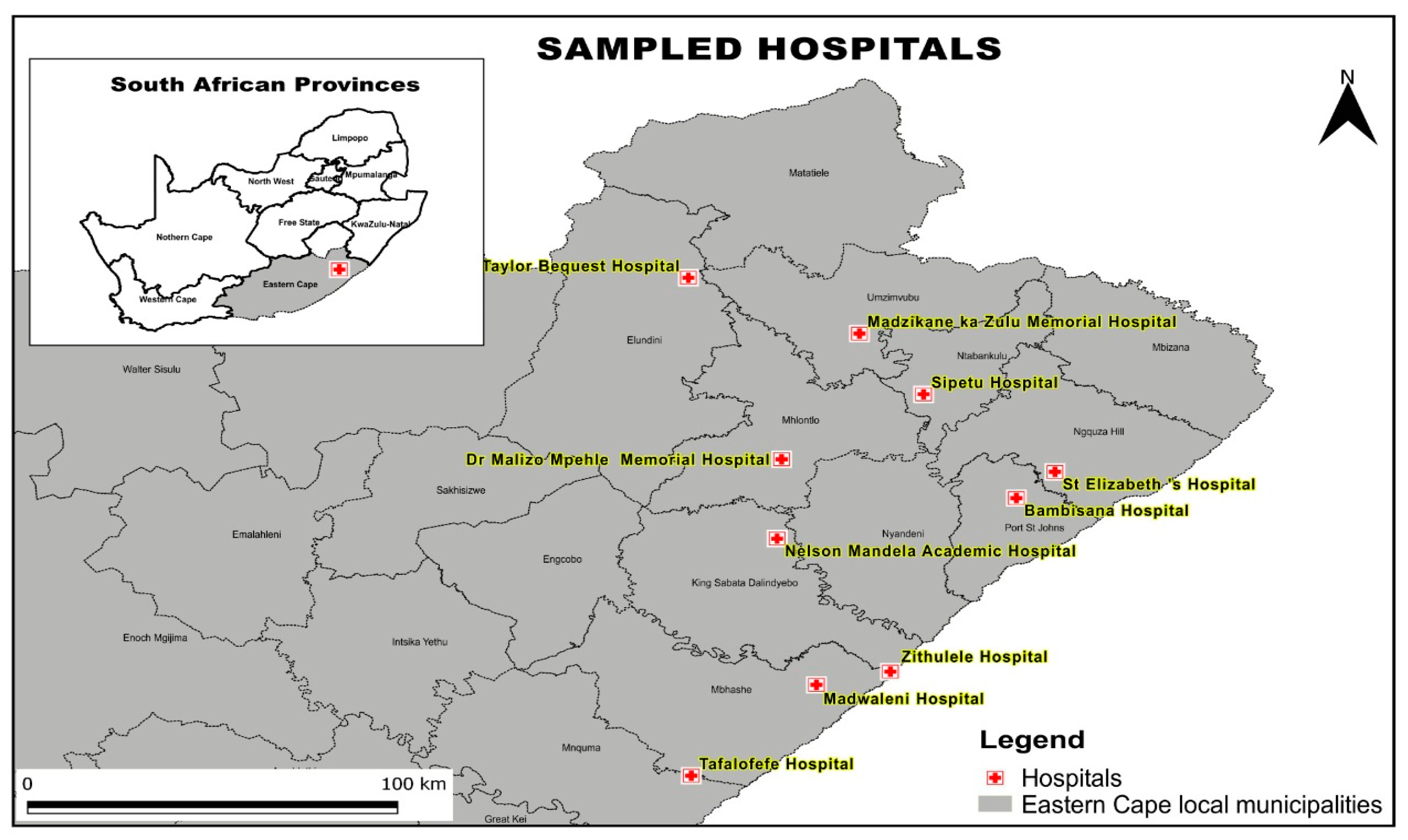
| CRE Isolates | Percentage |
|---|---|
| Klebsiella pneumoniae | 53% |
| Enterobacter cloacae | 17% |
| Escherichia coli | 8% |
| Serratia marcescens | 8% |
| Serratia fonticola | 2% |
| Enterobacter aerogenes | 2% |
| Klebsiella oxytoca | 2% |
| Citrobacter koseri | 2% |
| Citrobacter freundii | 2% |
| Group | Klebsiella Species (%) | Non Klebsiella Species (%) | p-Value |
|---|---|---|---|
| Neonates (<28 days and infants <1 year) | 10/53 (19%) | 11/53 (21%) | 1.00 |
| Pediatric (>13 months–≤14 years) | 0/53 (0%) | 3/53 (6%) | 0.24 |
| Young Adults (>14 years–≤19 years) | 3/53 (6%) | 2/53 (4%) | 1.00 |
| Adults (≥20 years–<65 years) | 9/53 (17%) | 10/53 (19%) | 1.00 |
| Elderly ≥65 years | 3/53 (6%) | 2/53 (4%) | 1.00 |
| Hospitals | Wards | Specimen | Organisms | Colistin MIC (µg/mL) | Interpretation |
|---|---|---|---|---|---|
| 1. NMAH | Female Medical Ward A | Urine | Enterobacter Cloacae | 8 | R |
| 2. NMAH | Female Medical Ward C | Urine | Enterobacter aerogenes | 16 | R |
| 3. NMAH | Neonatal High Care | Blood Culture | Serratia marcescens | 16 | R |
| 4. MRH | Neonatal Ward | Blood Culture | Klebsiella pneumoniae | 16 | R |
| 5. MRH | Neonatal Ward | Blood Culture | Serratia marcescens | 16 | R |
| 6. NMAH | Pediatric High Care | Pus Swab | Serratia marcescens | 16 | R |
| 7. NMAH | Neonatal Intensive Care | Pus Swab | Klebsiella pneumoniae | 8 | R |
| Current MIC Breakpoints (µg/mL) | |||
|---|---|---|---|
| MIC Interpretation | |||
| Carbapenems | Susceptible | Intermediate | Resistant |
| Doripenem | ≤1 | 2 | ≥4 |
| Ertapenem | ≤0.5 | 1 | ≥2 |
| Imipenem | ≤1 | 2 | ≥4 |
| Meropenem | ≤1 | 2 | ≥4 |
| Target Gene | Positive Control | Primer | Base Pair | Reference |
|---|---|---|---|---|
| mcr-1 gene | Escherichia coli 13846 | F:5/-AGTCCGTTTGTTCTTGTGGC-/3 R:5/-AGATCCTTGGTCTCGGCTTG-/3 | 320 bp | [20] |
| Step | Temperature (°C) | Time | Cycles |
|---|---|---|---|
| Activation | 94 | 15 min | 1 |
| Denaturation | 94 | 30 s | 25 |
| Annealing | 58 | 1 min 30 s | |
| Elongation | 72 | 1 min |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ndlela, S.; Singh, R.; Vasaikar, S. Determination of Colistin Resistance in Clinical Isolates from Healthcare Facilities in Mthatha and Surrounding Areas. Antibiotics 2025, 14, 505. https://doi.org/10.3390/antibiotics14050505
Ndlela S, Singh R, Vasaikar S. Determination of Colistin Resistance in Clinical Isolates from Healthcare Facilities in Mthatha and Surrounding Areas. Antibiotics. 2025; 14(5):505. https://doi.org/10.3390/antibiotics14050505
Chicago/Turabian StyleNdlela, Silindokuhle, Ravesh Singh, and Sandeep Vasaikar. 2025. "Determination of Colistin Resistance in Clinical Isolates from Healthcare Facilities in Mthatha and Surrounding Areas" Antibiotics 14, no. 5: 505. https://doi.org/10.3390/antibiotics14050505
APA StyleNdlela, S., Singh, R., & Vasaikar, S. (2025). Determination of Colistin Resistance in Clinical Isolates from Healthcare Facilities in Mthatha and Surrounding Areas. Antibiotics, 14(5), 505. https://doi.org/10.3390/antibiotics14050505







