Staphylococcus aureus: A Review of the Pathogenesis and Virulence Mechanisms
Abstract
1. Introduction
2. Pathogenesis of S. aureus
2.1. Colonization Dynamics
2.1.1. Nasal Cavity
Host–Microbiome Interactions
Risk Factors for Nasal Colonization
2.1.2. Skin Colonization
Role of Clumping Factor B (ClfB)
Role of Surface Protein G (SasG)
Immune Evasion and Vaccine Challenges
2.1.3. Throat and Oropharynx Colonization
Prevalence of S. aureus in the Throat/Oropharynx
Persistence and Challenges in Eradication
2.1.4. Gastrointestinal/Perineum and Urogenital Tract Colonization
Detection and Decolonization
2.2. Virulence Factors of S. aureus
2.2.1. Surface Proteins and Adhesion
2.2.2. Secreted Toxins and Immune Evasion
2.3. Staphylococcal PVL: Clinical Implications, Molecular Mechanisms, and Genetic Landscape
2.3.1. Importance of PVL in S. aureus Infections
2.3.2. Mechanism of Action of PVL
2.3.3. Genetics of the PVL in S. aureus
2.4. Metabolic Factors
2.4.1. Iron Acquisition by S. aureus
2.4.2. Carbon Metabolism in S. aureus
2.4.3. Amino Acid Metabolism
3. Regulation of Virulence Factors
3.1. The Accessory Gene Regulator (agr) System
3.2. The Staphylococcal Accessory Regulator (sar) System
4. Antibiotic Resistance and Mechanisms in S. aureus
4.1. Mechanisms of Antibiotic Resistance
4.1.1. Beta-Lactam Resistance
4.1.2. Mechanism of Methicillin Resistance in S. aureus
4.1.3. Glycopeptide Resistance
4.1.4. Aminoglycoside Resistance
4.1.5. Fluoroquinolone Resistance
4.1.6. Tetracycline and Macrolide Resistance
4.2. Epidemiology and Clinical Impact of MRSA
4.2.1. HA-MRSA
4.2.2. CA-MRSA
4.2.3. VRSA
| Region | Prevalence (%) | Countries with Notable Data | Country-Specific Prevalence (%) | Reference |
|---|---|---|---|---|
| Asia | 1.2% | India, Pakistan, Saudi Arabia | India (0.7%), Pakistan (0.1%), Saudi Arabia (18%) | [281] |
| Europe | 1.1% | Italy, Turkey, Germany, France, Belgium | Italy (1.1%), Turkey (2.7%), Germany (0.7%), France (2.2%), Belgium (2.5%) | [281] |
| America | 3.6% | Brazil | Brazil (3%) | [282] |
| Africa | 2.5% | Ethiopia, Egypt | Nigeria (29%), Egypt (Multiple reports) | [222,283] |
| Middle East | - | Saudi Arabia, Egypt | Saudi Arabia (18%), Egypt (210 isolates reported) | [283] |
5. Emerging Therapeutic Strategies for Mitigating S. aureus Biofilms, Antimicrobial Resistance, and Virulence Factors
5.1. Immunotherapeutic and Vaccination Strategies
5.2. CRISPR-Cas Systems for Genetic Targeting
5.3. Phage Therapy, Endolysins, and Antimicrobial Peptides
5.4. Nanotechnology and Nanoparticle Applications
5.5. Biofilm Disruption: Enzymatic, Physical, and Metabolic Strategies
5.6. QS Inhibition
5.7. Phytochemical and Small-Molecule Interventions
5.8. Anti-Virulence and Host–Pathogen Interaction Modulation
5.9. Combination Therapies and Surface Modifications
6. Conclusions and Future Directions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Getaneh, A. Staphylococcus aureus, ESKAPE Bacteria Challenging Current Health Care and Community Settings: A Literature Review. Clin. Lab. 2021, 67. [Google Scholar]
- Taylor, T.A.; Unakal, C.G. Staphylococcus aureus Infection. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2025. Available online: http://www.ncbi.nlm.nih.gov/books/NBK441868/ (accessed on 20 February 2025).
- Laux, C.; Peschel, A.; Krismer, B. Staphylococcus aureus Colonization of the Human Nose and Interaction with Other Microbiome Members. Microbiol. Spectr. 2019, 7. [Google Scholar] [CrossRef] [PubMed]
- Cheung, G.Y.C.; Bae, J.S.; Otto, M. Pathogenicity and virulence of Staphylococcus aureus. Virulence 2021, 12, 547–569. [Google Scholar] [CrossRef] [PubMed]
- Linz, M.S.; Mattappallil, A.; Finkel, D.; Parker, D. Clinical Impact of Staphylococcus aureus Skin and Soft Tissue Infections. Antibiotics 2023, 12, 557. [Google Scholar] [CrossRef]
- Peng, Q.; Tang, X.; Dong, W.; Sun, N.; Yuan, W. A Review of Biofilm Formation of Staphylococcus aureus and Its Regulation Mechanism. Antibiotics 2022, 12, 12. [Google Scholar] [CrossRef]
- Siddiqui, A.H.; Koirala, J. Methicillin-Resistant Staphylococcus aureus. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2023. Available online: http://www.ncbi.nlm.nih.gov/books/NBK482221/ (accessed on 9 July 2023).
- Fait, A.; Silva, S.F.; Abrahamsson, J.Å.H.; Ingmer, H. Staphylococcus aureus response and adaptation to vancomycin. In Advances in Microbial Physiology; Poole, R.K., Kelly, D.J., Eds.; Advances in Microbial Physiology; Academic Press: Cambridge, MA, USA, 2024; Volume 85, pp. 201–258. Available online: https://www.sciencedirect.com/science/article/pii/S0065291124000109 (accessed on 12 March 2025).
- Mlynarczyk-Bonikowska, B.; Kowalewski, C.; Krolak-Ulinska, A.; Marusza, W. Molecular Mechanisms of Drug Resistance in Staphylococcus aureus. Int. J. Mol. Sci. 2022, 23, 8088. [Google Scholar] [CrossRef]
- Rajput, P.; Nahar, K.S.; Rahman, K.M. Evaluation of Antibiotic Resistance Mechanisms in Gram-Positive Bacteria. Antibiotics 2024, 13, 1197. [Google Scholar] [CrossRef]
- Howden, B.P.; Giulieri, S.G.; Wong Fok Lung, T.; Baines, S.L.; Sharkey, L.K.; Lee, J.Y.H.; Hachani, A.; Monk, I.R.; Stinear, T.P. Staphylococcus aureus host interactions and adaptation. Nat. Rev. Microbiol. 2023, 21, 380–395. [Google Scholar] [CrossRef]
- Piewngam, P.; Otto, M. Staphylococcus aureus colonisation and strategies for decolonisation. Lancet Microbe 2024, 5, e606–e618. [Google Scholar] [CrossRef]
- Reategui Schwarz, E.; van de Guchte, A.; Dupper, A.C.; Caban, A.B.; Nadkarni, D.; Fox, L.; Mills, A.; Obla, A.; Chacko, K.I.; Oussenko, I.; et al. Everybody nose: Molecular and clinical characteristics of nasal colonization during active methicillin-resistant Staphylococcus aureus bloodstream infection. BMC Infect. Dis. 2022, 22, 400. [Google Scholar] [CrossRef]
- Gehrke, A.-K.E.; Giai, C.; Gómez, M.I. Staphylococcus aureus Adaptation to the Skin in Health and Persistent/Recurrent Infections. Antibiotics 2023, 12, 1520. [Google Scholar] [CrossRef] [PubMed]
- Sakr, A.; Brégeon, F.; Mège, J.-L.; Rolain, J.-M.; Blin, O. Staphylococcus aureus Nasal Colonization: An Update on Mechanisms, Epidemiology, Risk Factors, and Subsequent Infections. Front. Microbiol. 2018, 9, 2419. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Bitzer, A.; Power, J.J.; Belikova, D.; Torres-Salazar, B.; Adolf, L.A.; Gerlach, D.L.; Krismer, B.; Heilbronner, S. Nasal commensals reduce Staphylococcus aureus proliferation by restricting siderophore availability. bioRxiv 2024. [Google Scholar] [CrossRef] [PubMed]
- Hammer, N.D.; Skaar, E.P. Molecular mechanisms of Staphylococcus aureus iron acquisition. Annu. Rev. Microbiol. 2011, 65, 129–147. [Google Scholar] [CrossRef]
- Brozyna, J.R.; Sheldon, J.R.; Heinrichs, D.E. Growth promotion of the opportunistic human pathogen, Staphylococcus lugdunensis, by heme, hemoglobin, and coculture with Staphylococcus aureus. MicrobiologyOpen 2014, 3, 182–195. [Google Scholar] [CrossRef]
- Huang, S.; Hon, K.; Bennett, C.; Hu, H.; Menberu, M.; Wormald, P.-J.; Zhao, Y.; Vreugde, S.; Liu, S. Corynebacterium accolens inhibits Staphylococcus aureus induced mucosal barrier disruption. Front. Microbiol. 2022, 13, 984741. [Google Scholar] [CrossRef]
- Stubbendieck, R.M.; May, D.S.; Chevrette, M.G.; Temkin, M.I.; Wendt-Pienkowski, E.; Cagnazzo, J.; Carlson, C.M.; Gern, J.E.; Currie, C.R. Competition among Nasal Bacteria Suggests a Role for Siderophore-Mediated Interactions in Shaping the Human Nasal Microbiota. Appl. Environ. Microbiol. 2019, 85, e02406-18. [Google Scholar] [CrossRef]
- Jin, T.; Mohammad, M.; Pullerits, R.; Ali, A. Bacteria and Host Interplay in Staphylococcus aureus Septic Arthritis and Sepsis. Pathogens 2021, 10, 158. [Google Scholar] [CrossRef]
- Odunitan, T.T.; Oyaronbi, A.O.; Adebayo, F.A.; Adekoyeni, P.A.; Apanisile, B.T.; Oladunni, T.D.; Saibu, O.A. Antimicrobial peptides: A novel and promising arsenal against methicillin-resistant Staphylococcus aureus (MRSA) infections. Pharm. Sci. Adv. 2024, 2, 100034. [Google Scholar] [CrossRef]
- Han, W.; Camesano, T.A. LL37-Derived Fragments Improve the Antibacterial Potential of Penicillin G and Ampicillin against Methicillin-Resistant Staphylococcus aureus. Antibiotics 2023, 12, 1398. [Google Scholar] [CrossRef]
- Meade, K.G.; O’Farrelly, C. β-Defensins: Farming the Microbiome for Homeostasis and Health. Front. Immunol. 2019, 9, 3072. [Google Scholar] [CrossRef] [PubMed]
- Techasupaboon, T.; Vasikasin, V.; Varothai, N.; Raknaisil, N.; Nasomsong, W. Staphylococcus aureus nasal carriage and bloodstream infection among conventional hemodialysis patients in Thailand: A prospective multicenter cohort study. BMC Res. Notes 2022, 15, 290. [Google Scholar] [CrossRef] [PubMed]
- Hidron, A.I.; Kempker, R.; Moanna, A.; Rimland, D. Methicillin-resistant Staphylococcus aureus in HIV-infected patients. Infect. Drug Resist. 2010, 3, 73–86. [Google Scholar] [CrossRef] [PubMed]
- McNeil, J.C. Staphylococcus aureus—Antimicrobial resistance and the immunocompromised child. Infect. Drug Resist. 2014, 7, 117–127. [Google Scholar] [CrossRef]
- Congdon, S.T.; Guaglione, J.A.; Ricketts, O.M.A.; Murphy, K.V.; Anderson, M.G.; Trowbridge, D.A.; Al-Abduladheem, Y.; Phillips, A.M.; Beausoleil, A.M.; Stanley, A.J.; et al. Prevalence and antibiotic resistance of Staphylococcus aureus associated with a college-aged cohort: Life-style factors that contribute to nasal carriage. Front. Cell. Infect. Microbiol. 2023, 13, 1195758. [Google Scholar] [CrossRef]
- Sarkar, A.; McInroy, C.J.A.; Harty, S.; Raulo, A.; Ibata, N.G.O.; Valles-Colomer, M.; Johnson, K.V.-A.; Brito, I.L.; Henrich, J.; Archie, E.A.; et al. Microbial transmission in the social microbiome and host health and disease. Cell 2024, 187, 17–43. [Google Scholar] [CrossRef]
- Septimus, E.J.; Schweizer, M.L. Decolonization in Prevention of Health Care-Associated Infections. Clin. Microbiol. Rev. 2016, 29, 201–222. [Google Scholar] [CrossRef]
- Smith, M.; Herwaldt, L. Nasal decolonization: What antimicrobials and antiseptics are most effective before surgery and in the ICU. Am. J. Infect. Control 2023, 51, A64–A71. [Google Scholar] [CrossRef]
- Abie, S.; Tiruneh, M.; Abebe, W. Methicillin-resistant Staphylococcus aureus nasal carriage among janitors working in hospital and non-hospital areas: A comparative cross-sectional study. Ann. Clin. Microbiol. Antimicrob. 2020, 19, 47. [Google Scholar] [CrossRef]
- Conceição, T.; Martins, H.; Rodrigues, S.; de Lencastre, H.; Aires-de-Sousa, M. Staphylococcus aureus nasal carriage among homeless population in Lisbon, Portugal. Eur. J. Clin. Microbiol. Infect. Dis. 2019, 38, 2037–2044. [Google Scholar] [CrossRef]
- Tsang, S.T.J.; McHugh, M.P.; Guerendiain, D.; Gwynne, P.J.; Boyd, J.; Simpson, A.H.R.W.; Walsh, T.S.; Laurenson, I.F.; Templeton, K.E. Underestimation of Staphylococcus aureus (MRSA and MSSA) carriage associated with standard culturing techniques. Bone Jt. Res. 2018, 7, 79–84. [Google Scholar] [CrossRef] [PubMed]
- Ahmann, M.; Compton, J.; Pottinger, J.; Uhlenhopp, R.; Ward, M.; Haleem, A.; Willey, M.; Schweizer, M.; Herwaldt, L. Staphylococcus aureus colonization and surgical site infections among patients undergoing surgical fixation for acute fractures. Infect. Control Hosp. Epidemiol. 2025, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Zani, K.; Hobeika, J.; Sun, Y.; Kohler, C.; Cherian, A.; Fields, T.; Jia, Q.; Tang, L.; Hysmith, N.D.; Margolis, E.B. Nasal microbiota predictors for methicillin resistant Staphylococcus colonization in critically ill children. PLoS ONE 2025, 20, e0316460. [Google Scholar] [CrossRef]
- Boncompain, C.A.; Suárez, C.A.; Morbidoni, H.R. Staphylococcus aureus nasal Carriage in Health Care Workers: First Report from a Major Public Hospital in Argentina. Available online: https://www.elsevier.es/es-revista-revista-argentina-microbiologia-372-pdf-S032575411730010X (accessed on 6 March 2025).
- Bouiller, K.; Zeggay, A.; Gbaguidi-Haore, H.; Hocquet, D.; Chirouze, C.; Bertrand, X. Epidemiology and risk factors of nasal carriage of Staphylococcus aureus CC398 in two distinct cohorts in France. Front. Microbiol. 2022, 13, 1068420. [Google Scholar] [CrossRef] [PubMed]
- Kleine, L.M.; Kanu, E.M.; Grebe, T.; Sesay, D.M.; Loismann, H.; Sesay, M.; Theiler, T.; Rudolf, V.; Mellmann, A.; Kalkman, L.C.; et al. Nasopharyngeal carriage of Staphylococcus aureus in a rural population, Sierra Leone. Int. J. Med. Microbiol. 2025, 318, 151643. [Google Scholar] [CrossRef]
- Halablab, M.A.; Hijazi, S.M.; Fawzi, M.A.; Araj, G.F. Staphylococcus aureus nasal carriage rate and associated risk factors in individuals in the community. Epidemiol. Infect. 2010, 138, 702–706. [Google Scholar] [CrossRef]
- Chen, B.; Dai, X.; He, B.; Pan, K.; Li, H.; Liu, X.; Bao, Y.; Lao, W.; Wu, X.; Yao, Y.; et al. Differences in Staphylococcus aureus nasal carriage and molecular characteristics among community residents and healthcare workers at Sun Yat-Sen University, Guangzhou, Southern China. BMC Infect. Dis. 2015, 15, 303. [Google Scholar] [CrossRef]
- Al-Humaidan, O.S.; El-Kersh, T.A.; Al-Akeel, R.A. Risk factors of nasal carriage of Staphylococcus aureus and methicillin-resistant Staphylococcus aureus among health care staff in a teaching hospital in central Saudi Arabia. Saudi Med. J. 2015, 36, 1084–1090. [Google Scholar] [CrossRef]
- Agabou, A.; Ouchenane, Z.; Ngba Essebe, C.; Khemissi, S.; Chehboub, M.T.E.; Chehboub, I.B.; Sotto, A.; Dunyach-Remy, C.; Lavigne, J.-P. Emergence of Nasal Carriage of ST80 and ST152 PVL+ Staphylococcus aureus Isolates from Livestock in Algeria. Toxins 2017, 9, 303. [Google Scholar] [CrossRef]
- Feuillie, C.; Vitry, P.; McAleer, M.A.; Kezic, S.; Irvine, A.D.; Geoghegan, J.A.; Dufrêne, Y.F. Adhesion of Staphylococcus aureus to Corneocytes from Atopic Dermatitis Patients Is Controlled by Natural Moisturizing Factor Levels. mBio 2018, 9, e01184. [Google Scholar] [CrossRef]
- González-García, S.; Hamdan-Partida, A.; Valdez-Alarcón, J.J.; Bustos-Hamdan, A.; Bustos-Martínez, J.; González-García, S.; Hamdan-Partida, A.; Valdez-Alarcón, J.J.; Bustos-Hamdan, A.; Bustos-Martínez, J. Main Factors of Staphylococcus aureus Associated with the Interaction to the Cells for Their Colonization and Persistence. In Staphylococcal Infections—Recent Advances and Perspectives; IntechOpen: London, UK, 2022; ISBN 978-1-83768-206-5. Available online: https://www.intechopen.com/chapters/84091 (accessed on 14 March 2025).
- Sivori, F.; Cavallo, I.; Truglio, M.; De Maio, F.; Sanguinetti, M.; Fabrizio, G.; Licursi, V.; Francalancia, M.; Fraticelli, F.; La Greca, I.; et al. Staphylococcus aureus colonizing the skin microbiota of adults with severe atopic dermatitis exhibits genomic diversity and convergence in biofilm traits. Biofilm 2024, 8, 100222. [Google Scholar] [CrossRef] [PubMed]
- Fleury, O.M.; McAleer, M.A.; Feuillie, C.; Formosa-Dague, C.; Sansevere, E.; Bennett, D.E.; Towell, A.M.; McLean, W.H.I.; Kezic, S.; Robinson, D.A.; et al. Clumping Factor B Promotes Adherence of Staphylococcus aureus to Corneocytes in Atopic Dermatitis. Infect. Immun. 2017, 85, e00994-16. [Google Scholar] [CrossRef] [PubMed]
- Lacey, K.A.; Mulcahy, M.E.; Towell, A.M.; Geoghegan, J.A.; McLoughlin, R.M. Clumping factor B is an important virulence factor during Staphylococcus aureus skin infection and a promising vaccine target. PLoS Pathog. 2019, 15, e1007713. [Google Scholar] [CrossRef] [PubMed]
- Foster, T.J.; Geoghegan, J.A.; Ganesh, V.K.; Höök, M. Adhesion, invasion and evasion: The many functions of the surface proteins of Staphylococcus aureus. Nat. Rev. Microbiol. 2014, 12, 49–62. [Google Scholar] [CrossRef]
- Mills, K.B.; Maciag, J.J.; Wang, C.; Crawford, J.A.; Enroth, T.J.; Keim, K.C.; Dufrêne, Y.F.; Robinson, D.A.; Fey, P.D.; Herr, A.B.; et al. Staphylococcus aureus skin colonization is mediated by SasG lectin variation. bioRxiv 2023. [Google Scholar] [CrossRef]
- Reslane, I.; Watson, G.F.; Handke, L.D.; Fey, P.D. Regulatory dynamics of arginine metabolism in Staphylococcus aureus. Biochem. Soc. Trans. 2024, 52, 2513–2523. [Google Scholar] [CrossRef]
- Hajam, I.A.; Liu, G.Y. Linking S. aureus Immune Evasion Mechanisms to Staphylococcal Vaccine Failures. Antibiotics 2024, 13, 410. [Google Scholar] [CrossRef]
- Cho, J.S.; Pietras, E.M.; Garcia, N.C.; Ramos, R.I.; Farzam, D.M.; Monroe, H.R.; Magorien, J.E.; Blauvelt, A.; Kolls, J.K.; Cheung, A.L.; et al. IL-17 is essential for host defense against cutaneous Staphylococcus aureus infection in mice. J. Clin. Investig. 2010, 120, 1762–1773. [Google Scholar] [CrossRef]
- Teymournejad, O.; Montgomery, C.P. Evasion of Immunological Memory by S. aureus Infection: Implications for Vaccine Design. Front. Immunol. 2021, 12, 633672. [Google Scholar] [CrossRef]
- Miller, L.S.; Fowler, V.G.; Shukla, S.K.; Rose, W.E.; Proctor, R.A. Development of a vaccine against Staphylococcus aureus invasive infections: Evidence based on human immunity, genetics and bacterial evasion mechanisms. FEMS Microbiol. Rev. 2019, 44, 123–153. [Google Scholar] [CrossRef]
- Nilsson, P.; Ripa, T. Staphylococcus aureus Throat Colonization Is More Frequent than Colonization in the Anterior Nares. J. Clin. Microbiol. 2006, 44, 3334–3339. [Google Scholar] [CrossRef] [PubMed]
- Mertz, D.; Frei, R.; Periat, N.; Zimmerli, M.; Battegay, M.; Flückiger, U.; Widmer, A.F. Exclusive Staphylococcus aureus Throat Carriage: At-Risk Populations. Arch. Intern. Med. 2009, 169, 172–178. [Google Scholar] [CrossRef] [PubMed]
- Hamdan-Partida, A.; Sainz-Espuñes, T.; Bustos-Martínez, J. Characterization and Persistence of Staphylococcus aureus Strains Isolated from the Anterior Nares and Throats of Healthy Carriers in a Mexican Community. J. Clin. Microbiol. 2010, 48, 1701–1705. [Google Scholar] [CrossRef] [PubMed]
- Hanson, B.; Kates, A.; Mills, E.; Herwaldt, L.; Torner, J.; Dawson, J.; Smith, T. The Oropharynx as a Distinct Colonization Site for Staphylococcus aureus in the Community. bioRxiv 2017. [Google Scholar] [CrossRef]
- Hanson, B.M.; Kates, A.E.; O’Malley, S.M.; Mills, E.; Herwaldt, L.A.; Torner, J.C.; Dawson, J.D.; Farina, S.A.; Klostermann, C.; Wu, J.Y.; et al. Staphylococcus aureus in the nose and throat of Iowan families. Epidemiol. Infect. 2018, 146, 1777–1784. [Google Scholar] [CrossRef]
- Senn, L.; Basset, P.; Nahimana, I.; Zanetti, G.; Blanc, D.S. Which Anatomical Sites Should be Sampled for Screening of Methicillin-Resistant Staphylococcus aureus Carriage by Culture or by Rapid PCR Test? 2011. Available online: https://onlinelibrary.wiley.com/doi/10.1111/j.1469-0691.2011.03724.x (accessed on 15 March 2025).
- Batra, R.; Eziefula, A.; Wyncoll, D.; Edgeworth, J. Throat and rectal swabs may have an important role in MRSA screening of critically ill patients. Intensive Care Med. 2008, 34, 1703–1706. [Google Scholar] [CrossRef]
- Williamson, D.A.; Ritchie, S.; Keren, B.; Harrington, M.; Thomas, M.G.; Upton, A.; Lennon, D.; Leversha, A. Persistence, Discordance and Diversity of Staphylococcus aureus Nasal and Oropharyngeal Colonization in School-aged Children. Pediatr. Infect. Dis. J. 2016, 35, 744–748. [Google Scholar] [CrossRef]
- Ogawa, T.; Terao, Y.; Okuni, H.; Ninomiya, K.; Sakata, H.; Ikebe, K.; Maeda, Y.; Kawabata, S. Biofilm formation or internalization into epithelial cells enable Streptococcus pyogenes to evade antibiotic eradication in patients with pharyngitis. Microb. Pathog. 2011, 51, 58–68. [Google Scholar] [CrossRef]
- Turner, N.A.; Sharma-Kuinkel, B.K.; Maskarinec, S.A.; Eichenberger, E.M.; Shah, P.P.; Carugati, M.; Holland, T.L.; Fowler, V.G. Methicillin-resistant Staphylococcus aureus: An overview of basic and clinical research. Nat. Rev. Microbiol. 2019, 17, 203–218. [Google Scholar] [CrossRef]
- Liu, A.; Garrett, S.; Hong, W.; Zhang, J. Staphylococcus aureus Infections and Human Intestinal Microbiota. Pathogens 2024, 13, 276. [Google Scholar] [CrossRef]
- Tong, S.Y.C.; Chen, L.F.; Fowler, V.G. Colonization, pathogenicity, host susceptibility, and therapeutics for Staphylococcus aureus: What is the clinical relevance? Semin. Immunopathol. 2012, 34, 185–200. [Google Scholar] [CrossRef] [PubMed]
- Bhalla, A.; Aron, D.C.; Donskey, C.J. Staphylococcus aureus intestinal colonization is associated with increased frequency of S. aureus on skin of hospitalized patients. BMC Infect. Dis. 2007, 7, 105. [Google Scholar] [CrossRef] [PubMed]
- Kates, A.E.; Thapaliya, D.; Smith, T.C.; Chorazy, M.L. Prevalence and molecular characterization of Staphylococcus aureus from human stool samples. Antimicrob. Resist. Infect. Control 2018, 7, 42. [Google Scholar] [CrossRef] [PubMed]
- Raineri, E.J.M.; Altulea, D.; van Dijl, J.M. Staphylococcal trafficking and infection—From ‘nose to gut’ and back. FEMS Microbiol. Rev. 2022, 46, fuab041. [Google Scholar] [CrossRef]
- Shah, T.; Baloch, Z.; Shah, Z.; Cui, X.; Xia, X. The Intestinal Microbiota: Impacts of Antibiotics Therapy, Colonization Resistance, and Diseases. Int. J. Mol. Sci. 2021, 22, 6597. [Google Scholar] [CrossRef]
- Khetsuriani, S. Gastrointestinal Colonization by Staphylococcus aureus Strains as Risk Factor for Different Infections (Brief Review). Int. J. Multidiscip. Res. Publ. IJMRAP 2023, 5. [Google Scholar]
- Bwanga, F.; Mukashyaka, C.; Kateete, D.P.; Tumuhamye, J.; Okeng, A.; Aboce, E.; Namugga, O.; Kwizera, R.; Sommerfelt, H.; Nankabirwa, V. Vaginal colonization with virulent and methicillin resistant Staphylococcus aureus among Ugandan women in Labour. BMC Microbiol. 2024, 24, 307. [Google Scholar] [CrossRef]
- Deng, L.; Schilcher, K.; Burcham, L.R.; Kwiecinski, J.M.; Johnson, P.M.; Head, S.R.; Heinrichs, D.E.; Horswill, A.R.; Doran, K.S. Identification of Key Determinants of Staphylococcus aureus Vaginal Colonization. mBio 2019, e02321-19. [Google Scholar] [CrossRef]
- Nurjadi, D.; Eichel, V.M.; Tabatabai, P.; Klein, S.; Last, K.; Mutters, N.T.; Pöschl, J.; Zanger, P.; Heeg, K.; Boutin, S. Surveillance for Colonization, Transmission, and Infection With Methicillin-Susceptible Staphylococcus aureus in a Neonatal Intensive Care Unit. JAMA Netw. Open 2021, 4, e2124938. [Google Scholar] [CrossRef]
- Septimus, E.J. Universal MRSA/Staphylococcal Decolonization for Hospitalized Patients. In Infection Prevention: New Perspectives and Controversies; Bearman, G., Morgan, D.J., Murthy, R.K., Hota, S., Eds.; Springer International Publishing: Cham, Switzerland, 2022; pp. 101–110. ISBN 978-3-030-98427-4. [Google Scholar] [CrossRef]
- Yehia, F.A.A.; Yousef, N.; Askoura, M. Exploring Staphylococcus aureus Virulence Factors; Special Emphasis on Staphyloxanthin. Microbiol. Biotechnol. Lett. 2021, 49, 467–477. [Google Scholar] [CrossRef]
- Wang, D.; Wang, L.; Liu, Q.; Zhao, Y. Virulence factors in biofilm formation and therapeutic strategies for Staphylococcus aureus: A review. Anim. Zoonoses 2024, in press. Available online: https://www.sciencedirect.com/science/article/pii/S2950248924000130 (accessed on 9 March 2025). [CrossRef]
- Jenul, C.; Horswill, A.R. Regulation of Staphylococcus aureus Virulence. Microbiol. Spectr. 2019, 7. [Google Scholar] [CrossRef] [PubMed]
- Kong, C.; Neoh, H.; Nathan, S. Targeting Staphylococcus aureus Toxins: A Potential form of Anti-Virulence Therapy. Toxins 2016, 8, 72. [Google Scholar] [CrossRef]
- Risser, F.; López-Morales, J.; Nash, M.A. Adhesive Virulence Factors of Staphylococcus aureus Resist Digestion by Coagulation Proteases Thrombin and Plasmin. ACS Bio. Med. Chem. Au. 2022, 2, 586–599. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, M.; Horswill, A.R. The role of human extracellular matrix proteins in defining Staphylococcus aureus biofilm infections. FEMS Microbiol. Rev. 2024, 48, fuae002. [Google Scholar] [CrossRef] [PubMed]
- Speziale, P.; Pietrocola, G. The Multivalent Role of Fibronectin-Binding Proteins A and B (FnBPA and FnBPB) of Staphylococcus aureus in Host Infections. Front. Microbiol. 2020, 11, 2054. [Google Scholar] [CrossRef]
- Wójcik-Bojek, U.; Różalska, B.; Sadowska, B. Staphylococcus aureus—A Known Opponent against Host Defense Mechanisms and Vaccine Development—Do We Still Have a Chance to Win? Int. J. Mol. Sci. 2022, 23, 948. [Google Scholar] [CrossRef]
- Schulze, A.; Mitterer, F.; Pombo, J.P.; Schild, S. Biofilms by bacterial human pathogens: Clinical relevance—Development, composition and regulation—Therapeutical strategies. Microb. Cell 2021, 8, 28–56. [Google Scholar] [CrossRef]
- Moormeier, D.E.; Bayles, K.W. Staphylococcus aureus Biofilm: A Complex Developmental Organism. Mol. Microbiol. 2017, 104, 365–376. [Google Scholar] [CrossRef]
- Kaushik, A.; Kest, H.; Sood, M.; Thieman, C.; Steussy, B.W.; Padomek, M.; Gupta, S. Infective Endocarditis by Biofilm-Producing Methicillin-Resistant Staphylococcus aureus—Pathogenesis, Diagnosis, and Management. Antibiotics 2024, 13, 1132. [Google Scholar] [CrossRef]
- Ke, S.; Kil, H.; Roggy, C.; Shields, T.; Quinn, Z.; Quinn, A.P.; Small, J.M.; Towne, F.D.; Brooks, A.E.; Brooks, B.D. Potential Therapeutic Targets for Combination Antibody Therapy Against Staphylococcus aureus Infections. Antibiotics 2024, 13, 1046. [Google Scholar] [CrossRef] [PubMed]
- Bear, A.; Locke, T.; Rowland-Jones, S.; Pecetta, S.; Bagnoli, F.; Darton, T.C. The immune evasion roles of Staphylococcus aureus protein A and impact on vaccine development. Front. Cell. Infect. Microbiol. 2023, 13, 1242702. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, D.; Borges, A.; Simões, M. Staphylococcus aureus Toxins and Their Molecular Activity in Infectious Diseases. Toxins 2018, 10, 252. [Google Scholar] [CrossRef] [PubMed]
- Malak, H.A.; Abulreesh, H.H.; Organji, S.R.; Elbanna, K.; Shaaban, M.R.; Samreen; Ahmad, I.; Shami, A.; Alshehri, W.A.; Khalel, A.; et al. Immune System Evasion Mechanisms in Staphylococcus aureus: Current Understanding. J. Pure Appl. Microbiol. 2020, 14, 2219–2234. [Google Scholar] [CrossRef]
- Ahmad-Mansour, N.; Loubet, P.; Pouget, C.; Dunyach-Remy, C.; Sotto, A.; Lavigne, J.-P.; Molle, V. Staphylococcus aureus Toxins: An Update on Their Pathogenic Properties and Potential Treatments. Toxins 2021, 13, 677. [Google Scholar] [CrossRef]
- Tam, K.; Torres, V.J. Staphylococcus aureus Secreted Toxins & Extracellular Enzymes. Microbiol. Spectr. 2019, 7. [Google Scholar] [CrossRef]
- Alkuraythi, D. Virulence Factors and Pathogenicity of Staphylococcus aureus. In Advances and Perspectives of Infections Caused by Staphylococcus aureus; IntechOpen: London, UK, 2024; ISBN 978-0-85466-886-1. Available online: https://www.intechopen.com/chapters/1203878 (accessed on 7 March 2025).
- Xu, H.; Wang, S.; Liu, X.; Li, M.; Wang, X.; Chen, H.; Qu, C.; Liu, Y.; Liu, J. Strategies for Survival of Staphylococcus aureus in Host Cells. Int. J. Mol. Sci. 2025, 26, 720. [Google Scholar] [CrossRef]
- Bolon, B.; Uzal, F.A.; Schutten, M. Chapter 9—Bacterial Toxins. In Haschek and Rousseaux’ s Handbook of Toxicologic Pathology (Fourth Edition); Haschek, W.M., Rousseaux, C.G., Wallig, M.A., Bolon, B., Heinz-taheny, K.M., Rudmann, D.G., Mahler, B.W., Eds.; Academic Press: Cambridge, MA, USA, 2023; pp. 629–678. ISBN 978-0-443-16153-7. Available online: https://www.sciencedirect.com/science/article/pii/B9780443161537000095 (accessed on 15 March 2025).
- Vandenesch, F.; Lina, G.; Henry, T. Staphylococcus aureus Hemolysins, bi-component Leukocidins, and Cytolytic Peptides: A Redundant Arsenal of Membrane-Damaging Virulence Factors? Front. Cell. Infect. Microbiol. 2012, 2, 12. [Google Scholar] [CrossRef]
- Jarraud, S.; Mougel, C.; Thioulouse, J.; Lina, G.; Meugnier, H.; Forey, F.; Nesme, X.; Etienne, J.; Vandenesch, F. Relationships between Staphylococcus aureus Genetic Background, Virulence Factors, agr Groups (Alleles), and Human Disease. Infect. Immun. 2002, 70, 631–641. [Google Scholar] [CrossRef]
- Bukowski, M.; Wladyka, B.; Dubin, G. Exfoliative Toxins of Staphylococcus aureus. Toxins 2010, 2, 1148–1165. [Google Scholar] [CrossRef]
- Lindsay, J.A. Evolution of Staphylococcus aureus and MRSA during outbreaks. Infect. Genet. Evol. J. Mol. Epidemiol. Evol. Genet. Infect. Dis. 2014, 21, 548–553. [Google Scholar] [CrossRef] [PubMed]
- Pietrocola, G.; Nobile, G.; Rindi, S.; Speziale, P. Staphylococcus aureus Manipulates Innate Immunity through Own and Host-Expressed Proteases. Front. Cell. Infect. Microbiol. 2017, 7, 166. [Google Scholar] [CrossRef] [PubMed]
- Otto, M. Staphylococcal biofilms. Curr. Top. Microbiol. Immunol. 2008, 322, 207–228. [Google Scholar] [PubMed]
- Voyich, J.M.; Otto, M.; Mathema, B.; Braughton, K.R.; Whitney, A.R.; Welty, D.; Long, R.D.; Dorward, D.W.; Gardner, D.J.; Lina, G.; et al. Is Panton-Valentine leukocidin the major virulence determinant in community-associated methicillin-resistant Staphylococcus aureus disease? J. Infect. Dis. 2006, 194, 1761–1770. [Google Scholar] [CrossRef]
- Spaan, A.N.; Henry, T.; van Rooijen, W.J.M.; Perret, M.; Badiou, C.; Aerts, P.C.; Kemmink, J.; de Haas, C.J.C.; van Kessel, K.P.M.; Vandenesch, F.; et al. The Staphylococcal Toxin Panton-Valentine Leukocidin Targets Human C5a Receptors. Cell Host Microbe 2013, 13, 584–594. [Google Scholar] [CrossRef]
- Mairi, A.; Touati, A.; Lavigne, J.-P. Methicillin-Resistant Staphylococcus aureus ST80 Clone: A Systematic Review. Toxins 2020, 12, 119. [Google Scholar] [CrossRef]
- Jiang, J.-H.; Cameron, D.R.; Nethercott, C.; Aires-de-Sousa, M.; Peleg, A.Y. Virulence attributes of successful methicillin-resistant Staphylococcus aureus lineages. Clin. Microbiol. Rev. 2023, 36, e0014822. [Google Scholar] [CrossRef]
- He, H.; Wunderink, R.G. Staphylococcus aureus Pneumonia in the Community. Semin. Respir. Crit. Care Med. 2020, 41, 470–479. [Google Scholar] [CrossRef]
- Prista-Leão, B.; Abreu, I.; Duro, R.; Silva-Pinto, A.; Ceia, F.; Andrade, P.; Sobrinho-Simões, J.; Tavares, M.; Pereira, J.M.; Santos, L.; et al. Panton-Valentine Leukocidin-Producing Staphylococcus aureus Infection: A Case Series. Infect. Dis. Rep. 2020, 12, 14. [Google Scholar] [CrossRef]
- Shallcross, L.J.; Fragaszy, E.; Johnson, A.M.; Hayward, A.C. The role of the Panton-Valentine leucocidin toxin in staphylococcal disease: A systematic review and meta-analysis. Lancet Infect. Dis. 2013, 13, 43–54. [Google Scholar] [CrossRef]
- Leistner, R.; Hanitsch, L.G.; Krüger, R.; Lindner, A.K.; Stegemann, M.S.; Nurjadi, D. Skin Infections Due to Panton-Valentine Leukocidin-Producing S. aureus. Dtsch. Arzteblatt Int. 2022, 119, 775–784. [Google Scholar]
- Garbo, V.; Venuti, L.; Boncori, G.; Albano, C.; Condemi, A.; Natoli, G.; Frasca Polara, V.; Billone, S.; Canduscio, L.A.; Cascio, A.; et al. Severe Panton-Valentine-Leukocidin-Positive Staphylococcus aureus Infections in Pediatric Age: A Case Report and a Literature Review. Antibiot. Basel Switz. 2024, 13, 1192. [Google Scholar] [CrossRef] [PubMed]
- Castellazzi, M.L.; Bosis, S.; Borzani, I.; Tagliabue, C.; Pinzani, R.; Marchisio, P.; di Pietro, G.M. Panton-valentine leukocidin Staphylococcus aureus severe infection in an infant: A case report and a review of the literature. Ital. J. Pediatr. 2021, 47, 158. [Google Scholar] [CrossRef] [PubMed]
- Saeed, K.; Esposito, S.; Gould, I.; Ascione, T.; Bassetti, M.; Bonnet, E.; Bouza, E.; Chan, M.; Davis, J.S.; De Simone, G.; et al. Hot topics in necrotising skin and soft tissue infections. Int. J. Antimicrob. Agents 2018, 52, 1–10. [Google Scholar] [CrossRef]
- Tromp, A.T.; van Strijp, J.A.G. Studying Staphylococcal Leukocidins: A Challenging Endeavor. Front. Microbiol. 2020, 11, 611. [Google Scholar] [CrossRef]
- Bernabé, K.J.; Langendorf, C.; Ford, N.; Ronat, J.-B.; Murphy, R.A. Antimicrobial resistance in West Africa: A systematic review and meta-analysis. Int. J. Antimicrob. Agents 2017, 50, 629–639. [Google Scholar] [CrossRef]
- Hanratty, J.; Changez, H.; Smith, A.; Wales, C. Panton-Valentine leukocidin positive Staphylococcal aureus infections of the head and neck: Case series and brief review of literature. J. Oral Maxillofac. Surg. Off. J. Am. Assoc. Oral Maxillofac. Surg. 2015, 73, 666–670. [Google Scholar] [CrossRef]
- Nawrotek, P.; Karakulska, J.; Fijałkowski, K. Chapter 9—The Staphylococcal Panton-Valentine Leukocidin (PVL). In Pet-To-Man Travelling Staphylococci; Savini, V., Ed.; Academic Press Cambridge: Cambridge, MA, USA, 2018; pp. 117–125. ISBN 978-0-12-813547-1. Available online: https://www.sciencedirect.com/science/article/pii/B9780128135471000091 (accessed on 1 April 2025).
- Badarau, A.; Trstenjak, N.; Nagy, E. Structure and Function of the Two-Component Cytotoxins of Staphylococcus aureus—Learnings for Designing Novel Therapeutics. Adv. Exp. Med. Biol. 2017, 966, 15–35. [Google Scholar]
- Hulme, J. Staphylococcus Infection: Relapsing Atopic Dermatitis and Microbial Restoration. Antibiot. Basel Switz. 2023, 12, 222. [Google Scholar] [CrossRef]
- Okolie, C.E.; Cockayne, A.; Penfold, C.; James, R. Engineering of the LukS-PV and LukF-PV subunits of Staphylococcus aureus Panton-Valentine leukocidin for Diagnostic and Therapeutic Applications. BMC Biotechnol. 2013, 13, 103. [Google Scholar] [CrossRef]
- Humphrey, S.; Fillol-Salom, A.; Quiles-Puchalt, N.; Ibarra-Chávez, R.; Haag, A.F.; Chen, J.; Penadés, J.R. Bacterial chromosomal mobility via lateral transduction exceeds that of classical mobile genetic elements. Nat. Commun. 2021, 12, 6509. [Google Scholar] [CrossRef] [PubMed]
- Cervera-Alamar, M.; Guzmán-Markevitch, K.; Žiemytė, M.; Ortí, L.; Bernabé-Quispe, P.; Pineda-Lucena, A.; Pemán, J.; Tormo-Mas, M.Á. Mobilisation Mechanism of Pathogenicity Islands by Endogenous Phages in Staphylococcus aureus clinical strains. Sci. Rep. 2018, 8, 16742. [Google Scholar] [CrossRef] [PubMed]
- Boakes, E.; Kearns, A.M.; Ganner, M.; Perry, C.; Hill, R.L.; Ellington, M.J. Distinct Bacteriophages Encoding Panton-Valentine Leukocidin (PVL) among International Methicillin-Resistant Staphylococcus aureus Clones Harboring PVL. J. Clin. Microbiol. 2011, 49, 684–692. [Google Scholar] [CrossRef] [PubMed]
- Arunachalam, K.; Pandurangan, P.; Shi, C.; Lagoa, R. Regulation of Staphylococcus aureus Virulence and Application of Nanotherapeutics to Eradicate S. aureus Infection. Pharmaceutics 2023, 15, 310. [Google Scholar] [CrossRef]
- Melles, D.C.; van Leeuwen, W.B.; Boelens, H.A.M.; Peeters, J.K.; Verbrugh, H.A.; van Belkum, A. Panton-Valentine leukocidin genes in Staphylococcus aureus. Emerg. Infect. Dis. 2006, 12, 1174–1175. [Google Scholar] [CrossRef]
- Mccarthy, A.J.; Witney, A.A.; Lindsay, J.A. Staphylococcus aureus Temperate Bacteriophage: Carriage and Horizontal Gene Transfer is Lineage Associated. Front. Cell. Infect. Microbiol. 2012, 2, 6. [Google Scholar] [CrossRef]
- Mohamad Farook, N.A.; Argimón, S.; Abdul Samat, M.N.; Salleh, S.A.; Sulaiman, S.; Tan, T.L.; Periyasamy, P.; Lau, C.L.; Ismail, Z.; Muhammad Azami, N.A.; et al. Diversity and Dissemination of Methicillin-Resistant Staphylococcus aureus (MRSA) Genotypes in Southeast Asia. Trop. Med. Infect. Dis. 2022, 7, 438. [Google Scholar] [CrossRef]
- Coombs, G.W.; Baines, S.L.; Howden, B.P.; Swenson, K.M.; O’Brien, F.G. Diversity of bacteriophages encoding Panton-Valentine leukocidin in temporally and geographically related Staphylococcus aureus. PLoS ONE 2020, 15, e0228676. [Google Scholar] [CrossRef]
- Arumugam, P.; Kielian, T. Metabolism Shapes Immune Responses to Staphylococcus aureus. J. Innate Immun. 2023, 16, 12. [Google Scholar] [CrossRef]
- van Dijk, M.C.; de Kruijff, R.M.; Hagedoorn, P.-L. The Role of Iron in Staphylococcus aureus Infection and Human Disease: A Metal Tug of War at the Host—Microbe Interface. Front. Cell Dev. Biol. 2022, 10, 857237. [Google Scholar] [CrossRef]
- Carlson, S.K.; Erickson, D.L.; Wilson, E. Staphylococcus aureus metal acquisition in the mastitic mammary gland. Microb. Pathog. 2020, 144, 104179. [Google Scholar] [CrossRef] [PubMed]
- Ghssein, G.; Ezzeddine, Z. The Key Element Role of Metallophores in the Pathogenicity and Virulence of Staphylococcus aureus: A Review. Biology 2022, 11, 1525. [Google Scholar] [CrossRef] [PubMed]
- Marchetti, M.; De Bei, O.; Bettati, S.; Campanini, B.; Kovachka, S.; Gianquinto, E.; Spyrakis, F.; Ronda, L. Iron Metabolism at the Interface between Host and Pathogen: From Nutritional Immunity to Antibacterial Development. Int. J. Mol. Sci. 2020, 21, 2145. [Google Scholar] [CrossRef] [PubMed]
- Pietrocola, G.; Pellegrini, A.; Alfeo, M.J.; Marchese, L.; Foster, T.J.; Speziale, P. The iron-regulated surface determinant B (IsdB) protein from Staphylococcus aureus acts as a receptor for the host protein vitronectin. J. Biol. Chem. 2020, 295, 10008–10022. [Google Scholar] [CrossRef]
- Balasubramanian, D.; Harper, L.; Shopsin, B.; Torres, V.J. Staphylococcus aureus pathogenesis in diverse host environments. Pathog. Dis. 2017, 75, ftx005. [Google Scholar] [CrossRef]
- Potter, A.D.; Butrico, C.E.; Ford, C.A.; Curry, J.M.; Trenary, I.A.; Tummarakota, S.S.; Hendrix, A.S.; Young, J.D.; Cassat, J.E. Host nutrient milieu drives an essential role for aspartate biosynthesis during invasive Staphylococcus aureus infection. Proc. Natl. Acad. Sci. USA 2020, 117, 12394–12401. [Google Scholar] [CrossRef]
- Zeden, M.S.; Gallagher, L.A.; Bueno, E.; Nolan, A.C.; Ahn, J.; Shinde, D.; Razvi, F.; Sladek, M.; Burke, Ó.; O’Neill, E.; et al. Metabolic reprogramming and flux to cell envelope precursors in a pentose phosphate pathway mutant increases MRSA resistance to β-lactam antibiotics. bioRxiv 2023, 2023.03.03.530734. [Google Scholar]
- Eichelberger, K.R.; Cassat, J.E. Metabolic Adaptations During Staphylococcus aureus and Candida albicans Co-Infection. Front. Immunol. 2021, 12, 797550. [Google Scholar] [CrossRef]
- Wu, K.; Conly, J.; McClure, J.-A.; Kurwa, H.A.; Zhang, K. Arginine Catabolic Mobile Element in Evolution and Pathogenicity of the Community-Associated Methicillin-Resistant Staphylococcus aureus Strain USA300. Microorganisms 2020, 8, 275. [Google Scholar] [CrossRef]
- Bianco, C.M.; Moustafa, A.M.; O’Brien, K.; Martin, M.A.; Read, T.D.; Kreiswirth, B.N.; Planet, P.J. Pre-epidemic evolution of the MRSA USA300 clade and a molecular key for classification. Front. Cell. Infect. Microbiol. 2023, 13, 1081070. [Google Scholar] [CrossRef]
- Shokrollahi, P.; Hasani, A.; Aghazadeh, M.; Memar, M.Y.; Hasani, A.; Zaree, M.; Rezaee, M.A.; Sadeghi, J. Contribution of Arginine Catabolic Mobile Element and Copper and Mercury Resistance Element in Methicillin-Resistant Staphylococcus aureus: A Vantage Point. Can. J. Infect. Dis. Med. Microbiol. 2022, 2022, 9916255. [Google Scholar] [CrossRef] [PubMed]
- Giulieri, S.G.; Guérillot, R.; Duchene, S.; Hachani, A.; Daniel, D.; Seemann, T.; Davis, J.S.; Tong, S.Y.; Young, B.C.; Wilson, D.J.; et al. Niche-specific genome degradation and convergent evolution shaping Staphylococcus aureus adaptation during severe infections. eLife 2022, 11, e77195. [Google Scholar] [CrossRef] [PubMed]
- Langhanki, L.; Berger, P.; Treffon, J.; Catania, F.; Kahl, B.C.; Mellmann, A. In vivo competition and horizontal gene transfer among distinct Staphylococcus aureus lineages as major drivers for adaptational changes during long-term persistence in humans. BMC Microbiol. 2018, 18, 152. [Google Scholar] [CrossRef] [PubMed]
- Tsimbalyuk, S.; Shornikov, A.; Srivastava, P.; Le, V.T.B.; Warren, I.; Khandokar, Y.B.; Kuhn, M.L.; Forwood, J.K. Structural and Kinetic Characterization of the SpeG Spermidine/Spermine N-acetyltransferase from Methicillin-Resistant Staphylococcus aureus USA300. Cells 2023, 12, 1829. [Google Scholar] [CrossRef]
- Nassar, R.; Hachim, M.; Nassar, M.; Kaklamanos, E.G.; Jamal, M.; Williams, D.; Senok, A. Microbial Metabolic Genes Crucial for S. aureus Biofilms: An Insight From Re-analysis of Publicly Available Microarray Datasets. Front. Microbiol. 2021, 11, 607002. [Google Scholar] [CrossRef]
- Shibamura-Fujiogi, M.; Wang, X.; Maisat, W.; Koutsogiannaki, S.; Li, Y.; Chen, Y.; Lee, J.C.; Yuki, K. GltS regulates biofilm formation in methicillin-resistant Staphylococcus aureus. Commun. Biol. 2022, 5, 1284. [Google Scholar] [CrossRef]
- Gray, B.; Hall, P.; Gresham, H. Targeting agr- and agr-Like Quorum Sensing Systems for Development of Common Therapeutics to Treat Multiple Gram-Positive Bacterial Infections. Sensors 2013, 13, 5130. [Google Scholar] [CrossRef]
- Shen, Y.; Gao, S.; Fan, Q.; Zuo, J.; Wang, Y.; Yi, L.; Wang, Y. New antibacterial targets: Regulation of quorum sensing and secretory systems in zoonotic bacteria. Microbiol. Res. 2023, 274, 127436. [Google Scholar] [CrossRef]
- Williams, P.; Hill, P.; Bonev, B.; Chan, W.C. Quorum-sensing, intra- and inter-species competition in the staphylococci. Microbiology 2023, 169, 001381. [Google Scholar] [CrossRef]
- Xu, T.; Wang, X.-Y.; Cui, P.; Zhang, Y.-M.; Zhang, W.-H.; Zhang, Y. The Agr Quorum Sensing System Represses Persister Formation through Regulation of Phenol Soluble Modulins in Staphylococcus aureus. Front. Microbiol. 2017, 8, 2189. [Google Scholar] [CrossRef]
- Kaushik, A.; Kest, H.; Sood, M.; Steussy, B.W.; Thieman, C.; Gupta, S. Biofilm Producing Methicillin-Resistant Staphylococcus aureus (MRSA) Infections in Humans: Clinical Implications and Management. Pathogens 2024, 13, 76. [Google Scholar] [CrossRef] [PubMed]
- McCarthy, H.; Rudkin, J.K.; Black, N.S.; Gallagher, L.; O’Neill, E.; O’Gara, J.P. Methicillin resistance and the biofilm phenotype in Staphylococcus aureus. Front. Cell. Infect. Microbiol. 2015, 5, 1. [Google Scholar] [CrossRef] [PubMed]
- Fang, L.; Cosgriff, C.; Alonzo, F. Determinants of maturation of the Staphylococcus aureus autoinducing peptide. J. Bacteriol. 2024, 206, e00195-24. [Google Scholar] [CrossRef] [PubMed]
- Novick, R.P.; Muir, T.W. Virulence gene regulation by peptides in staphylococci and other Gram-positive bacteria. Curr. Opin. Microbiol. 1999, 2, 40–45. [Google Scholar] [CrossRef]
- Vasquez, J.K.; Blackwell, H.E. Simplified Autoinducing Peptide Mimetics with Single-Nanomolar Activity Against the Staphylococcus aureus AgrC Quorum Sensing Receptor. ACS Infect. Dis. 2019, 5, 484–492. [Google Scholar] [CrossRef]
- Kim, M.K.; Zhao, A.; Wang, A.; Brown, Z.Z.; Muir, T.W.; Stone, H.A.; Bassler, B.L. Surface-Attached Molecules Control Staphylococcus aureus Quorum Sensing and Biofilm Development. Nat. Microbiol. 2017, 2, 17080. [Google Scholar] [CrossRef]
- Oliveira, D.; Borges, A.; Ruiz, R.M.; Negrín, Z.R.; Distinto, S.; Borges, F.; Simões, M. 2-(2-Methyl-2-nitrovinyl)furan but Not Furvina Interfere with Staphylococcus aureus Agr Quorum-Sensing System and Potentiate the Action of Fusidic Acid against Biofilms. Int. J. Mol. Sci. 2021, 22, 613. [Google Scholar] [CrossRef]
- Pietrocola, G.; Campoccia, D.; Motta, C.; Montanaro, L.; Arciola, C.R.; Speziale, P. Colonization and Infection of Indwelling Medical Devices by Staphylococcus aureus with an Emphasis on Orthopedic Implants. Int. J. Mol. Sci. 2022, 23, 5958. [Google Scholar] [CrossRef]
- Yamazaki, Y.; Ito, T.; Tamai, M.; Nakagawa, S.; Nakamura, Y. The role of Staphylococcus aureus quorum sensing in cutaneous and systemic infections. Inflamm. Regen. 2024, 44, 9. [Google Scholar] [CrossRef]
- Snowden, J.N.; Beaver, M.; Beenken, K.; Smeltzer, M.; Horswill, A.R.; Kielian, T. Staphylococcus aureus sarA Regulates Inflammation and Colonization during Central Nervous System Biofilm Formation. PLoS ONE 2013, 8, e84089. [Google Scholar] [CrossRef]
- Manna, A.C.; Ingavale, S.S.; Maloney, M.; van Wamel, W.; Cheung, A.L. Identification of sarV (SA2062), a New Transcriptional Regulator, Is Repressed by SarA and MgrA (SA0641) and Involved in the Regulation of Autolysis in Staphylococcus aureus. J. Bacteriol. 2004, 186, 5267–5280. [Google Scholar] [CrossRef] [PubMed]
- Trotonda, M.P.; Manna, A.C.; Cheung, A.L.; Lasa, I.; Penadés, J.R. SarA Positively Controls Bap-Dependent Biofilm Formation in Staphylococcus aureus. J. Bacteriol. 2005, 187, 5790–5798. [Google Scholar] [CrossRef] [PubMed]
- Balamurugan, P.; Praveen Krishna, V.; Bharath, D.; Lavanya, R.; Vairaprakash, P.; Adline Princy, S. Staphylococcus aureus Quorum Regulator SarA Targeted Compound, 2-[(Methylamino)methyl]phenol Inhibits Biofilm and Down-Regulates Virulence Genes. Front. Microbiol. 2017, 8, 1290. [Google Scholar] [CrossRef] [PubMed]
- Fujimoto, D.F.; Higginbotham, R.H.; Maleki, S.J.; Segall1, A.M.; Smeltzer, M.S.; Hurlburt, B.K. Staphylococcus aureus SarA is a Regulatory Protein Responsive to Redox and pH that can Support Bacteriophage Lambda Integrase-Mediated Excision/Recombination. Mol. Microbiol. 2009, 74, 1445–1458. [Google Scholar] [CrossRef]
- Tamber, S.; Cheung, A.L. SarZ promotes the expression of virulence factors and represses biofilm formation by modulating SarA and agr in Staphylococcus aureus. Infect. Immun. 2009, 77, 419–428. [Google Scholar] [CrossRef]
- Wang, M.; Buist, G.; van Dijl, J.M. Staphylococcus aureus cell wall maintenance—The multifaceted roles of peptidoglycan hydrolases in bacterial growth, fitness, and virulence. FEMS Microbiol. Rev. 2022, 46, fuac025. [Google Scholar] [CrossRef]
- Chan, P.F.; Foster, S.J. Role of SarA in Virulence Determinant Production and Environmental Signal Transduction in Staphylococcus aureus. J. Bacteriol. 1998, 180, 6232–6241. [Google Scholar] [CrossRef]
- Chien, Y.; Manna, A.C.; Projan, S.J.; Cheung, A.L. SarA, a global regulator of virulence determinants in Staphylococcus aureus, binds to a conserved motif essential for sar-dependent gene regulation. J. Biol. Chem. 1999, 274, 37169–37176. [Google Scholar] [CrossRef]
- Pratten, J.; Foster, S.J.; Chan, P.F.; Wilson, M.; Nair, S.P. Staphylococcus aureus accessory regulators: Expression within biofilms and effect on adhesion. Microbes Infect. 2001, 3, 633–637. [Google Scholar] [CrossRef]
- Oriol, C.; Cengher, L.; Manna, A.C.; Mauro, T.; Pinel-Marie, M.-L.; Felden, B.; Cheung, A.; Rouillon, A. Expanding the Staphylococcus aureus SarA Regulon to Small RNAs. mSystems 2021, 6, e00713-21. [Google Scholar] [CrossRef]
- Beenken, K.E.; Campbell, M.J.; Smeltzer, M.S. The ability of sarA to limit protease production plays a key role in the pathogenesis of Staphylococcus aureus osteomyelitis irrespective of the functional status of agr. Infect. Immun. 2025, 93, e0047324. [Google Scholar] [CrossRef] [PubMed]
- Bashabsheh, R.H.F.; AL-Fawares, O.; Natsheh, I.; Bdeir, R.; Al-Khreshieh, R.O.; Bashabsheh, H.H.F. Staphylococcus aureus epidemiology, pathophysiology, clinical manifestations and application of nano-therapeutics as a promising approach to combat methicillin resistant Staphylococcus aureus. Pathog. Glob. Health 2023, 118, 209–231. [Google Scholar] [CrossRef] [PubMed]
- Lade, H.; Kim, J.-S. Bacterial Targets of Antibiotics in Methicillin-Resistant Staphylococcus aureus. Antibiotics 2021, 10, 398. [Google Scholar] [CrossRef] [PubMed]
- Pulithara Sebastian, A.S.; Vaiyapuri, M.; Badireddy, M.R. Vancomycin and Methicillin Resistance in Staphylococcus aureus: What Is the Next? In Handbook on Antimicrobial Resistance: Current Status, Trends in Detection and Mitigation Measures; Mothadaka, M.P., Vaiyapuri, M., Rao Badireddy, M., Nagarajrao Ravishankar, C., Bhatia, R., Jena, J., Eds.; Springer Nature: Singapore, 2023; pp. 1–19. ISBN 978-981-16-9723-4. [Google Scholar] [CrossRef]
- Brdová, D.; Ruml, T.; Viktorová, J. Mechanism of staphylococcal resistance to clinically relevant antibiotics. Drug Resist. Updat. 2024, 77, 101147. [Google Scholar] [CrossRef]
- Chambers, H.F.; DeLeo, F.R. Waves of Resistance: Staphylococcus aureus in the Antibiotic Era. Nat. Rev. Microbiol. 2009, 7, 629–641. [Google Scholar] [CrossRef]
- Lowy, F.D. Antimicrobial resistance: The example of Staphylococcus aureus. J. Clin. Investig. 2003, 111, 1265–1273. [Google Scholar] [CrossRef]
- Schito, G.C. The importance of the development of antibiotic resistance in Staphylococcus aureus. Clin. Microbiol. Infect. 2006, 12, 3–8. [Google Scholar] [CrossRef]
- Gaynes, R. The Discovery of Penicillin—New Insights After More Than 75 Years of Clinical Use. Emerg. Infect. Dis. 2017, 23, 849–853. [Google Scholar] [CrossRef]
- Hutchings, M.I.; Truman, A.W.; Wilkinson, B. Antibiotics: Past, present and future. Curr. Opin. Microbiol. 2019, 51, 72–80. [Google Scholar] [CrossRef]
- Lobanovska, M.; Pilla, G. Penicillin’s Discovery and Antibiotic Resistance: Lessons for the Future? Yale J. Biol. Med. 2017, 90, 135–145. [Google Scholar]
- Selvarajan, R.; Obize, C.; Sibanda, T.; Abia, A.L.K.; Long, H. Evolution and Emergence of Antibiotic Resistance in Given Ecosystems: Possible Strategies for Addressing the Challenge of Antibiotic Resistance. Antibiotics 2023, 12, 28. [Google Scholar] [CrossRef] [PubMed]
- Landecker, H. Antibiotic Resistance and the Biology of History. Body Soc. 2016, 22, 19–52. [Google Scholar] [CrossRef] [PubMed]
- Foster, T.J. Antibiotic resistance in Staphylococcus aureus. Current status and future prospects. FEMS Microbiol. Rev. 2017, 41, 430–449. [Google Scholar] [CrossRef] [PubMed]
- Kirby, W.M. Extraction of a Highly Potent Penicillin Inactivator from Penicillin Resistant Staphylococci. Science 1944, 99, 452–453. [Google Scholar] [CrossRef]
- Kirby, W.M.M. Properties of a Penicillin Inactivator Extracted from Penicillin-Resistant Staphylococci 1. J. Clin. Investig. 1945, 24, 170–174. [Google Scholar] [CrossRef]
- Smith, T.L.; Jarvis, W.R. Antimicrobial resistance in Staphylococcus aureus. Microbes Infect. 1999, 1, 795–805. [Google Scholar] [CrossRef]
- Abraham, E.P.; Chain, E. An enzyme from bacteria able to destroy penicillin. 1940. Rev. Infect. Dis. 1988, 10, 677–678. [Google Scholar]
- Ferraz, M.P. Antimicrobial Resistance: The Impact from and on Society According to One Health Approach. Societies 2024, 14, 187. [Google Scholar] [CrossRef]
- Muteeb, G.; Rehman, M.T.; Shahwan, M.; Aatif, M. Origin of Antibiotics and Antibiotic Resistance, and Their Impacts on Drug Development: A Narrative Review. Pharmaceuticals 2023, 16, 1615. [Google Scholar] [CrossRef]
- Oliveira, M.; Antunes, W.; Mota, S.; Madureira-Carvalho, Á.; Dinis-Oliveira, R.J.; Dias da Silva, D. An Overview of the Recent Advances in Antimicrobial Resistance. Microorganisms 2024, 12, 1920. [Google Scholar] [CrossRef]
- Jian, Z.; Zeng, L.; Xu, T.; Sun, S.; Yan, S.; Yang, L.; Huang, Y.; Jia, J.; Dou, T. Antibiotic resistance genes in bacteria: Occurrence, spread, and control. J. Basic Microbiol. 2021, 61, 1049–1070. [Google Scholar] [CrossRef] [PubMed]
- Maeda, T.; Furusawa, C. Laboratory Evolution of Antimicrobial Resistance in Bacteria to Develop Rational Treatment Strategies. Antibiotics 2024, 13, 94. [Google Scholar] [CrossRef] [PubMed]
- Salam, M.A.; Al-Amin, M.Y.; Salam, M.T.; Pawar, J.S.; Akhter, N.; Rabaan, A.A.; Alqumber, M.A.A. Antimicrobial Resistance: A Growing Serious Threat for Global Public Health. Healthcare 2023, 11, 1946. [Google Scholar] [CrossRef] [PubMed]
- Ali Alghamdi, B.; Al-Johani, I.; Al-Shamrani, J.M.; Musamed Alshamrani, H.; Al-Otaibi, B.G.; Almazmomi, K.; Yusnoraini Yusof, N. Antimicrobial resistance in methicillin-resistant Staphylococcus aureus. Saudi J. Biol. Sci. 2023, 30, 103604. [Google Scholar] [CrossRef]
- Morell, E.A.; Balkin, D.M. Methicillin-Resistant Staphylococcus aureus: A Pervasive Pathogen Highlights the Need for New Antimicrobial Development. Yale J. Biol. Med. 2010, 83, 223–233. [Google Scholar]
- Alanzi, T.K.; Alhazmi, O.A.; Alanezi, K.; Alammari, W.M.; Alrwily, A.A.; Alshammari, M.M.; Albuhairan, R. Prevalence of Methicillin-Resistant Staphylococcus aureus in Saudi Arabia: A Systematic Review and Meta-Analysis. Cureus 2024, 16, e70230. [Google Scholar] [CrossRef]
- Hou, Z.; Liu, L.; Wei, J.; Xu, B. Progress in the Prevalence, Classification and Drug Resistance Mechanisms of Methicillin-Resistant Staphylococcus aureus. Infect. Drug Resist. 2023, 16, 3271–3292. [Google Scholar] [CrossRef]
- Marciniak, K.; Tyczewska, A.; Grzywacz, K. Genetics of antibiotic resistance in methicillin-resistant Staphylococcus aureus (MRSA). Biotechnologia 2024, 105, 169–177. [Google Scholar] [CrossRef]
- Abebe, A.A.; Birhanu, A.G. Methicillin Resistant Staphylococcus aureus: Molecular Mechanisms Underlying Drug Resistance Development and Novel Strategies to Combat. Infect. Drug Resist. 2023, 16, 7641–7662. [Google Scholar] [CrossRef]
- García-Castellanos, R.; Mallorquí-Fernández, G.; Marrero, A.; Potempa, J.; Coll, M.; Gomis-Rüth, F.X. On the Transcriptional Regulation of Methicillin Resistance: MecI repressor in complex with its operator. J. Biol. Chem. 2004, 279, 17888–17896. [Google Scholar] [CrossRef]
- Petinaki, E.; Arvaniti, A.; Dimitracopoulos, G.; Spiliopoulou, I. Detection of mecA, mecR1 and mecI genes among clinical isolates of methicillin-resistant staphylococci by combined polymerase chain reactions. J. Antimicrob. Chemother. 2001, 47, 297–304. [Google Scholar] [CrossRef] [PubMed]
- Karaman, R.; Jubeh, B.; Breijyeh, Z. Resistance of Gram-Positive Bacteria to Current Antibacterial Agents and Overcoming Approaches. Mol. Basel Switz. 2020, 25, 2888. [Google Scholar]
- Douglas, E.J.A.; Laabei, M. Staph wars: The antibiotic pipeline strikes back. Microbiology 2023, 169, 001387. [Google Scholar] [CrossRef] [PubMed]
- Appelbaum, P.C. The emergence of vancomycin-intermediate and vancomycin-resistant Staphylococcus aureus. Clin. Microbiol. Infect. 2006, 12, 16–23. [Google Scholar] [CrossRef]
- Gardete, S.; Tomasz, A. Mechanisms of vancomycin resistance in Staphylococcus aureus. J. Clin. Invest. 2014, 124, 2836–2840. [Google Scholar] [CrossRef]
- Aminzadeh, Z.; Yadegarynia, D.; Fatemi, A.; Tahmasebian Dehkordi, E.; Azad Armaki, S. Vancomycin Minimum Inhibitory Concentration for Methicillin-Resistant Staphylococcus aureus Infections; Is There Difference in Mortality Between Patients? Jundishapur J. Microbiol. 2014, 7, e12831. [Google Scholar] [CrossRef]
- Cong, Y.; Yang, S.; Rao, X. Vancomycin resistant Staphylococcus aureus infections: A review of case updating and clinical features. J. Adv. Res. 2019, 21, 169–176. [Google Scholar] [CrossRef]
- Haas, W.; Singh, N.; Lainhart, W.; Mingle, L.; Nazarian, E.; Mitchell, K.; Nattanmai, G.; Kohlerschmidt, D.; Dickinson, M.C.; Kacica, M.; et al. Genomic Analysis of Vancomycin-Resistant Staphylococcus aureus Isolates from the 3rd Case Identified in the United States Reveals Chromosomal Integration of the vanA Locus. Microbiol. Spectr. 2023, 11, e04317-22. [Google Scholar] [CrossRef]
- Kumar, P. 33—Pharmacology of Specific Drug Groups: Antibiotic Therapy∗∗The author wishes to recognize Dr. Thomas J. Pallasch for his past contributions to this chapter. In Pharmacology and Therapeutics for Dentistry (Seventh Edition); Dowd, F.J., Johnson, B.S., Mariotti, A.J., Eds.; Mosby: Maryland Heights, MI, USA, 2017; pp. 457–487. ISBN 978-0-323-39307-2. Available online: https://www.sciencedirect.com/science/article/pii/B9780323393072000333 (accessed on 30 March 2025).
- van der Aart, L.T.; Lemmens, N.; van Wamel, W.J.; van Wezel, G.P. Substrate Inhibition of VanA by d-Alanine Reduces Vancomycin Resistance in a VanX-Dependent Manner. Antimicrob. Agents Chemother. 2016, 60, 4930–4939. [Google Scholar] [CrossRef]
- Azimian, A.; Havaei, S.A.; Fazeli, H.; Naderi, M.; Ghazvini, K.; Samiee, S.M.; Soleimani, M.; Peerayeh, S.N. Genetic Characterization of a Vancomycin-Resistant Staphylococcus aureus Isolate from the Respiratory Tract of a Patient in a University Hospital in Northeastern Iran. J. Clin. Microbiol. 2012, 50, 3581–3585. [Google Scholar] [CrossRef]
- Zhu, W.; Clark, N.C.; McDougal, L.K.; Hageman, J.; McDonald, L.C.; Patel, J.B. Vancomycin-Resistant Staphylococcus aureus Isolates Associated with Inc18-Like vanA Plasmids in Michigan. Antimicrob. Agents Chemother. 2008, 52, 452–457. [Google Scholar] [CrossRef] [PubMed]
- Nelson, J.L.; Rice, K.C.; Slater, S.R.; Fox, P.M.; Archer, G.L.; Bayles, K.W.; Fey, P.D.; Kreiswirth, B.N.; Somerville, G.A. Vancomycin-Intermediate Staphylococcus aureus Strains Have Impaired Acetate Catabolism: Implications for Polysaccharide Intercellular Adhesin Synthesis and Autolysis. Antimicrob. Agents Chemother. 2007, 51, 616–622. [Google Scholar] [CrossRef] [PubMed]
- Cui, L.; Ma, X.; Sato, K.; Okuma, K.; Tenover, F.; Mamizuka, E.; Gemmell, C.; Kim, M.-N.; Ploy, M.-C.; Solh, N.; et al. Cell Wall Thickening Is a Common Feature of Vancomycin Resistance in Staphylococcus aureus. J. Clin. Microbiol. 2003, 41, 5–14. [Google Scholar] [CrossRef] [PubMed]
- Howden, B.P.; Davies, J.K.; Johnson, P.D.R.; Stinear, T.P.; Grayson, M.L. Reduced Vancomycin Susceptibility in Staphylococcus aureus, Including Vancomycin-Intermediate and Heterogeneous Vancomycin-Intermediate Strains: Resistance Mechanisms, Laboratory Detection, and Clinical Implications. Clin. Microbiol. Rev. 2010, 23, 99–139. [Google Scholar] [CrossRef]
- Howden, B.P.; Johnson, P.D.R.; Ward, P.B.; Stinear, T.P.; Davies, J.K. Isolates with low-level vancomycin resistance associated with persistent methicillin-resistant Staphylococcus aureus bacteremia. Antimicrob. Agents Chemother. 2006, 50, 3039–3047. [Google Scholar] [CrossRef]
- Hu, Q.; Peng, H.; Rao, X. Molecular Events for Promotion of Vancomycin Resistance in Vancomycin Intermediate Staphylococcus aureus. Front. Microbiol. 2016, 7, 1601. [Google Scholar] [CrossRef]
- Alexander, E.L.; Gardete, S.; Bar, H.Y.; Wells, M.T.; Tomasz, A.; Rhee, K.Y. Intermediate-type vancomycin resistance (VISA) in genetically-distinct Staphylococcus aureus isolates is linked to specific, reversible metabolic alterations. PloS One 2014, 9, e97137. [Google Scholar] [CrossRef]
- Gardete, S.; Kim, C.; Hartmann, B.M.; Mwangi, M.; Roux, C.M.; Dunman, P.M.; Chambers, H.F.; Tomasz, A. Genetic Pathway in Acquisition and Loss of Vancomycin Resistance in a Methicillin Resistant Staphylococcus aureus (MRSA) Strain of Clonal Type USA300. PLOS Pathog. 2012, 8, e1002505. [Google Scholar] [CrossRef]
- Howden, B.P.; Peleg, A.Y.; Stinear, T.P. The evolution of vancomycin intermediate Staphylococcus aureus (VISA) and heterogenous-VISA. Infect. Genet. Evol. J. Mol. Epidemiol. Evol. Genet. Infect. Dis. 2014, 21, 575–582. [Google Scholar] [CrossRef]
- Belete, M.A.; Gedefie, A.; Alemayehu, E.; Debash, H.; Mohammed, O.; Gebretsadik, D.; Ebrahim, H.; Tilahun, M. The prevalence of vancomycin-resistant Staphylococcus aureus in Ethiopia: A systematic review and meta-analysis. Antimicrob. Resist. Infect. Control 2023, 12, 86. [Google Scholar] [CrossRef]
- Gajic, I.; Tomic, N.; Lukovic, B.; Jovicevic, M.; Kekic, D.; Petrovic, M.; Jankovic, M.; Trudic, A.; Mitic Culafic, D.; Milenkovic, M.; et al. A Comprehensive Overview of Antibacterial Agents for Combating Multidrug-Resistant Bacteria: The Current Landscape, Development, Future Opportunities, and Challenges. Antibiotics 2025, 14, 221. [Google Scholar] [CrossRef] [PubMed]
- Fathi, J.; Hashemizadeh, Z.; Dehkordi, R.S.; Bazargani, A.; Javadi, K.; Hosseini-Nave, H.; Hadadi, M. Evaluation of aminoglycoside modifying enzymes, SCCmec, coagulase gene and PCR-RFLP coagulase gene typing of Staphylococcus aureus isolates from hospitals in Shiraz, southwest of Iran. Heliyon 2022, 8, e10230. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, N.; Wang, M.; Luo, M.; Peng, Y.; Li, Z.; Xu, J.; Ou, M.; Kan, B.; Li, X.; et al. The prevalence and distribution of aminoglycoside resistance genes. Biosaf. Health 2023, 5, 14–20. [Google Scholar] [CrossRef]
- Naderi, M.; Yousefi Nojookambari, N.; Talebi, S.; Mohammadi, M.R.; Yazdansetad, S. Molecular Characterization of Aminoglycoside-modifying Enzymes (AMEs)in Aminoglycoside-Resistant Staphylococcus aureus: A Cross-sectional Study in Northeastern Iran. Iran. J. Pathol. 2025, 20, 118–125. [Google Scholar] [CrossRef] [PubMed]
- Shokravi, Z.; Mehrad, L.; Ramazani, A. Detecting the frequency of aminoglycoside modifying enzyme encoding genes among clinical isolates of methicillin-resistant Staphylococcus aureus. BioImpacts BI 2015, 5, 87–91. [Google Scholar] [CrossRef][Green Version]
- Yadegar, A.; Sattari, M.; Mozafari, N.A.; Goudarzi, G.R. Prevalence of the genes encoding aminoglycoside-modifying enzymes and methicillin resistance among clinical isolates of Staphylococcus aureus in Tehran, Iran. Microb. Drug Resist. Larchmt. N 2009, 15, 109–113. [Google Scholar] [CrossRef]
- Garneau-Tsodikova, S.; Labby, K.J. Mechanisms of Resistance to Aminoglycoside Antibiotics: Overview and Perspectives. MedChemComm 2015, 7, 11. [Google Scholar] [CrossRef]
- Ramirez, M.S.; Tolmasky, M.E. Aminoglycoside Modifying Enzymes. Drug Resist. Updat. Rev. Comment. Antimicrob. Anticancer Chemother. 2010, 13, 151–171. [Google Scholar] [CrossRef]
- Smith, C.A.; Bhattacharya, M.; Toth, M.; Stewart, N.K.; Vakulenko, S.B. Aminoglycoside resistance profile and structural architecture of the aminoglycoside acetyltransferase AAC(6’)-Im. Microb. Cell 2017, 4, 402–410. [Google Scholar] [CrossRef]
- Elshobary, M.E.; Badawy, N.K.; Ashraf, Y.; Zatioun, A.A.; Masriya, H.H.; Ammar, M.M.; Mohamed, N.A.; Mourad, S.; Assy, A.M. Combating Antibiotic Resistance: Mechanisms, Multidrug-Resistant Pathogens, and Novel Therapeutic Approaches: An Updated Review. Pharmaceuticals 2025, 18, 402. [Google Scholar] [CrossRef]
- Shayista, H.; Prasad, M.N.N.; Raj, S.N.; Prasad, A.; Lakshmi, S.; Ranjini, H.K.; Manju, K.; Ravikumara; Chouhan, R.S.; Khohlova, O.Y.; et al. Complexity of antibiotic resistance and its impact on gut microbiota dynamics. Eng. Microbiol. 2025, 5, 100187. [Google Scholar] [CrossRef]
- Dafale, N.A.; Srivastava, S.; Purohit, H.J. Zoonosis: An Emerging Link to Antibiotic Resistance Under “One Health Approach”. Indian J. Microbiol. 2020, 60, 139–152. [Google Scholar] [CrossRef] [PubMed]
- Krause, K.M.; Serio, A.W.; Kane, T.R.; Connolly, L.E. Aminoglycosides: An Overview. Cold Spring Harb. Perspect. Med. 2016, 6, a027029. [Google Scholar] [CrossRef] [PubMed]
- Tang, K.W.K.; Millar, B.C.; Moore, J.E. Antimicrobial Resistance (AMR). Br. J. Biomed. Sci. 2023, 80, 11387. [Google Scholar] [CrossRef]
- Zhang, T.; Nickerson, R.; Zhang, W.; Peng, X.; Shang, Y.; Zhou, Y.; Luo, Q.; Wen, G.; Cheng, Z. The impacts of animal agriculture on One Health—Bacterial zoonosis, antimicrobial resistance, and beyond. One Health 2024, 18, 100748. [Google Scholar] [CrossRef]
- Vestergaard, M.; Frees, D.; Ingmer, H. Antibiotic Resistance and the MRSA Problem. Microbiol. Spectr. 2019, 7. [Google Scholar] [CrossRef]
- Geremia, N.; Giovagnorio, F.; Colpani, A.; De Vito, A.; Botan, A.; Stroffolini, G.; Toc, D.-A.; Zerbato, V.; Principe, L.; Madeddu, G.; et al. Fluoroquinolones and Biofilm: A Narrative Review. Pharmaceuticals 2024, 17, 1673. [Google Scholar] [CrossRef]
- Akgün, K.B.; Ceylan, C. Trend of Use of Quinolone Antibiotics in Community-Acquired Pneumonia. Available online: https://mjima.org/articles/trend-of-use-of-quinolone-antibiotics-in-community-acquired-pneumoni (accessed on 30 March 2025).
- Thompson, D.; Xu, J.; Ischia, J.; Bolton, D. Fluoroquinolone resistance in urinary tract infections: Epidemiology, mechanisms of action and management strategies. BJUI Compass 2023, 5, 5–11. [Google Scholar] [CrossRef]
- Hooper, D.C.; Jacoby, G.A. Topoisomerase Inhibitors: Fluoroquinolone Mechanisms of Action and Resistance. Cold Spring Harb. Perspect. Med. 2016, 6, a025320. [Google Scholar] [CrossRef]
- Fàbrega, A.; Madurga, S.; Giralt, E.; Vila, J. Mechanism of action of and resistance to quinolones. Microb. Biotechnol. 2009, 2, 40–61. [Google Scholar] [CrossRef]
- Fournier, B.; Hooper, D.C. Mutations in Topoisomerase IV and DNA Gyrase of Staphylococcus aureus: Novel Pleiotropic Effects on Quinolone and Coumarin Activity. Antimicrob. Agents Chemother. 1998, 42, 121. [Google Scholar] [CrossRef] [PubMed]
- Dalhoff, A. Global Fluoroquinolone Resistance Epidemiology and Implictions for Clinical Use. Interdiscip. Perspect. Infect. Dis. 2012, 2012, 976273. [Google Scholar] [CrossRef] [PubMed]
- Vittorakis, E.; Vica, M.L.; Zervaki, C.O.; Vittorakis, E.; Maraki, S.; Mavromanolaki, V.E.; Schürger, M.E.; Neculicioiu, V.S.; Papadomanolaki, E.; Junie, L.M. A Comparative Analysis of MRSA: Epidemiology and Antibiotic Resistance in Greece and Romania. Int. J. Mol. Sci. 2024, 25, 7535. [Google Scholar] [CrossRef] [PubMed]
- Alseqely, M.; Newton-Foot, M.; Khalil, A.; El-Nakeeb, M.; Whitelaw, A.; Abouelfetouh, A. Association between fluoroquinolone resistance and MRSA genotype in Alexandria, Egypt. Sci. Rep. 2021, 11, 4253. [Google Scholar] [CrossRef]
- Holden, M.T.G.; Hsu, L.-Y.; Kurt, K.; Weinert, L.A.; Mather, A.E.; Harris, S.R.; Strommenger, B.; Layer, F.; Witte, W.; de Lencastre, H.; et al. A genomic portrait of the emergence, evolution, and global spread of a methicillin-resistant Staphylococcus aureus pandemic. Genome Res. 2013, 23, 653–664. [Google Scholar] [CrossRef]
- Dashtbani-Roozbehani, A.; Brown, M.H. Efflux Pump Mediated Antimicrobial Resistance by Staphylococci in Health-Related Environments: Challenges and the Quest for Inhibition. Antibiotics 2021, 10, 1502. [Google Scholar] [CrossRef]
- Shariati, A.; Arshadi, M.; Khosrojerdi, M.A.; Abedinzadeh, M.; Ganjalishahi, M.; Maleki, A.; Heidary, M.; Khoshnood, S. The resistance mechanisms of bacteria against ciprofloxacin and new approaches for enhancing the efficacy of this antibiotic. Front. Public Health 2022, 10, 1025633. [Google Scholar] [CrossRef]
- Kounatidis, D.; Dalamaga, M.; Grivakou, E.; Karampela, I.; Koufopoulos, P.; Dalopoulos, V.; Adamidis, N.; Mylona, E.; Kaziani, A.; Vallianou, N.G. Third-Generation Tetracyclines: Current Knowledge and Therapeutic Potential. Biomolecules 2024, 14, 783. [Google Scholar] [CrossRef]
- Pearson, J.C.; Gillett, E.; Gadri, N.D.; Dionne, B. Tetracyclines, the old and the new: A narrative review. CMI Commun. 2025, 2, 105059. [Google Scholar] [CrossRef]
- Shutter, M.C.; Akhondi, H. Tetracycline. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2025. Available online: http://www.ncbi.nlm.nih.gov/books/NBK549905/ (accessed on 30 March 2025).
- Grossman, T.H. Tetracycline Antibiotics and Resistance. Cold Spring Harb. Perspect. Med. 2016, 6, a025387. [Google Scholar] [CrossRef]
- Nguyen, F.; Starosta, A.L.; Arenz, S.; Sohmen, D.; Dönhöfer, A.; Wilson, D.N. Tetracycline antibiotics and resistance mechanisms. Biol. Chem. 2014, 395, 559–575. [Google Scholar] [CrossRef] [PubMed]
- Blake, K.S.; Xue, Y.-P.; Gillespie, V.J.; Fishbein, S.R.S.; Tolia, N.H.; Wencewicz, T.A.; Dantas, G. The tetracycline resistome is shaped by selection for specific resistance mechanisms by each antibiotic generation. Nat. Commun. 2025, 16, 1452. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Atkinson, G.C.; Thakor, N.S.; Allas, Ü.; Lu, C.; Chan, K.-Y.; Tenson, T.; Schulten, K.; Wilson, K.S.; Hauryliuk, V.; et al. Mechanism of Tetracycline Resistance by Ribosomal Protection Protein Tet(O). Nat. Commun. 2013, 4, 1477. [Google Scholar] [CrossRef]
- Spahn, C.M.T.; Blaha, G.; Agrawal, R.K.; Penczek, P.; Grassucci, R.A.; Trieber, C.A.; Connell, S.R.; Taylor, D.E.; Nierhaus, K.H.; Frank, J. Localization of the Ribosomal Protection Protein Tet(O) on the Ribosome and the Mechanism of Tetracycline Resistance. Mol. Cell 2001, 7, 1037–1045. [Google Scholar] [CrossRef]
- Bukowski, M.; Piwowarczyk, R.; Madry, A.; Zagorski-Przybylo, R.; Hydzik, M.; Wladyka, B. Prevalence of Antibiotic and Heavy Metal Resistance Determinants and Virulence-Related Genetic Elements in Plasmids of Staphylococcus aureus. Front. Microbiol. 2019, 10, 805. [Google Scholar] [CrossRef] [PubMed]
- Dinos, G.P. The macrolide antibiotic renaissance. Br. J. Pharmacol. 2017, 174, 2967–2983. [Google Scholar] [CrossRef]
- Patel, P.H.; Hashmi, M.F. Macrolides. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2025. [Google Scholar]
- Fyfe, C.; Grossman, T.H.; Kerstein, K.; Sutcliffe, J. Resistance to Macrolide Antibiotics in Public Health Pathogens. Cold Spring Harb. Perspect. Med. 2016, 6, a025395. [Google Scholar] [CrossRef]
- Leclercq, R. Mechanisms of Resistance to Macrolides and Lincosamides: Nature of the Resistance Elements and Their Clinical Implications. Clin. Infect. Dis. 2002, 34, 482–492. [Google Scholar] [CrossRef]
- Mahfouz, A.A.; Said, H.S.; Elfeky, S.M.; Shaaban, M.I. Inhibition of Erythromycin and Erythromycin-Induced Resistance among Staphylococcus aureus Clinical Isolates. Antibiotics 2023, 12, 503. [Google Scholar] [CrossRef]
- Miklasińska-Majdanik, M. Mechanisms of Resistance to Macrolide Antibiotics among Staphylococcus aureus. Antibiotics 2021, 10, 1406. [Google Scholar] [CrossRef]
- Nor Amdan, N.A.; Shahrulzamri, N.A.; Hashim, R.; Mohamad Jamil, N. Understanding the evolution of macrolides resistance: A mini review. J. Glob. Antimicrob. Resist. 2024, 38, 368–375. [Google Scholar] [CrossRef] [PubMed]
- Gaurav, A.; Bakht, P.; Saini, M.; Pandey, S.; Pathania, R. Role of bacterial efflux pumps in antibiotic resistance, virulence, and strategies to discover novel efflux pump inhibitors. Microbiology 2023, 169, 001333. [Google Scholar] [CrossRef] [PubMed]
- Feßler, A.T.; Wang, Y.; Wu, C.; Schwarz, S. Mobile macrolide resistance genes in staphylococci. Plasmid 2018, 99, 2–10. [Google Scholar] [CrossRef] [PubMed]
- Golkar, T.; Zieliński, M.; Berghuis, A.M. Look and Outlook on Enzyme-Mediated Macrolide Resistance. Front. Microbiol. 2018, 9, 1942. [Google Scholar] [CrossRef]
- Shoaib, M.; Aqib, A.I.; Muzammil, I.; Majeed, N.; Bhutta, Z.A.; Kulyar, M.F.-A.; Fatima, M.; Zaheer, C.-N.F.; Muneer, A.; Murtaza, M.; et al. MRSA compendium of epidemiology, transmission, pathophysiology, treatment, and prevention within one health framework. Front. Microbiol. 2023, 13, 1067284. [Google Scholar] [CrossRef]
- Nikaido, H. Multidrug Resistance in Bacteria. Annu. Rev. Biochem. 2009, 78, 119. [Google Scholar] [CrossRef]
- Kumar, S.; Mahato, R.P.; Ch, S.; Kumbham, S. Current strategies against multidrug-resistant Staphylococcus aureus and advances toward future therapy. The Microbe 2025, 6, 100281. [Google Scholar] [CrossRef]
- Raygada, J.L.; Levine, D.P. Methicillin-Resistant Staphylococcus aureus: A Growing Risk in the Hospital and in the Community. Am. Health Drug Benefits 2009, 2, 86–95. [Google Scholar]
- Uhlemann, A.-C.; Otto, M.; Lowy, F.D.; DeLeo, F.R. Evolution of community- and healthcare-associated methicillin-resistant Staphylococcus aureus. Infect. Genet. Evol. 2014, 21, 563–574. [Google Scholar] [CrossRef]
- Lade, H.; Kim, J.-S. Molecular Determinants of β-Lactam Resistance in Methicillin-Resistant Staphylococcus aureus (MRSA): An Updated Review. Antibiotics 2023, 12, 1362. [Google Scholar] [CrossRef]
- David, M.Z.; Daum, R.S. Community-Associated Methicillin-Resistant Staphylococcus aureus: Epidemiology and Clinical Consequences of an Emerging Epidemic. Clin. Microbiol. Rev. 2010, 23, 616–687. [Google Scholar] [CrossRef] [PubMed]
- Friães, A.; Resina, C.; Manuel, V.; Lito, L.; Ramirez, M.; Melo-Cristino, J. Epidemiological survey of the first case of vancomycin-resistant Staphylococcus aureus infection in Europe. Epidemiol. Infect. 2014, 143, 745. [Google Scholar] [CrossRef] [PubMed]
- Choo, E.J.; Chambers, H.F. Treatment of Methicillin-Resistant Staphylococcus aureus Bacteremia. Infect. Chemother. 2016, 48, 267–273. [Google Scholar] [CrossRef] [PubMed]
- Jones, R.N. Key considerations in the treatment of complicated staphylococcal infections. Clin. Microbiol. Infect. 2008, 14, 3–9. [Google Scholar] [CrossRef]
- Micek, S. Alternatives to Vancomycin for the Treatment of Methicillin-Resistant Staphylococcus aureus Infections. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2007, 45 (Suppl. 3), S184–S190. [Google Scholar] [CrossRef]
- Shariati, A.; Dadashi, M.; Moghadam, M.T.; van Belkum, A.; Yaslianifard, S.; Darban-Sarokhalil, D. Global prevalence and distribution of vancomycin resistant, vancomycin intermediate and heterogeneously vancomycin intermediate Staphylococcus aureus clinical isolates: A systematic review and meta-analysis. Sci. Rep. 2020, 10, 12689. [Google Scholar] [CrossRef]
- CIDRAP. Report Describes Rare US Vancomycin-Resistant Staph Case. 2024. Available online: https://www.cidrap.umn.edu/antimicrobial-stewardship/report-describes-rare-us-vancomycin-resistant-staph-case (accessed on 31 March 2025).
- Taha, A.E. A Review of Vancomycin-Resistant Staphylococcus aureus Prevalence in Egypt and Saudi Arabia. Asian J. Biol. Life Sci. 2019, 8, 94. [Google Scholar] [CrossRef]
- Deng, J.; Wang, X.; Zhang, B.-Z.; Gao, P.; Lin, Q.; Kao, R.Y.-T.; Gustafsson, K.; Yuen, K.-Y.; Huang, J.-D. Broad and Effective Protection against Staphylococcus aureus Is Elicited by a Multivalent Vaccine Formulated with Novel Antigens. mSphere 2019, 4, e00362-19. [Google Scholar] [CrossRef]
- Raafat, D.; Otto, M.; Reppschläger, K.; Iqbal, J.; Holtfreter, S. Fighting Staphylococcus aureus Biofilms with Monoclonal Antibodies. Trends Microbiol. 2019, 27, 303–322. [Google Scholar] [CrossRef]
- Sause, W.E.; Buckley, P.T.; Strohl, W.R.; Lynch, A.S.; Torres, V.J. Antibody-based Biologics and Their Promise to Combat Staphylococcus aureus Infections. Trends Pharmacol. Sci. 2015, 37, 231. [Google Scholar] [CrossRef]
- Schaffer, A.; Lee, J. Vaccine-Based Strategies for Prevention of Staphylococcus aureus Infection. In Staphylococci in Human Disease; Wiley: Hoboken, NJ, USA, 2009; pp. 594–611. ISBN 978-1-4051-6332-3. [Google Scholar]
- Chand, U.; Priyambada, P.; Kushawaha, P.K. Staphylococcus aureus vaccine strategy: Promise and challenges. Microbiol. Res. 2023, 271, 127362. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Liu, F.; Long, J.; Jin, Y.; Chen, S.; Duan, G.; Yang, H. The application of CRISPR-Cas system in Staphylococcus aureus infection. Heliyon 2024, 10, e34383. [Google Scholar] [CrossRef] [PubMed]
- Vivekanandan, K.E.; Kumar, P.V.; Jaysree, R.C.; Rajeshwari, T. Exploring molecular mechanisms of drug resistance in bacteria and progressions in CRISPR/Cas9-based genome expurgation solutions. Glob. Med. Genet. 2025, 12, 100042. [Google Scholar] [CrossRef] [PubMed]
- Ali Agha, A.S.A.; Al-Samydai, A.; Aburjai, T. New frontiers in CRISPR: Addressing antimicrobial resistance with Cas9, Cas12, Cas13, and Cas14. Heliyon 2025, 11, e42013. [Google Scholar] [CrossRef]
- Tao, S.; Chen, H.; Li, N.; Liang, W. The Application of the CRISPR-Cas System in Antibiotic Resistance. Infect. Drug Resist. 2022, 15, 4155–4168. [Google Scholar] [CrossRef]
- Lu, Y.; Lu, Y.; Li, B.; Liu, J.; Wang, L.; Zhang, L.; Li, Y.; Zhong, Q. StAP1 phage: An effective tool for treating methicillin-resistant Staphylococcus aureus infections. Front. Microbiol. 2023, 14, 1267786. [Google Scholar] [CrossRef]
- Belete, M.A.; Tadesse, S.; Tilahun, M.; Alemayehu, E.; Saravanan, M. Phage endolysins as new therapeutic options for multidrug resistant Staphylococcus aureus: An emerging antibiotic-free way to combat drug resistant infections. Front. Cell. Infect. Microbiol. 2024, 14, 1397935. [Google Scholar] [CrossRef]
- Atshan, S.S.; Hamat, R.A.; Aljaberi, M.A.; Chen, J.-S.; Huang, S.-W.; Lin, C.-Y.; Mullins, B.J.; Kicic, A. Phage Therapy as an Alternative Treatment Modality for Resistant Staphylococcus aureus Infections. Antibiotics 2023, 12, 286. [Google Scholar] [CrossRef]
- Liu, K.; Wang, C.; Zhou, X.; Guo, X.; Yang, Y.; Liu, W.; Zhao, R.; Song, H. Bacteriophage therapy for drug-resistant Staphylococcus aureus infections. Front. Cell. Infect. Microbiol. 2024, 14, 1336821. [Google Scholar] [CrossRef]
- Bi, Y.; Xia, G.; Shi, C.; Wan, J.; Liu, L.; Chen, Y.; Wu, Y.; Zhang, W.; Zhou, M.; He, H.; et al. Therapeutic strategies against bacterial biofilms. Fundam. Res. 2021, 1, 193–212. [Google Scholar] [CrossRef]
- Kadirvelu, L.; Sivaramalingam, S.S.; Jothivel, D.; Chithiraiselvan, D.D.; Karaiyagowder Govindarajan, D.; Kandaswamy, K. A review on antimicrobial strategies in mitigating biofilm-associated infections on medical implants. Curr. Res. Microb. Sci. 2024, 6, 100231. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.; Hu, Z.; Jin, Q.; Chen, Q.; Zhao, C.; Qiang, R.; Xie, Z.; Li, L.; Zhang, H. Structural characteristics, functions, and counteracting strategies of biofilms in Staphylococcus aureus. Comput. Struct. Biotechnol. J. 2025, 27, 488–500. [Google Scholar] [CrossRef] [PubMed]
- Parastan, R.; Kargar, M.; Solhjoo, K.; Kafilzadeh, F. Staphylococcus aureus biofilms: Structures, antibiotic resistance, inhibition, and vaccines. Gene Rep. 2020, 20, 100739. [Google Scholar] [CrossRef]
- Wang, X.; Chen, C.; Hu, J.; Liu, C.; Ning, Y.; Lu, F. Current strategies for monitoring and controlling bacterial biofilm formation on medical surfaces. Ecotoxicol. Environ. Saf. 2024, 282, 116709. [Google Scholar] [CrossRef]
- Pérez-Flores, J.G.; García-Curiel, L.; Pérez-Escalante, E.; Contreras-López, E.; Aguilar-Lira, G.Y.; Ángel-Jijón, C.; González-Olivares, L.G.; Baena-Santillán, E.S.; Ocampo-Salinas, I.O.; Guerrero-Solano, J.A.; et al. Plant Antimicrobial Compounds and Their Mechanisms of Action on Spoilage and Pathogenic Bacteria: A Bibliometric Study and Literature Review. Appl. Sci. 2025, 15, 3516. [Google Scholar] [CrossRef]
- Barbarossa, A.; Rosato, A.; Tardugno, R.; Carrieri, A.; Corbo, F.; Limongelli, F.; Fumarola, L.; Fracchiolla, G.; Carocci, A. Antibiofilm Effects of Plant Extracts Against Staphylococcus aureus. Microorganisms 2025, 13, 454. [Google Scholar] [CrossRef]
- Angelini, P. Plant-Derived Antimicrobials and Their Crucial Role in Combating Antimicrobial Resistance. Antibiot. Basel Switz. 2024, 13, 746. [Google Scholar] [CrossRef]
- Zouine, N.; Ghachtouli, N.E.; Abed, S.E.; Koraichi, S.I. A comprehensive review on medicinal plant extracts as antibacterial agents: Factors, mechanism insights and future prospects. Sci. Afr. 2024, 26, e02395. [Google Scholar] [CrossRef]
- Gao, P.; Ho, P.L.; Yan, B.; Sze, K.H.; Davies, J.; Kao, R.Y.T. Suppression of Staphylococcus aureus virulence by a small-molecule compound. Proc. Natl. Acad. Sci. USA 2018, 115, 8003–8008. [Google Scholar] [CrossRef]
- Kobayashi, S.D.; DeLeo, F.R. Towards a Monoclonal Antibody-Based Therapy for Prevention and Treatment of Staphylococcus aureus Infections. J. Infect. Dis. 2019, 219, 848–850. [Google Scholar] [CrossRef]
- Douglas, E.J.A.; Wulandari, S.W.; Lovell, S.D.; Laabei, M. Novel antimicrobial strategies to treat multi-drug resistant Staphylococcus aureus infections. Microb. Biotechnol. 2023, 16, 1456–1474. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Wang, L.; Zhang, J.; Liu, Q.; Wang, B.; Liu, D.; Gao, F.; Lanzi, G.; Zhao, Y.; Shi, Y. Thwarting resistance: MgrA inhibition with methylophiopogonanone a unveils a new battlefront against S. aureus. NPJ Biofilms Microbiomes 2024, 10, 15. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhang, D.; Zhang, X.; Zhang, X. Artificial intelligence applications in the diagnosis and treatment of bacterial infections. Front. Microbiol. 2024, 15, 1449844. [Google Scholar] [CrossRef] [PubMed]
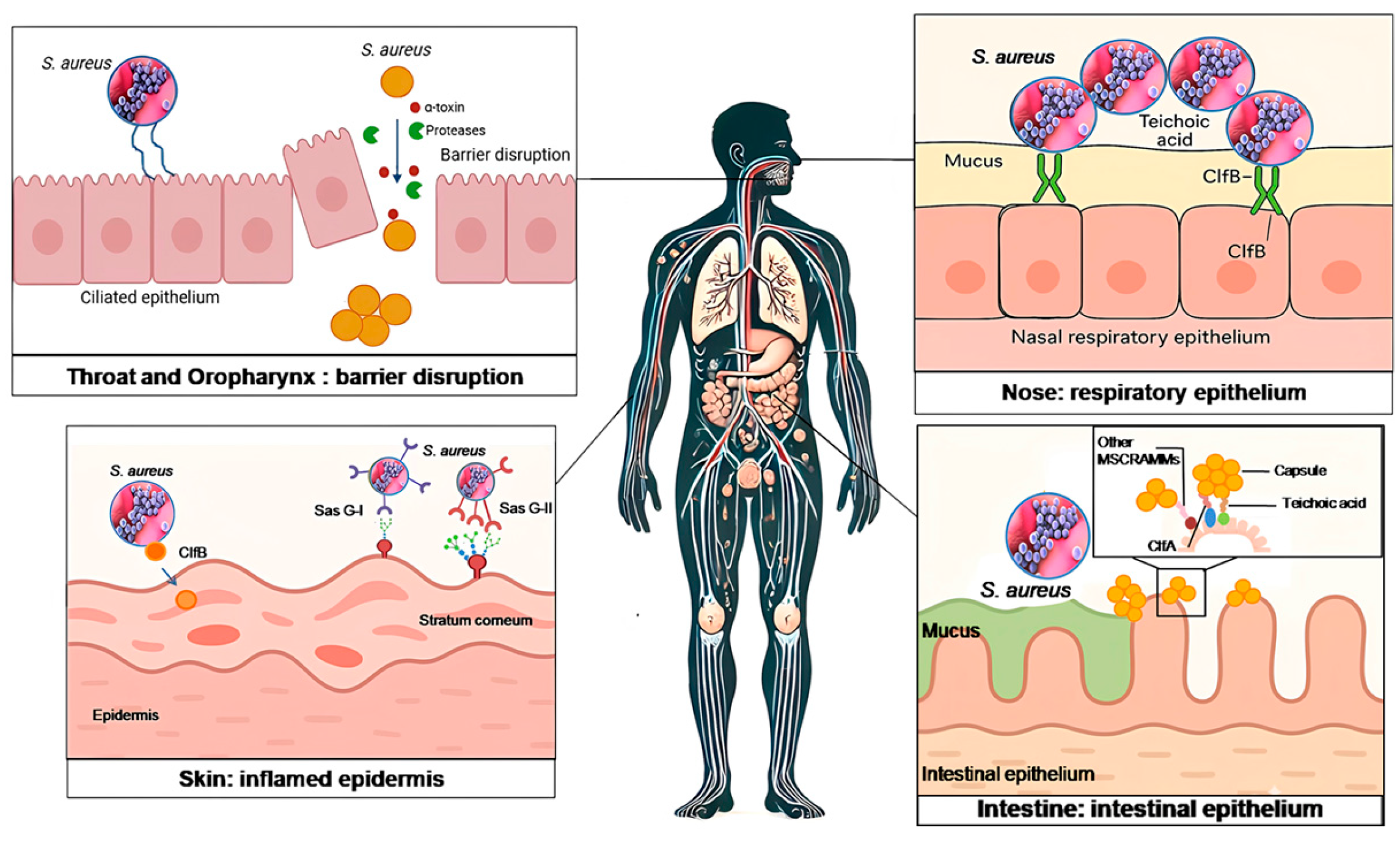
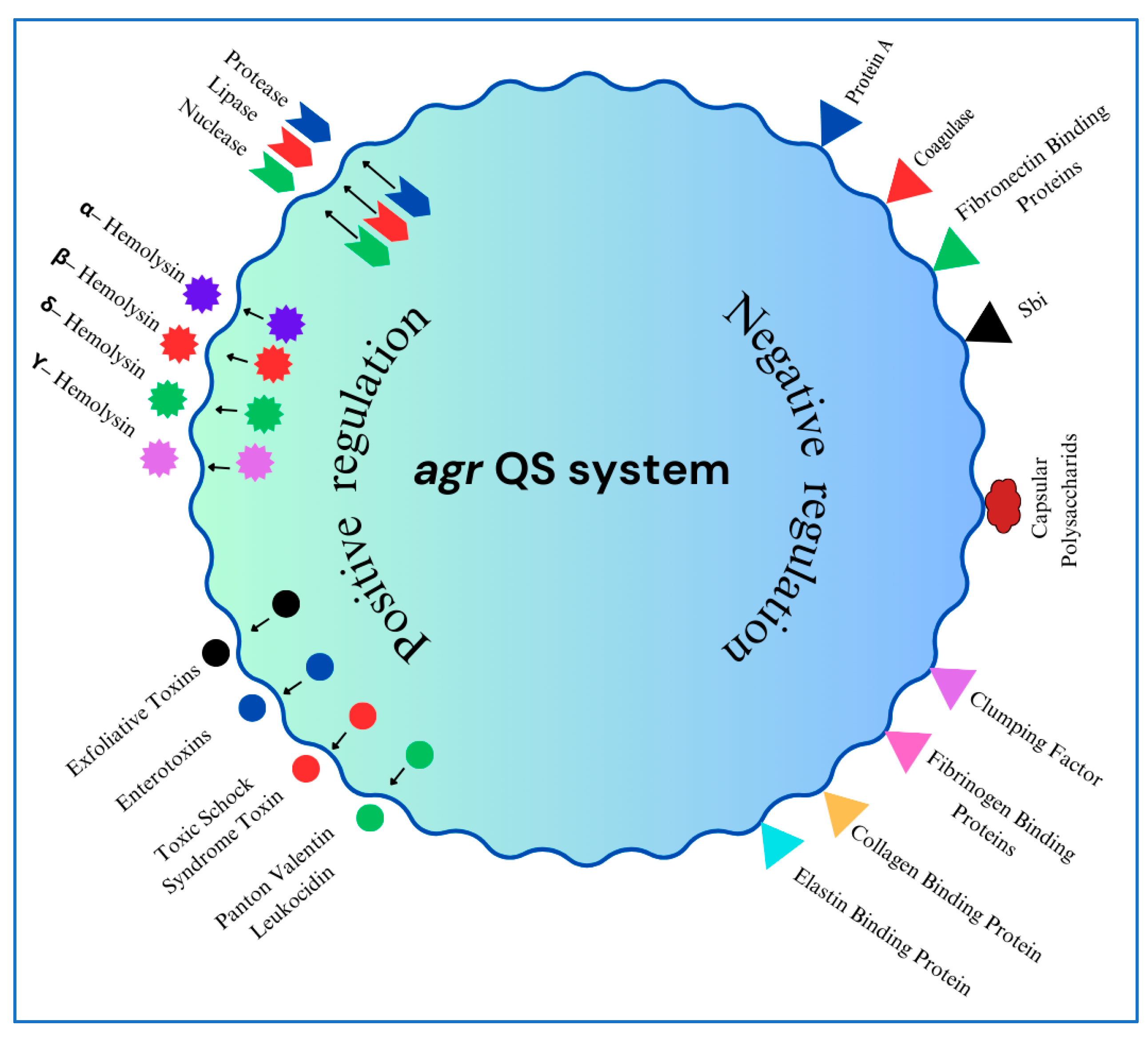
| Country | Population Studied | MSSA Carriage Rate | MRSA Carriage Rate | Source |
|---|---|---|---|---|
| United Kingdom | Orthopedic outpatients | 22.4–35.6% | 1.2–4.3% | [34] |
| USA | Patients undergoing fracture fixation | 20.18% | 4.70% | [35] |
| USA | Critically ill pediatric patients | Not specified | 6.5% | [36] |
| Ethiopia | Hospital janitors | 22.2% | 8.1% | [32] |
| Ethiopia | Non-hospital janitors | 14.4% | 1.4% | [32] |
| Argentina | Healthcare workers | 23.7% | 6.3% | [37] |
| France | Healthy Blood Donors | 29.3% | 0.3% | [38] |
| France | Hospitalized Patients | 20.2% | 1.1% | [38] |
| Sierra Leone | General community | 42.7% | 14% | [39] |
| Lebanon | General community | 38.4% | 1.6% | [40] |
| China | Medical students | 15.4–23.1% | 3.0–9.4% | [41] |
| Saudi Arabia | Healthcare workers | 40% | 18% | [42] |
| Algeria | Livestock and humans in contact | 50% (Humans) | 7.6% (Livestock) | [43] |
| Portugal | Homeless individuals (Lisbon | 50% | 1.2% | [33] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Touaitia, R.; Mairi, A.; Ibrahim, N.A.; Basher, N.S.; Idres, T.; Touati, A. Staphylococcus aureus: A Review of the Pathogenesis and Virulence Mechanisms. Antibiotics 2025, 14, 470. https://doi.org/10.3390/antibiotics14050470
Touaitia R, Mairi A, Ibrahim NA, Basher NS, Idres T, Touati A. Staphylococcus aureus: A Review of the Pathogenesis and Virulence Mechanisms. Antibiotics. 2025; 14(5):470. https://doi.org/10.3390/antibiotics14050470
Chicago/Turabian StyleTouaitia, Rahima, Assia Mairi, Nasir Adam Ibrahim, Nosiba S. Basher, Takfarinas Idres, and Abdelaziz Touati. 2025. "Staphylococcus aureus: A Review of the Pathogenesis and Virulence Mechanisms" Antibiotics 14, no. 5: 470. https://doi.org/10.3390/antibiotics14050470
APA StyleTouaitia, R., Mairi, A., Ibrahim, N. A., Basher, N. S., Idres, T., & Touati, A. (2025). Staphylococcus aureus: A Review of the Pathogenesis and Virulence Mechanisms. Antibiotics, 14(5), 470. https://doi.org/10.3390/antibiotics14050470









