Diagnostic Stewardship Trends and Antimicrobial Resistance Profiles of Bacteria Isolated in Zambia: A Five-Year Retrospective Study (2020–2024)
Abstract
1. Introduction
2. Results
2.1. Bacteriology Specimens Analysed per Sentinel Site over Five Years
2.2. Specimen Collection Trends
2.3. Isolation of Priority Pathogens
2.4. Antimicrobial Susceptibility Patterns of Bacteria
2.5. Important Antimicrobial Resistance (AMR) Alerts
2.6. Non-Susceptibility Trend Analysis Among Priority Pathogens
3. Discussion
3.1. Implications for Policy and Practice
3.2. Study Limitations
4. Materials and Methods
4.1. Study Design and Setting
4.2. Specimen Collection and Microbiological Procedures
4.3. Antimicrobial Susceptibility Testing (AST)
- β-lactams: ampicillin (AMP), Amoxicillin–Clavulanate (AMC), ceftriaxone (CRO), cefotaxime (CTX), cefepime (FEP), Ceftazidime (CAZ), cefuroxime (CXM), cefazolin (CZ), and cefoxitin (FOX);
- Carbapenems: imipenem (IPM), meropenem (MEM), and doripenem (DOR);
- Aminoglycosides: gentamicin (GEN), tobramycin (TOB), and amikacin (AK);
- Fluoroquinolones: ciprofloxacin (CIP), levofloxacin (LEV), ofloxacin (OFX), norfloxacin (NOR), and nalidixic acid (NA);
- Macrolides and others: erythromycin (ERY), azithromycin (AZM), clarithromycin (CLA), clindamycin (CLI), linezolid (LZD), vancomycin (VAN), doxycycline (DOX), tetracycline (TET), Chloramphenicol (CHL), nitrofurantoin (NIT), trimethoprim-sulfamethoxazole (SXT), and Quinupristin/Dalfopristin (RP).
4.4. Data Management
4.5. Data Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Disclaimer
Abbreviations
| AMC | Amoxicillin/Clavulanic acid |
| AMP | Ampicillin |
| AMR | Antimicrobial resistance |
| AMS | Antimicrobial stewardship |
| AMX | Amoxicillin |
| AZM | Azithromycin |
| CAZ | Ceftazidime |
| CCH | Chipata Central Hospital |
| CHL | Chloramphenicol |
| CIP | Ciprofloxacin |
| CLI | Clindamycin |
| CMH | Chilonga Mission Hospital |
| COL | Colistin |
| COVID-19 | Corona Virus Disease 2019 |
| CRO | Ceftriaxone |
| CSF | Cerebrospinal fluid |
| CTX | Cefotaxime |
| CZO | Cefazolin |
| DOX | Doxycycline |
| DS | Diagnostic stewardship |
| ERY | Erythromycin |
| ETP | Ertapenem |
| FEP | Cefepime |
| FOX | Cefoxitin |
| GEN | Gentamicin |
| GLASS | Global Antimicrobial Resistance and Use Surveillance System |
| IMP | Imipenem |
| LUTHLUTH | Livingstone University Teaching Hospital |
| LGH | Lewanika General Hospital |
| LNZ | Linezolid |
| MEM | Meropenem |
| MGH | Mansa General Hospital |
| NAL | Nalidixic acid |
| NAP | National Action Plan |
| NIT | Nitrofurantoin |
| NOR | Norfloxacin |
| NTH | Ndola Teaching Hospital |
| OXA | Oxacillin |
| PEN | Penicillin G |
| QDA | Quinupristin/Dalfopristin |
| RIF | Rifampicin |
| SPT | Spectinomycin |
| SXT | Trimethoprim/Sulfamethoxazole |
| TCY | Tetracycline |
| TGC | Tigecycline |
| TMP | Trimethoprim |
| UTHs | University Teaching Hospitals |
| VAN | Vancomycin |
| ZNPHI | Zambia National Public Health Institute |
References
- Prestinaci, F.; Patrizio, P.; Pantosti, A. Antimicrobial resistance: A global multifaceted phenomenon. Pathog. Glob. Health 2015, 109, 309–318. [Google Scholar] [CrossRef] [PubMed]
- Salam, M.A.; Al-Amin, M.Y.; Salam, M.T.; Pawar, J.S.; Akhter, N.; Rabaan, A.A.; Alqumber, M.A.A. Antimicrobial Resistance: A Growing Serious Threat for Global Public Health. Healthcare 2023, 11, 1946. [Google Scholar] [CrossRef]
- Murray, C.J.L.; Ikuta, K.S.; Sharara, F.; Swetschinski, L.; Robles Aguilar, G.; Gray, A.; Han, C.; Bisignano, C.; Rao, P.; Wool, E.; et al. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef]
- Godman, B.; Egwuenu, A.; Wesangula, E.; Schellack, N.; Kalungia, A.C.; Tiroyakgosi, C.; Kgatlwane, J.; Mwita, J.C.; Patrick, O.; Niba, L.L.; et al. Tackling antimicrobial resistance across sub-Saharan Africa: Current challenges and implications for the future. Expert Opin. Drug Saf. 2022, 21, 1089–1111. [Google Scholar] [CrossRef]
- Saleem, Z.; Godman, B.; Cook, A.; Khan, M.A.; Campbell, S.M.; Seaton, R.A.; Siachalinga, L.; Haseeb, A.; Amir, A.; Kurdi, A.; et al. Ongoing Efforts to Improve Antimicrobial Utilization in Hospitals among African Countries and Implications for the Future. Antibiotics 2022, 11, 1824. [Google Scholar] [CrossRef] [PubMed]
- Sartorius, B.; Gray, A.P.; Davis Weaver, N.; Robles Aguilar, G.; Swetschinski, L.R.; Ikuta, K.S.; Mestrovic, T.; Chung, E.; Wool, E.E.; Han, C.; et al. The burden of bacterial antimicrobial resistance in the WHO African region in 2019: A cross-country systematic analysis. Lancet Glob. Health 2024, 12, e201–e216. [Google Scholar] [CrossRef]
- Zakhour, J.; Haddad, S.F.; Kerbage, A.; Wertheim, H.; Tattevin, P.; Voss, A.; Ünal, S.; Ouedraogo, A.S.; Kanj, S.S. Diagnostic stewardship in infectious diseases: A continuum of antimicrobial stewardship in the fight against antimicrobial resistance. Int. J. Antimicrob. Agents 2023, 62, 106816. [Google Scholar] [CrossRef] [PubMed]
- Claeys, K.C.; Johnson, M.D. Leveraging diagnostic stewardship within antimicrobial stewardship programmes. Drugs Context 2023, 12, 2022-9-5. [Google Scholar] [CrossRef]
- Singh, H.K.; Claeys, K.C.; Advani, S.D.; Ballam, Y.J.; Penney, J.; Schutte, K.M.; Baliga, C.; Milstone, A.M.; Hayden, M.K.; Morgan, D.J.; et al. Diagnostic stewardship to improve patient outcomes and healthcare-associated infection (HAI) metrics. Infect. Control Hosp. Epidemiol. 2024, 45, 405–411. [Google Scholar] [CrossRef]
- World Health Organization. Diagnostic Stewardship: A Guide to Implementation in Antimicrobial Resistance Surveillance Sites. 2016. Available online: https://www.who.int/publications/i/item/WHO-DGO-AMR-2016.3 (accessed on 20 June 2025).
- Kapona, O. Zambia Successfully Launches the First Multi-Sectoral National Action Plan on Antimicrobial Resistance (AMR). Health Press Zamb. Bull. 2017, 1, 5–7. [Google Scholar]
- World Health Organization. Global Action Plan on Antimicrobial Resistance. 2015. Available online: https://www.who.int/antimicrobial-resistance/publications/global-action-plan/en/ (accessed on 20 October 2020).
- World_Health_Organization. Monitoring and Evaluation of the Global Action Plan on Antimicrobial Resistance. 2019. Available online: https://apps.who.int/iris/bitstream/handle/10665/325006/9789241515665-eng.pdf?sequence=1&isAllowed=y (accessed on 17 June 2025).
- Mudenda, S.; Mufwambi, W.; Mohamed, S. The Burden of Antimicrobial Resistance in Zambia, a Sub-Saharan African Country: A One Health Review of the Current Situation, Risk Factors, and Solutions. Pharmacol. Pharm. 2024, 15, 403–465. [Google Scholar] [CrossRef]
- World Health Organization. Global Antimicrobial Resistance Surveillance System: Manual for Early Implementation. 2015. Available online: https://www.who.int/publications/i/item/9789241549400 (accessed on 17 June 2025).
- World Health Organization. Global Antimicrobial Resistance and Use Surveillance System (GLASS) Report 2021; World Health Organization: Geneva, Switzerland, 2021; pp. 1–180. [Google Scholar]
- Matee, M.; Mshana, S.E.; Mtebe, M.; Komba, E.V.; Moremi, N.; Lutamwa, J.; Kapona, O.; Sekamatte, M.; Mboera, L.E.G. Mapping and gap analysis on antimicrobial resistance surveillance systems in Kenya, Tanzania, Uganda and Zambia. Bull. Natl. Res. Cent. 2023, 47, 12. [Google Scholar] [CrossRef]
- Yamba, K.; Chizimu, J.Y.; Mudenda, S.; Lukwesa, C.; Chanda, R.; Nakazwe, R.; Simunyola, B.; Shawa, M.; Kalungia, A.C.; Chanda, D.; et al. Assessment of Antimicrobial Resistance Laboratory-based Surveillance Capacity of Hospitals in Zambia: Findings and Implications for System Strengthening. J. Hosp. Infect. 2024, 148, 129–137. [Google Scholar] [CrossRef]
- Okolie, O.J.; Igwe, U.; Ismail, S.U.; Ighodalo, U.L.; Adukwu, E.C. Systematic review of surveillance systems for AMR in Africa. J. Antimicrob. Chemother. 2022, 78, 31–51. [Google Scholar] [CrossRef] [PubMed]
- Malania, L.; Wagenaar, I.; Karatuna, O.; Tambic Andrasevic, A.; Tsereteli, D.; Baidauri, M.; Imnadze, P.; Nahrgang, S.; Ruesen, C. Setting up laboratory-based antimicrobial resistance surveillance in low- and middle-income countries: Lessons learned from Georgia. Clin. Microbiol. Infect. 2021, 27, 1409–1413. [Google Scholar] [CrossRef]
- Essack, P.S.; Essack, S.Y. AMR Surveillance in Africa: Are We There Yet? Int. J. Infect. Dis. 2025, 152, 107828. [Google Scholar] [CrossRef]
- Kajumbula, H.M.; Amoako, D.G.; Tessema, S.K.; Aworh, M.K.; Chikuse, F.; Okeke, I.N.; Okomo, U.; Jallow, S.; Egyir, B.; Kanzi, A.M.; et al. Enhancing clinical microbiology for genomic surveillance of antimicrobial resistance implementation in Africa. Antimicrob. Resist. Infect. Control 2024, 13, 135. [Google Scholar] [CrossRef] [PubMed]
- Musa, K.; Okoliegbe, I.; Abdalaziz, T.; Aboushady, A.T.; Stelling, J.; Gould, I.M. Laboratory Surveillance, Quality Management, and Its Role in Addressing Antimicrobial Resistance in Africa: A Narrative Review. Antibiotics 2023, 12, 1313. [Google Scholar] [CrossRef]
- Yao, K.; Luman, E.T.; Nkengasong, J.N.; Maruta, T. The SLMTA programme: Transforming the laboratory landscape in developing countries. Afr. J. Lab. Med. 2016, 5, 1–8. [Google Scholar]
- Lappan, R.; Chown, S.L.; French, M.; Perlaza-Jiménez, L.; Macesic, N.; Davis, M.; Brown, R.; Cheng, A.; Clasen, T.; Conlan, L.; et al. Towards integrated cross-sectoral surveillance of pathogens and antimicrobial resistance: Needs, approaches, and considerations for linking surveillance to action. Environ. Int. 2024, 192, 109046. [Google Scholar] [CrossRef]
- Dacombe, R.; Bates, I.; Bhardwaj, M.; Wallis, S.; Pulford, J. Fleming Fund: Supporting Surveillance Capacity for Antimicrobial Resistance An Analysis of Approaches to Laboratory Capacity Strengthening for Drug Resistant Infections in Low and Middle Income Countries. 2016. Available online: http://researchonline.ljmu.ac.uk/id/eprint/6678/1/FF%20An%20analysis%20of%20approaches%20to%20lab%20cap%20strenghtening%20for%20drug%20resistant.pdf (accessed on 17 June 2025).
- Gopolang, F.; Zulu-Mwamba, F.; Nsama, D.; Kruuner, A.; Nsofwa, D.; Kasvosve, I.; Gomo, R.; Motlhabane, T.; Chohan, B.; Soge, O.; et al. Improving laboratory quality and capacity through leadership and management training: Lessons from Zambia 2016–2018. Afr. J. Lab. Med. 2021, 10, 1–9. [Google Scholar] [CrossRef]
- Shempela, D.M.; Mudenda, S.; Kasanga, M.; Daka, V.; Kangongwe, M.H.; Kamayani, M.; Sikalima, J.; Yankonde, B.; Kasonde, C.B.; Nakazwe, R. A Situation Analysis of the Capacity of Laboratories in Zambia to Conduct Surveillance of Antimicrobial Resistance: Strengths, Weaknesses, Opportunities, and Challenges. 2024. Available online: https://www.preprints.org/frontend/manuscript/9d35f7af5f7132cc69b149b7521cf6f0/download_pub (accessed on 17 June 2025). [CrossRef]
- Kasanga, M.; Mudenda, S.; Siyanga, M.; Chileshe, M.; Mwiikisa, M.J.; Kasanga, M.; Solochi, B.B.; Gondwe, T.; Kantenga, T.; LShibemba, A.; et al. Antimicrobial susceptibility Patterns of Bacteria that Commonly Cause Bacteremia at a Tertiary Hospital in Zambia. Future Microbiol. 2020, 15, 1735–1745. [Google Scholar] [CrossRef]
- Kasanga, M.; Mukosha, R.; Kasanga, M.; Siyanga, M.; Mudenda, S.; Solochi, B.B.; Chileshe, M.; Mwiikisa, M.J.; Gondwe, T.; Kantenga, T.; et al. Antimicrobial Resistance Patterns of Bacterial Pathogens: Their Distribution in University Teaching Hospitals in Zambia. Future Microbiol. 2021, 16, 811–824. [Google Scholar] [CrossRef]
- Kasanga, M.; Gajdács, M.; Muleya, W.; Ikhimiukor, O.O.; Mudenda, S.; Kasanga, M.; Chizimu, J.; Shempela, D.M.; Solochi, B.B.; Mwikisa, M.J.; et al. Genotypic Characterisation and Antimicrobial Resistance of Extended-Spectrum β-Lactamase-Producing Escherichia coli in Humans, Animals, and the Environment from Lusaka, Zambia: Public Health Implications and One Health Surveillance. Antibiotics 2024, 13, 951. [Google Scholar] [CrossRef] [PubMed]
- Kasanga, M.; Shempela, D.M.; Daka, V.; Mwikisa, M.J.; Sikalima, J.; Chanda, D.; Mudenda, S. Antimicrobial resistance profiles of Escherichia coli isolated from clinical and environmental samples: Findings and implications. JAC-Antimicrob. Resist. 2024, 6, dlae061. [Google Scholar] [CrossRef]
- Bumbangi, F.N.; Llarena, A.-K.; Skjerve, E.; Hang’ombe, B.M.; Mpundu, P.; Mudenda, S.; Mutombo, P.B.; Muma, J.B. Evidence of Community-Wide Spread of Multi-Drug Resistant Escherichia coli in Young Children in Lusaka and Ndola Districts, Zambia. Microorganisms 2022, 10, 1684. [Google Scholar] [CrossRef]
- Chanda, W.; Manyepa, M.; Chikwanda, E.; Daka, V.; Chileshe, J.; Tembo, M.; Kasongo, J.; Chipipa, A.; Handema, R.; Mulemena, J.A. Evaluation of antibiotic susceptibility patterns of pathogens isolated from routine laboratory specimens at Ndola Teaching Hospital: A retrospective study. PLoS ONE 2019, 14, e0226676. [Google Scholar] [CrossRef]
- Mwansa, T.N.; Kamvuma, K.; Mulemena, J.A.; Phiri, C.N.; Chanda, W. Antibiotic susceptibility patterns of pathogens isolated from laboratory specimens at Livingstone Central Hospital in Zambia. PLoS Glob. Public Health 2022, 2, e0000623. [Google Scholar] [CrossRef] [PubMed]
- Chiyangi, H.; Muma, J.B.; Malama, S.; Manyahi, J.; Abade, A.; Kwenda, G.; Matee, M.I. Identification and antimicrobial resistance patterns of bacterial enteropathogens from children aged 0–59 months at the University Teaching Hospital, Lusaka, Zambia: A prospective cross sectional study. BMC Infect. Dis. 2017, 17, 117. [Google Scholar] [CrossRef] [PubMed]
- Siame, A.; Yamba, K.; Samutela, M.; Mukubesa, A.; Mulundu, G. Carriage and antimicrobial susceptibility patterns of rectal ESBL E. coli in surgical patients at the University Teaching Hospitals in Lusaka, Zambia. JAC-Antimicrob. Resist. 2024, 6, dlae159. [Google Scholar] [CrossRef]
- Yamba, K.; Mudenda, S.; Mpabalwani, E.; Mainda, G.; Mukuma, M.; Samutela, M.T.; Lukwesa, C.; Chizimu, J.; Kaluba, C.K.; Mutalange, M.; et al. Antibiotic prescribing patterns and carriage of antibiotic-resistant Escherichia coli and Enterococcus species in healthy individuals from selected communities in Lusaka and Ndola districts, Zambia. JAC-Antimicrob. Resist. 2024, 6, dlae027. [Google Scholar] [CrossRef]
- Mudenda, S.; Focus, L.A.; Shazia, J.; Bibian, B.; Racheal, S.; Mervis, C.; Lorraine, K.; Twaambo, M.L.; Nsoni, B.F.; Yamweka, C.J.; et al. Point Prevalence Survey of Antibiotic Use in Level 1 hospitals in Zambia: Future Prospects for Antimicrobial Stewardship Programs. Infect. Drug Resist. 2025, 18, 887–902. [Google Scholar] [CrossRef]
- Kalungia, A.C.; Mukosha, M.; Mwila, C.; Banda, D.; Mwale, M.; Kagulura, S.; Ogunleye, O.O.; Meyer, J.C.; Godman, B. Antibiotic Use and Stewardship Indicators in the First- and Second-Level Hospitals in Zambia: Findings and Implications for the Future. Antibiotics 2022, 11, 1626. [Google Scholar] [CrossRef]
- Chizimu, J.Y.; Mudenda, S.; Yamba, K.; Lukwesa, C.; Chanda, R.; Nakazwe, R.; Shawa, M.; Chambaro, H.; Kamboyi, H.K.; Kalungia, A.C.; et al. Antibiotic use and adherence to the WHO AWaRe guidelines across 16 hospitals in Zambia: A point prevalence survey. JAC-Antimicrob. Resist. 2024, 6, dlae170. [Google Scholar] [CrossRef] [PubMed]
- Mudenda, S.; Chilimboyi, R.; Matafwali, S.K.; Daka, V.; Mfune, R.L.; Kemgne, L.A.M.; Bumbangi, F.N.; Hangoma, J.; Chabalenge, B.; Mweetwa, L.; et al. Hospital prescribing patterns of antibiotics in Zambia using the WHO prescribing indicators post-COVID-19 pandemic: Findings and implications. JAC-Antimicrob. Resist. 2024, 6, dlae023. [Google Scholar] [CrossRef] [PubMed]
- Mudenda, S.; Nsofu, E.; Chisha, P.; Daka, V.; Chabalenge, B.; Mufwambi, W.; Kainga, H.; Kanaan, M.H.G.; Mfune, R.L.; Mwaba, F.; et al. Prescribing Patterns of Antibiotics According to the WHO AWaRe Classification during the COVID-19 Pandemic at a Teaching Hospital in Lusaka, Zambia: Implications for Strengthening of Antimicrobial Stewardship Programmes. Pharmacoepidemiology 2023, 2, 42–53. [Google Scholar] [CrossRef]
- Mudenda, S.; Chomba, M.; Chabalenge, B.; Hikaambo, C.N.a.; Banda, M.; Daka, V.; Zulu, A.; Mukesela, A.; Kasonde, M.; Lukonde, P.; et al. Antibiotic Prescribing Patterns in Adult Patients According to the WHO AWaRe Classification: A Multi-Facility Cross-Sectional Study in Primary Healthcare Hospitals in Lusaka, Zambia. Pharmacol. Pharm. 2022, 13, 379–392. [Google Scholar] [CrossRef]
- Masich, A.M.; Vega, A.D.; Callahan, P.; Herbert, A.; Fwoloshi, S.; Zulu, P.M.; Chanda, D.; Chola, U.; Mulenga, L.; Hachaambwa, L.; et al. Antimicrobial usage at a large teaching hospital in Lusaka, Zambia. PLoS ONE 2020, 15, e0228555. [Google Scholar] [CrossRef]
- Ngoma, M.T.; Sitali, D.; Mudenda, S.; Mukuma, M.; Bumbangi, F.N.; Bunuma, E.; Skjerve, E.; Muma, J.B. Community antibiotic consumption and associated factors in Lusaka district of Zambia: Findings and implications for antimicrobial resistance and stewardship. JAC-Antimicrob. Resist. 2024, 6, dlae034. [Google Scholar] [CrossRef]
- Makiko, F.; Kalungia, A.C.; Kampamba, M.; Mudenda, S.; Schellack, N.; Meyer, J.C.; Bumbangi, F.N.; Okorie, M.; Banda, D.; Munkombwe, D.; et al. Current status and future direction of antimicrobial stewardship programs and antibiotic prescribing in primary care hospitals in Zambia. JAC-Antimicrob. Resist. 2025, 7, dlaf085. [Google Scholar] [CrossRef]
- Kalonga, J.; Hangoma, J.; Banda, M.; Munkombwe, D.; Mudenda, S. Antibiotic Prescribing Patterns in Paediatric Patients at Levy Mwanawasa University Teaching Hospital in Lusaka, Zambia. J. Pharm. Res. Sci. Technol. 2020, 4, 1–9. [Google Scholar] [CrossRef]
- Mudenda, S.; Simbaya, R.; Moonga, G.; Mwaba, F.; Zulu, M.; Tembo, R.; Chiyangi, H.K.; Vlahakis, P.; Mohamed, S.; Lubanga, A.F. Surveillance of Antibiotic Use and Adherence to the WHO/INRUD Core Prescribing Indicators at a Primary Healthcare Hospital in Southern Zambia: Opportunities for Antimicrobial Stewardship Programs. J. Pharmacol. Pharm. 2025, 16, 1–19. [Google Scholar] [CrossRef]
- Kalungia, A.C.; Kampamba, M.; Banda, D.; Bambala, A.M.; Marshall, S.; Newport, M.; Clair-Jones, A.S.; Alutuli, L.; Chambula, E.; Munsaka, L.; et al. Impact of a hub-and-spoke approach to hospital antimicrobial stewardship programmes on antibiotic use in Zambia. JAC-Antimicrob. Resist. 2024, 6, dlae178. [Google Scholar] [CrossRef]
- Mudenda, S.; Kapolowe, K.; Chirwa, U.; Chanda, M.; Chanda, R.; Kalaba, R.; Fwoloshi, S.; Phiri, C.; Mwamba, M.; Chirwa, R.K.; et al. Antimicrobial Stewardship Impact on Antibiotic Use in Three Tertiary Hospitals in Zambia: A Comparative Point Prevalence Survey. Antibiotics 2025, 14, 284. [Google Scholar] [CrossRef]
- Zambia National Public Health Institute. Zambia’s Integrated Antimicrobial Resistance Surveillance Framework. 2020. Available online: https://www.afro.who.int/publications/zambias-integrated-antimicrobial-resistance-surveillance-framework (accessed on 17 June 2025).
- Ashley, E.A.; Recht, J.; Chua, A.; Dance, D.; Dhorda, M.; Thomas, N.V.; Ranganathan, N.; Turner, P.; Guerin, P.J.; White, N.J.; et al. An inventory of supranational antimicrobial resistance surveillance networks involving low- and middle-income countries since 2000. J. Antimicrob. Chemother. 2018, 73, 1737–1749. [Google Scholar] [CrossRef]
- Yopa, D.S.; Anya, P.; Mendjime, P.; Elouga, T.; Nnanga-Nga, E.; Nguefack-Tsague, G. Evaluation of the Antimicrobial Resistance Surveillance System in Sentinel Sites in Cameroon. Cureus 2023, 15, e40779. [Google Scholar] [CrossRef]
- Seale, A.C.; Gordon, N.C.; Islam, J.; Peacock, S.J.; Scott, J.A.G. AMR Surveillance in low and middle-income settings—A roadmap for participation in the Global Antimicrobial Surveillance System (GLASS). Wellcome Open Res. 2017, 2, 92. [Google Scholar] [CrossRef]
- Omulo, S.; Thumbi, S.M.; Njenga, M.K.; Call, D.R. A review of 40 years of enteric antimicrobial resistance research in Eastern Africa: What can be done better? Antimicrob. Resist. Infect. Control 2015, 4, 1. [Google Scholar] [CrossRef]
- Zwane, T.; Shuping, L.; Perovic, O. Etiology and Antimicrobial Susceptibility of Pathogens Associated with Urinary Tract Infections among Women Attending Antenatal Care in Four South African Tertiary-Level Facilities, 2015–2019. Antibiotics 2021, 10, 669. [Google Scholar] [CrossRef] [PubMed]
- Ngong, I.N.; Fru-Cho, J.; Yung, M.A.; Akoachere, J.-F.K.T. Prevalence, antimicrobial susceptibility pattern and associated risk factors for urinary tract infections in pregnant women attending ANC in some integrated health centers in the Buea Health District. BMC Pregnancy Childbirth 2021, 21, 673. [Google Scholar] [CrossRef] [PubMed]
- Kebede, D.; Shiferaw, Y.; Kebede, E.; Demsiss, W. Antimicrobial susceptibility and risk factors of uropathogens in symptomatic urinary tract infection cases at Dessie Referral Hospital, Ethiopia. BMC Microbiol. 2025, 25, 126. [Google Scholar] [CrossRef]
- Tansarli Giannoula, S.; Athanasiou, S.; Falagas Matthew, E. Evaluation of Antimicrobial Susceptibility of Enterobacteriaceae Causing Urinary Tract Infections in Africa. Antimicrob. Agents Chemother. 2013, 57, 3628–3639. [Google Scholar] [CrossRef] [PubMed]
- Flores-Mireles, A.L.; Walker, J.N.; Caparon, M.; Hultgren, S.J. Urinary tract infections: Epidemiology, mechanisms of infection and treatment options. Nat. Rev. Microbiol. 2015, 13, 269–284. [Google Scholar] [CrossRef] [PubMed]
- Agyepong, N.; Govinden, U.; Owusu-Ofori, A.; Essack, S.Y. Multidrug-resistant gram-negative bacterial infections in a teaching hospital in Ghana. Antimicrob. Resist. Infect. Control 2018, 7, 37. [Google Scholar] [CrossRef]
- Mouanga Ndzime, Y.; Richard, O.; Fabrice, K.K.R.; Michelle, B.; Philippe, M.N.P.; Amahani, G.; Wenceslas, L.R.; Kelly, M.M.; Bisseye, C. Epidemiology of Community Origin Escherichia coli and Klebsiella pneumoniae Uropathogenic Strains Resistant to Antibiotics in Franceville, Gabon. Infect. Drug Resist. 2021, 14, 585–594. [Google Scholar] [CrossRef] [PubMed]
- Kot, B.; Grużewska, A.; Szweda, P.; Wicha, J.; Parulska, U. Antibiotic Resistance of Uropathogens Isolated from Patients Hospitalized in District Hospital in Central Poland in 2020. Antibiotics 2021, 10, 447. [Google Scholar] [CrossRef]
- Hope, D.; Ampaire, L.; Oyet, C.; Muwanguzi, E.; Twizerimana, H.; Apecu, R.O. Antimicrobial resistance in pathogenic aerobic bacteria causing surgical site infections in Mbarara regional referral hospital, Southwestern Uganda. Sci. Rep. 2019, 9, 17299. [Google Scholar] [CrossRef]
- Bora, P.; Datta, P.; Gupta, V.; Singhal, L.; Chander, J. Characterization and antimicrobial susceptibility of coagulase-negative staphylococci isolated from clinical samples. J. Lab. Physicians 2018, 10, 414–419. [Google Scholar] [CrossRef]
- Michels, R.; Last, K.; Becker, S.L.; Papan, C. Update on Coagulase-Negative Staphylococci—What the Clinician Should Know. Microorganisms 2021, 9, 830. [Google Scholar] [CrossRef]
- Al-enizi, H.F.M.; Al-Shammari, A.A.; Alshammari, M.S.; Aldhafiry, E.A.; Alshammeri, G.M.; Kandeel, A.Y. Understanding the Practices and Challenges in Antimicrobial Susceptibility Testing: A Qualitative Study of Medical Laboratory Technicians in Saudi Arabia. J. Int. Crisis Risk Commun. Res. 2024, 7, 1073. [Google Scholar]
- Halstead, D.C.; Sautter, R.L. A Literature Review on How We Can Address Medical Laboratory Scientist Staffing Shortages. Lab. Med. 2022, 54, e31–e36. [Google Scholar] [CrossRef] [PubMed]
- Dumm, R.E.; Marlowe, E.M.; Patterson, L.; Larkin, P.M.K.; She, R.C.; Filkins, L.M. The foundation for the microbiology laboratory’s essential role in diagnostic stewardship: An ASM Laboratory Practices Subcommittee report. J. Clin. Microbiol. 2024, 62, e00960-24. [Google Scholar] [CrossRef] [PubMed]
- Fabre, V.; Davis, A.; Diekema, D.J.; Granwehr, B.; Hayden, M.K.; Lowe, C.F.; Pfeiffer, C.D.; Sick-Samuels, A.C.; Sullivan, K.V.; Van Schooneveld, T.C.; et al. Principles of diagnostic stewardship: A practical guide from the Society for Healthcare Epidemiology of America Diagnostic Stewardship Task Force. Infect. Control Hosp. Epidemiol. 2023, 44, 178–185. [Google Scholar] [CrossRef]
- Osei Sekyere, J.; Schneiders, T.; Majewski, P. Molecular mechanisms of resistance to “last resort” antimicrobials in Enterobacterales. Front. Cell. Infect. Microbiol. 2024, 14, 1429200. [Google Scholar] [CrossRef]
- Verkaik, N.J.; Wielders, C.C.H.; Boer, H.d.; Langerak, D.; Vogel, M.; Witteveen, S.; Haan, A.d.; Bos, J.; Westreenen, M.v.; Notermans, D.W.; et al. Antimicrobial susceptibility to last-resort antibiotics in carbapenemase-producing bacteria from Ukrainian patients. Microbiol. Spectr. 2024, 12, e01142-24. [Google Scholar] [CrossRef]
- Moja, L.; Zanichelli, V.; Mertz, D.; Gandra, S.; Cappello, B.; Cooke, G.S.; Chuki, P.; Harbarth, S.; Pulcini, C.; Mendelson, M.; et al. WHO’s essential medicines and AWaRe: Recommendations on first- and second-choice antibiotics for empiric treatment of clinical infections. Clin. Microbiol. Infect. 2024, 30, S1–S51. [Google Scholar] [CrossRef]
- Saleem, Z.; Sheikh, S.; Godman, B.; Haseeb, A.; Afzal, S.; Qamar, M.; Imam, M.; Abuhussain, S.; Sharland, M. Increasing the use of the WHO AWaRe system in Antibiotic Surveillance and Stewardship Programs in Low- and Middle-Income Countries. JAC-Antimicrob. Resist. 2025, 7, dlaf031. [Google Scholar] [CrossRef]
- Sharland, M.; Gandra, S.; Huttner, B.; Moja, L.; Pulcini, C.; Zeng, M.; Mendelson, M.; Cappello, B.; Cooke, G.; Magrini, N.; et al. Encouraging AWaRe-ness and discouraging inappropriate antibiotic use:—The new 2019 Essential Medicines List becomes a global antibiotic stewardship tool. Lancet Infect. Dis. 2019, 19, 1278–1280. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. The WHO AWaRe (Access, Watch, Reserve) Antibiotic Book; World Health Organization: Geneva, Switzerland, 2022; p. 697. [Google Scholar]
- Zanichelli, V.; Sharland, M.; Cappello, B.; Moja, L.; Getahun, H.; Pessoa-Silva, C.; Sati, H.; van Weezenbeek, C.; Balkhy, H.; Simão, M.; et al. The WHO AWaRe (Access, Watch, Reserve) antibiotic book and prevention of antimicrobial resistance. Bull. World Health Organ. 2023, 101, 290–296. [Google Scholar] [CrossRef]
- Mendelson, M.; Lewnard, J.A.; Sharland, M.; Cook, A.; Pouwels, K.B.; Alimi, Y.; Mpundu, M.; Wesangula, E.; Weese, J.S.; Røttingen, J.-A.; et al. Ensuring progress on sustainable access to effective antibiotics at the 2024 UN General Assembly: A target-based approach. Lancet 2024, 403, 2551–2564. [Google Scholar] [CrossRef]
- Godman, B.; Egwuenu, A.; Haque, M.; Malande, O.O.; Schellack, N.; Kumar, S.; Saleem, Z.; Sneddon, J.; Hoxha, I.; Islam, S.; et al. Strategies to Improve Antimicrobial Utilization with a Special Focus on Developing Countries. Life 2021, 11, 528. [Google Scholar] [CrossRef] [PubMed]
- Moriyama, Y.; Ishikane, M.; Mezaki, K.; Ohmagari, N. Comparison of penicillins (penicillin G and ampicillin) and cefazolin as a definitive therapy against penicillin-susceptible Staphylococcus aureus (PSSA) bacteremia in Japan: A retrospective cohort study. J. Infect. Chemother. 2020, 26, 358–362. [Google Scholar] [CrossRef]
- Meesters, K.; Alemayehu, T.; Benou, S.; Buonsenso, D.; Decloedt, E.H.; Pillay-Fuentes Lorente, V.; Downes, K.J.; Allegaert, K. Pharmacokinetics of Antimicrobials in Children with Emphasis on Challenges Faced by Low and Middle Income Countries, a Clinical Review. Antibiotics 2023, 12, 17. [Google Scholar] [CrossRef]
- Bos, J.C.; van Hest, R.M.; Mistício, M.C.; Nunguiane, G.; Lang, C.N.; Beirão, J.C.; Mathôt, R.A.A.; Prins, J.M. Pharmacokinetics and Pharmacodynamic Target Attainment of Benzylpenicillin in an Adult Severely Ill Sub-Saharan African Patient Population. Clin. Infect. Dis. 2017, 66, 1261–1269. [Google Scholar] [CrossRef]
- Williams, P.; Cotta, M.O.; Tabah, A.; Sandaradura, I.; Kanji, S.; Scheetz, M.H.; Imani, S.; Elhadi, M.; Pardos, S.L.; Schellack, N.; et al. Antimicrobial therapeutic drug monitoring in critically ill adult patients—An international perspective on access, utilisation, and barriers. Int. J. Antimicrob. Agents 2024, 64, 107192. [Google Scholar] [CrossRef]
- Lagacé-Wiens, P.R.S.; Adam, H.J.; Poutanen, S.; Baxter, M.R.; Denisuik, A.J.; Golden, A.R.; Nichol, K.A.; Walkty, A.; Karlowsky, J.A.; Mulvey, M.R.; et al. Trends in antimicrobial resistance over 10 years among key bacterial pathogens from Canadian hospitals: Results of the CANWARD study 2007–16. J. Antimicrob. Chemother. 2019, 74 (Suppl. S4), iv22–iv31. [Google Scholar] [CrossRef]
- Tomczyk, S.; Taylor, A.; Brown, A.; de Kraker, M.E.A.; El-Saed, A.; Alshamrani, M.; Hendriksen, R.S.; Jacob, M.; Löfmark, S.; Perovic, O.; et al. Impact of the COVID-19 pandemic on the surveillance, prevention and control of antimicrobial resistance: A global survey. J. Antimicrob. Chemother. 2021, 76, 3045–3058. [Google Scholar] [CrossRef]
- Hindler, J.F.; Simner, P.J.; Abbott, A.; Benahmed, F.H.; Bhowmick, T.; Das, S.; Erdman, S.M.; Ferrell, A.L.; Johnson, K.; Lubbers, B.V. Analysis and Presentation of Cumulative Antimicrobial Susceptibility Test Data; Clinical and Laboratory Standards Institute: Malvern, PA, USA, 2022. [Google Scholar]
- Centres for Disease Control & Prevention. National Healthcare Safety Network (NHSN) Patient Safety Component Manual; Centres for Disease Control & Prevention: Atlanta, GA, USA, 2019. [Google Scholar]
- Magiorakos, A.P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef] [PubMed]
- Sirijatuphat, R.; Sripanidkulchai, K.; Boonyasiri, A.; Rattanaumpawan, P.; Supapueng, O.; Kiratisin, P.; Thamlikitkul, V. Implementation of global antimicrobial resistance surveillance system (GLASS) in patients with bacteremia. PLoS ONE 2018, 13, e0190132. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Manual for Laboratory Diagnosis of Bacterial Infections in Humans: Enterobacteriaceae and Other Gram-Negative Rods; WHO: Geneva, Switzerland, 2003. [Google Scholar]
- O’Hara, C.M. Manual and Automated Instrumentation for Identification of Enterobacteriaceae and Other Aerobic Gram-Negative Bacilli. Clin. Microbiol. Rev. 2005, 18, 147–162. [Google Scholar] [CrossRef] [PubMed]
- Humphries, R.; Bobenchik, A.M.; Hindler, J.A.; Schuetz, A.N. Overview of Changes to the Clinical and Laboratory Standards Institute, Performance Standards for Antimicrobial Susceptibility Testing, M100, 31st Edition. J. Clin. Microbiol. 2021, 59, 10–1128. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Global Antimicrobial Resistance Surveillance System; WHO: Geneva, Switzerland, 2015; Volume 36. [Google Scholar]
- Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing. In CLSI Supplement M100, 33rd ed.; Clinical and Laboratory Standards Institute (CLSI): Malvern, PA, USA, 2023. [Google Scholar]

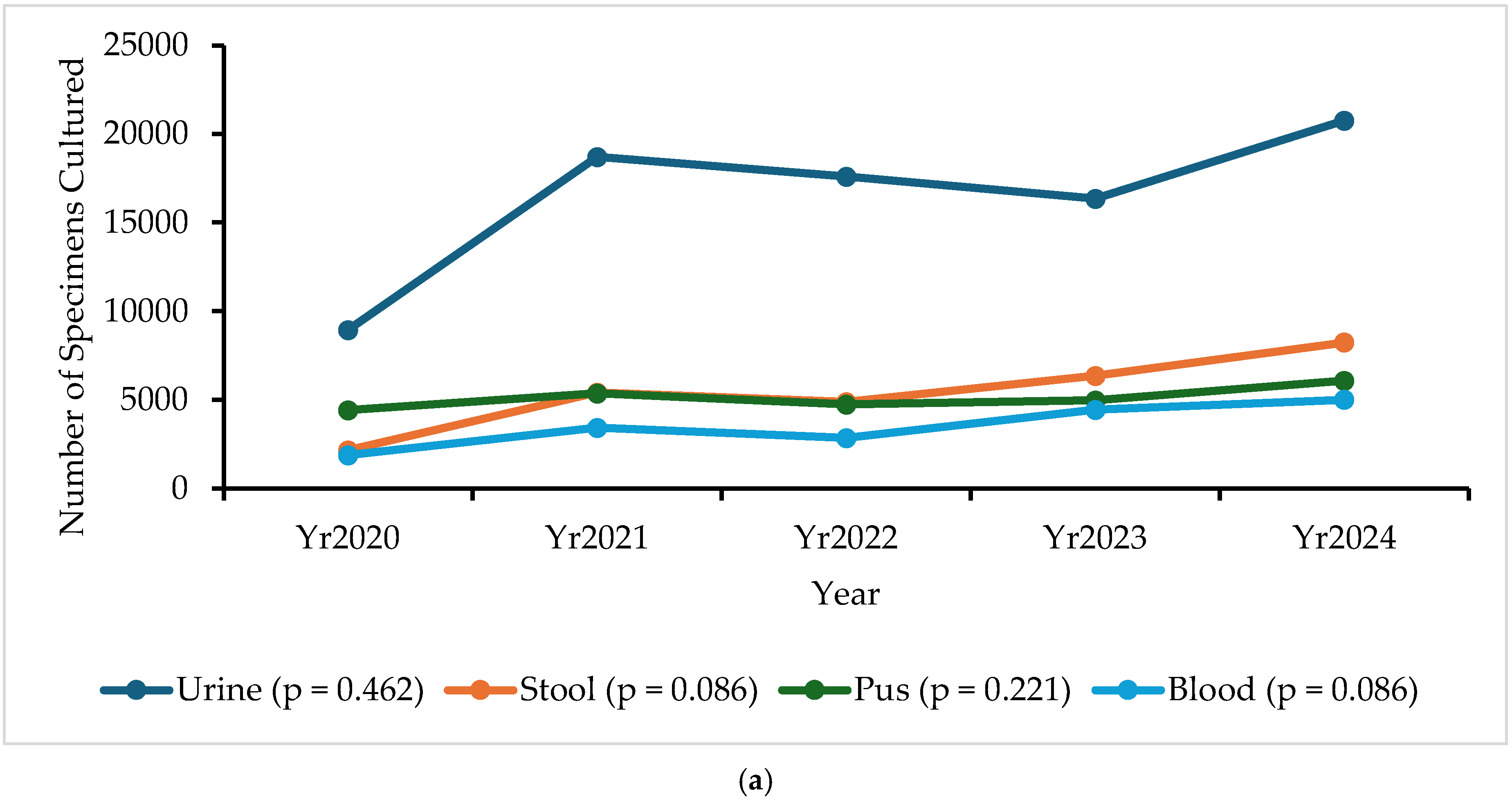
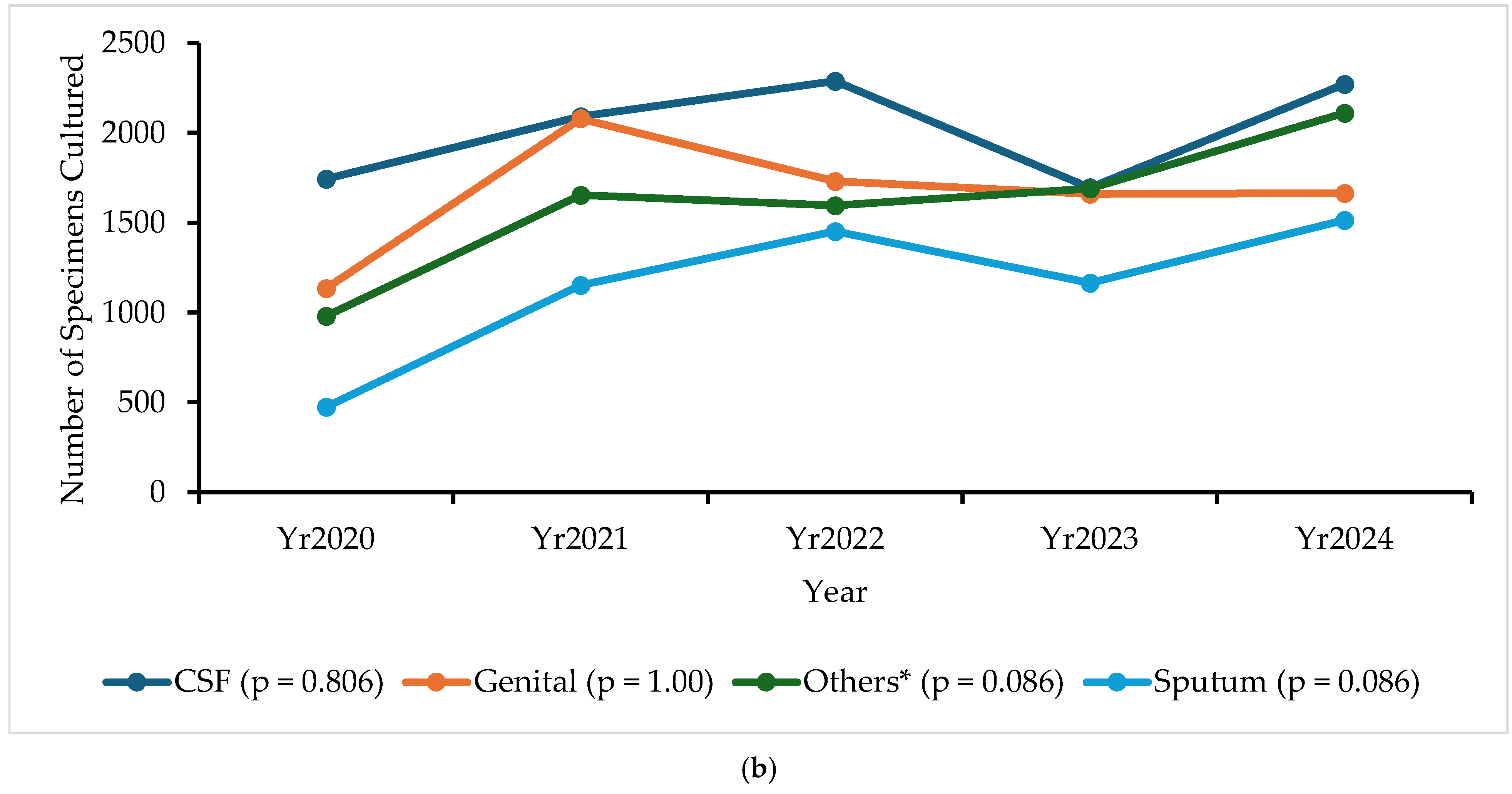
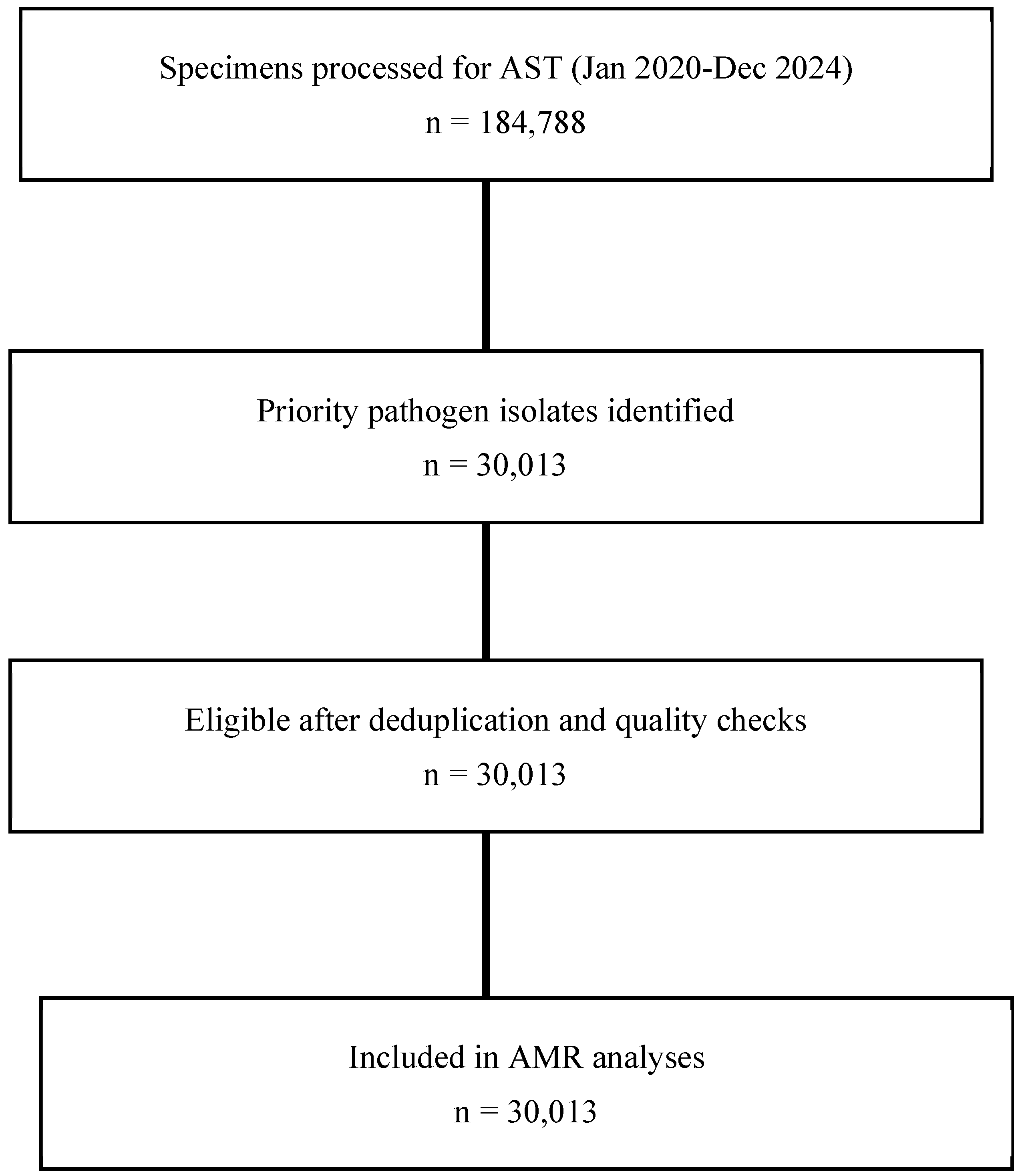
| Sentinel Site | Total Specimens Cultured, n (%) | Total | ||||
|---|---|---|---|---|---|---|
| 2020 | 2021 | 2022 | 2023 | 2024 | ||
| UTHs | 11,719 (53.8) | 23,329 (58.4) | 19,335 (52.1) | 16,265 (42.4) | 21,798 (45.7) | 92,443 |
| NTH | 4673 (21.5) | 5449 (13.7) | 5504 (14.2) | 5988 (15.6) | 8296 (17.4) | 29,910 |
| LCH | 2556 (11.7) | 4401 (11.0) | 4937 (13.3) | 5256 (13.7) | 5442 (11.4) | 22,592 |
| MGH | 1253 (5.8) | 2601 (6.5) | 3070 (8.3) | 3618 (9.4) | 4310 (9.0) | 14,852 |
| CCH | 840 (3.9) | 2535 (6.4) | 2996 (8.1) | 3242 (8.5) | 3138 (6.6) | 12,751 |
| CMH | 633 (2.9) | 794 (2.0) | 651 (1.8) | 2454 (6.4) | 3039 (6.4) | 7571 |
| LGH | 82 (0.4) | 793 (2.0) | 636 (1.7) | 1509 (3.9) | 1649 (3.5) | 4669 |
| Grand Total | 21,756 (100) | 39,899 (100) | 37,129 (100) | 38,332 (100) | 47,672 (100) | 184,788 |
| Organisms | Total | Period By Years (2020 to 2024) | p-Value of Trend per Organism | ||||
|---|---|---|---|---|---|---|---|
| Year 2020 | Year 2021 | Year 2022 | Year 2023 | Year 2024 | |||
| Escherichia coli | 7766 | 825 | 1275 | 1638 | 1771 | 2257 | 0.003 |
| Staphylococcus aureus | 4854 | 754 | 1111 | 924 | 982 | 1083 | 0.0001 |
| Klebsiella pneumoniae | 4517 | 455 | 819 | 855 | 963 | 1425 | 0.004 |
| Enterobacter sp. | 4409 | 598 | 977 | 1008 | 774 | 1052 | <0.0001 |
| Enterococcus sp. | 2790 | 232 | 501 | 595 | 609 | 853 | 0.005 |
| Acinetobacter sp. | 1655 | 178 | 296 | 303 | 359 | 519 | 0.004 |
| Pseudomonas aeruginosa | 1443 | 243 | 340 | 246 | 232 | 382 | 0.001 |
| Candida albicans | 723 | 107 | 157 | 71 | 113 | 275 | 0.015 |
| Salmonella sp. | 564 | 30 | 131 | 84 | 155 | 164 | 0.011 |
| Vibrio cholerae | 537 | 9 | 117 | 411 | 0.275 | ||
| Shigella sp. | 395 | 28 | 70 | 70 | 87 | 140 | 0.012 |
| Streptococcus pneumoniae | 237 | 27 | 42 | 33 | 52 | 83 | 0.009 |
| Neisseria gonorrhoeae | 85 | 15 | 25 | 14 | 20 | 11 | 0.002 |
| Haemophilus influenzae | 29 | 8 | 1 | 3 | 8 | 9 | 0.022 |
| Neisseria meningitidis | 9 | 4 | 4 | 1 | 0.095 | ||
| Antibiotic Name | WHO AWaRe Classification | No. of Isolates | %R | %I | %S |
|---|---|---|---|---|---|
| Oxacillin | Access | 484 | 90% | 0% | 10% |
| Ampicillin | Access | 10,989 | 82% | 4% | 14% |
| Penicillin G | Access | 8756 | 82% | 0% | 17% |
| Spectinomycin | Access | 20 | 80% | 10% | 10% |
| Cefazolin | Access | 4566 | 77% | 9% | 14% |
| Trimethoprim/Sulfamethoxazole | Access | 11,362 | 74% | 3% | 23% |
| Trimethoprim | Access | 87 | 72% | 3% | 24% |
| Azithromycin | Watch | 3879 | 63% | 4% | 33% |
| Nalidixic acid | Access | 6395 | 62% | 7% | 31% |
| Amoxicillin/Clavulanic acid | Access | 7385 | 61% | 14% | 26% |
| Cefotaxime | Watch | 6922 | 59% | 5% | 36% |
| Tetracycline | Access | 14,913 | 57% | 7% | 37% |
| Ceftriaxone | Watch | 7822 | 57% | 5% | 39% |
| Ceftazidime | Watch | 8087 | 56% | 9% | 35% |
| Cefoxitin | Watch | 6601 | 55% | 0% | 44% |
| Cefepime | Watch | 7757 | 53% | 4% | 43% |
| Amoxicillin | Access | 312 | 53% | 13% | 35% |
| Doxycycline | Access | 2585 | 52% | 8% | 40% |
| Erythromycin | Access | 10,388 | 49% | 17% | 34% |
| Quinupristin/Dalfopristin | Reserve | 1335 | 46% | 3% | 51% |
| Ciprofloxacin | Watch | 19,094 | 45% | 11% | 44% |
| Norfloxacin | Watch | 2449 | 44% | 6% | 50% |
| Gentamicin | Access | 17,000 | 40% | 9% | 50% |
| Vancomycin | Watch | 2944 | 38% | 11% | 51% |
| Clindamycin | Watch | 6132 | 31% | 15% | 54% |
| Rifampicin | Watch | 571 | 31% | 2% | 67% |
| Chloramphenicol | Access | 12,983 | 23% | 6% | 70% |
| Linezolid | Reserve | 5081 | 19% | 11% | 71% |
| Ertapenem | Watch | 3098 | 15% | 7% | 78% |
| Colistin | Reserve | 162 | 15% | 0% | 85% |
| Meropenem | Watch | 6907 | 13% | 8% | 79% |
| Imipenem | Watch | 7104 | 11% | 8% | 81% |
| Tigecycline | Reserve | 84 | 2% | 0% | 98% |
| Important Resistance Alerts | UTH | LGH | NTH | CCH | MGH | CMH | LCH | Total |
|---|---|---|---|---|---|---|---|---|
| n (%) | n (%) | n (%) | n (%) | n (%) | n (%) | n (%) | ||
| Carbapenems = non-susceptible | 1018 (44) | 93 (4) | 418 (18) | 166 (7) | 250 (11) | 35 (2) | 346 (15) | 2326 |
| Cephalosporin III = non-susceptible | 32 (28) | 1 (1) | 18 (16) | 3 (3) | 42 (37) | 3 (3) | 15 (13) | 114 |
| Fluoroquinolones = non-susceptible | 2896 (41) | 145 (2) | 170 (3) | 1200 (17) | 1449 (20) | 318 (4) | 912 (13) | 7090 |
| Imipenem = Possible decreased susceptibility | 167 (46) | 2 (1) | 110 (30) | 16 (4) | 48 (13) | 8 (2) | 14 (4) | 365 |
| Linezolid = non-susceptible | 941 (72) | 60 (5) | 41 (3) | 45 (3) | 61 (5) | 61 (5) | 89 (7) | 1298 |
| Meropenem = Possible decreased susceptibility | 87 (19) | 21 (5) | 56 (13) | 44 (10) | 57 (13) | 13 (3) | 170 (38) | 448 |
| Methicillin-resistant Staphylococcus aureus | 726 (57) | 59 (5) | 9 (1) | 231 (18) | 117 (9) | 51 (4) | 70 (6) | 1263 |
| Oxacillin = Tested by disk diffusion | 144 (61) | 15 (6) | - | - | 48 (20) | 12 (5) | 47 (20) | 236 |
| Penicillin-non-susceptible S. pneumoniae | 4 (57) | 1 (14) | - | - | 2 (29) | - | - | 7 |
| Penicillins = non-susceptible | 23 (11) | 4 (2) | 22 (11) | 11 (5) | 127 (62) | 10 (5) | 9 (4) | 206 |
| Possible ESBL-producing Enterobacterales | 2826 (36) | 140 (2) | 1204 (15) | 821 (11) | 1802 (23) | 161 (2) | 855 (11) | 7809 |
| Quinupristin/Dalfopristin = non-susceptible | 51 (60) | 5 (6) | - | 29 (34) | - | - | - | 85 |
| Vancomycin or Teicoplanin = non-susceptible | 92 (64) | - | 22 (15) | 11 (8) | 12 (8) | - | 7 (5) | 144 |
| Vancomycin or Teicoplanin = Non-susceptible (Disk diffusion) | 24 (100) | - | - | - | - | - | - | 24 |
| Vancomycin-resistant Enterococcus | 505 (65) | 2 (0.3) | 82 (11) | 74 (9) | 38 (5) | 7 (1) | 72 (9) | 780 |
| Grand Total | 11,340 (51) | 603 (3) | 2873 (13) | 3065 (14) | 4487 (20) | 788 (4) | 2907 (13) | 22,195 |
| Bacteria Isolate and Priority Antibiotics Tested | Proportion (%) of Non-Susceptible Isolates by Year | Results of the Mann-Kendall Test | ||||||
|---|---|---|---|---|---|---|---|---|
| 2020 | 2021 | 2022 | 2023 | 2024 | Kendall’s Tau | p-Value | Sen’s Slope | |
| Escherichia coli (n = 8052) | 864 | 1299 | 1645 | 1777 | 2467 | |||
| CIP (W) | 61.9 | 67.1 | 68.4 | 56.2 | 66.3 | 0.000 | 1.000 | 0.417 |
| FEP (W) | 45.4 | 52.2 | 58.6 | 47.5 | 49.9 | 0.200 | 0.806 | 0.913 |
| CTX (W) | 73.3 | 62.4 | 60.2 | 58.0 | 62.4 | −0.527 | 0.312 | −2.200 |
| CRO (W) | 60.0 | 70.0 | 53.0 | 48.3 | 57.6 | −0.400 | 0.462 | −3.700 |
| IPM (W) | 9.2 | 8.4 | 4.6 | 10.8 | 10.0 | 0.200 | 0.806 | 0.367 |
| MEM (W) | 9.7 | 20.5 | 12.2 | 32.6 | 8.8 | 0.000 | 1.000 | 0.513 |
| SXT (A) | 82.7 | 88.9 | 91.8 | 78.4 | 79.6 | −0.200 | 0.806 | −1.104 |
| NIT (A) | 30.9 | 21.6 | 19.8 | 31.3 | 24.0 | 0.000 | 1.000 | −0.796 |
| Klebsiella pneumoniae (n = 4647) | 469 | 834 | 856 | 971 | 1517 | |||
| CIP (W) | 70.7 | 77.3 | 65.4 | 70.9 | 64.8 | −0.400 | 0.462 | −2.063 |
| FEP (W) | 68.6 | 62.9 | 73.2 | 77.0 | 64.0 | 0.200 | 0.806 | 1.333 |
| CTX (W) | 75.5 | 86.8 | 74.2 | 78.6 | 74.8 | −0.200 | 0.806 | −0.413 |
| CRO (W) | 75.8 | 85.7 | 73.6 | 70.6 | 74.4 | −0.400 | 0.462 | −1.417 |
| IPM (W) | 21.6 | 23.3 | 9.5 | 11.7 | 16.8 | −0.200 | 0.806 | −1.683 |
| MEM (W) | 33.3 | 29.1 | 20.4 | 39.9 | 14.7 | −0.400 | 0.462 | −4.425 |
| SXT (A) | 78.7 | 88.8 | 85.7 | 78.6 | 80.1 | −0.200 | 0.806 | −1.417 |
| NIT (A) | 68.8 | 51.6 | 49.8 | 59.0 | 55.5 | −0.200 | 0.806 | −2.533 |
| Acinetobacter sp. (n = 1709) | 179 | 297 | 305 | 364 | 564 | |||
| CIP (W) | 57.6 | 58.0 | 56.5 | 54.7 | 53.5 | −0.800 | 0.086 | −1.350 |
| FEP (W) | 74.2 | 57.4 | 53.8 | 51.6 | 60.2 | −0.400 | 0.462 | −3.200 |
| CTX (W) | 83.3 | 70.4 | 75.9 | 81.7 | 77.8 | 0.000 | 1.000 | 0.208 |
| CRO (W) | 70.0 | 79.4 | 63.0 | 83.3 | 78.9 | 0.200 | 0.806 | 2.088 |
| CAZ (W) | 70.9 | 74.3 | 76.1 | 76.4 | 65.0 | 0.200 | 0.806 | 0.675 |
| GEN (A) | 53.4 | 52.6 | 38.0 | 52.1 | 45.5 | −0.600 | 0.221 | −1.388 |
| IPM (W) | 31.4 | 19.5 | 18.9 | 25.0 | 20.0 | −0.200 | 0.806 | −1.367 |
| MEM (W) | 0.0 | 26.1 | 25.0 | 42.6 | 28.2 | 0.600 | 0.221 | 7.650 |
| TCY (A) | 71.4 | 65.6 | 30.0 | 47.3 | 55.3 | −0.400 | 0.462 | −4.913 |
| SXT (A) | 83.8 | 74.8 | 71.4 | 57.5 | 65.7 | −0.800 | 0.086 | −5.363 |
| Pseudomonas aeruginosa (n = 1488) | 253 | 343 | 247 | 238 | 407 | |||
| CIP (W) | 24.2 | 28.3 | 18.6 | 21.5 | 25.6 | 0.000 | 1.000 | −0.275 |
| FEP (W) | 30.4 | 41.3 | 29.2 | 29.3 | 35.9 | 0.000 | 1.000 | −0.133 |
| CAZ (W) | 56.6 | 35.0 | 45.7 | 47.5 | 29.7 | −0.400 | 0.462 | −4.242 |
| GEN (A) | 45.4 | 38.1 | 35.3 | 18.0 | 34.8 | −0.800 | 0.086 | −3.925 |
| IPM (W) | 11.6 | 9.3 | 0.0 | 15.0 | 9.6 | 0.000 | 1.000 | −0.200 |
| MEM (W) | 7.7 | 21.1 | 28.6 | 25.0 | 17.3 | 0.200 | 0.806 | 2.175 |
| AMK (A) | 0.0 | 18.4 | 8.3 | 16.2 | 12.9 | 0.200 | 0.806 | 2.763 |
| TZP (W) | 49.3 | 36.0 | 21.4 | 18.2 | 26.0 | −0.600 | 0.221 | −7.363 |
| Salmonella sp. (n = 547) | 30 | 119 | 73 | 156 | 169 | |||
| AMP (A) | 100.0 | 62.3 | 70.6 | 67.4 | 56.3 | −0.600 | 0.221 | −9.008 |
| AZM (W) | 0.0 | 5.3 | 0.0 | 7.1 | 0.0 | 0.120 | 1.000 | 0.000 |
| CIP (W) | 100.0 | 66.7 | 40.5 | 37.5 | 57.9 | −0.600 | 0.221 | −12.563 |
| CTX (W) | 50.0 | 16.0 | 14.3 | 10.7 | 25.6 | −0.400 | 0.462 | −3.125 |
| CRO (W) | 33.3 | 20.0 | 25.0 | 16.0 | 9.2 | −0.800 | 0.086 | −5.896 |
| IPM (W) | 0.0 | 7.4 | 5.0 | 9.1 | 21.1 | 0.800 | 0.086 | 4.333 |
| TCY (A) | 60.0 | 22.4 | 60.0 | 35.8 | 40.0 | −0.105 | 1.000 | −2.500 |
| SXT (A) | 57.1 | 60.0 | 45.0 | 53.3 | 49.2 | −0.400 | 0.462 | −2.663 |
| Shigella sp. (n = 401) | 30 | 70 | 70 | 88 | 143 | |||
| AMP (A) | 69.2 | 55.6 | 60.7 | 76.0 | 81.0 | 0.600 | 0.221 | 5.050 |
| AZM (W) | 0.0 | 22.9 | 24.1 | 35.7 | 37.0 | 1.000 | 0.027 * | 7.850 |
| CIP (W) | 25.0 | 28.9 | 32.4 | 30.8 | 28.0 | 0.200 | 0.806 | 0.850 |
| CTX (W) | 0.0 | 19.2 | 11.1 | 8.3 | 11.5 | 0.200 | 0.806 | 1.483 |
| CRO (W) | 50.0 | 6.7 | 12.5 | 7.4 | 8.5 | −0.200 | 0.806 | −3.550 |
| IPM (W) | 0.0 | 16.7 | 10.5 | 16.7 | 6.7 | 0.105 | 1.000 | 0.838 |
| TCY (A) | 90.9 | 76.7 | 53.8 | 82.4 | 76.7 | −0.316 | 0.613 | −3.192 |
| SXT (A) | 75.0 | 85.2 | 100.0 | 88.2 | 85.7 | 0.400 | 0.462 | 2.088 |
| Enterobacter sp. (n = 4481) | 615 | 982 | 1019 | 776 | 1089 | |||
| CIP (W) | 63.6 | 60.2 | 57.4 | 65.1 | 65.4 | 0.400 | 0.462 | 0.475 |
| FEP (W) | 50.5 | 51.9 | 47.3 | 58.1 | 47.3 | −0.105 | 1.000 | −0.400 |
| CTX (W) | 56.9 | 66.3 | 67.0 | 70.3 | 65.2 | 0.400 | 0.462 | 2.038 |
| CRO (W) | 70.5 | 65.6 | 59.4 | 62.3 | 66.3 | −0.200 | 0.806 | −1.350 |
| CAZ (W) | 69.8 | 64.3 | 65.2 | 59.0 | 68.3 | −0.200 | 0.806 | −1.338 |
| IPM (W) | 17.7 | 21.0 | 13.2 | 13.1 | 16.5 | −0.400 | 0.462 | −0.900 |
| MEM (W) | 6.7 | 18.1 | 12.7 | 33.1 | 14.8 | 0.400 | 0.462 | 2.513 |
| TCY (A) | 65.5 | 66.1 | 79.5 | 62.7 | 68.8 | 0.200 | 0.806 | 0.712 |
| SXT (A) | 77.6 | 82.1 | 81.9 | 74.4 | 77.9 | −0.200 | 0.806 | −0.633 |
| NIT (A) | 40.7 | 29.5 | 26.3 | 34.6 | 36.2 | 0.000 | 1.000 | 0.238 |
| Staphylococcus aureus (n = 4984) | 781 | 1117 | 928 | 1005 | 1153 | |||
| PEN (A) | 93.2 | 91.4 | 90.4 | 93.5 | 93.0 | 0.000 | 1.000 | 0.025 |
| CIP (W) | 39.3 | 41.9 | 35.4 | 29.7 | 36.5 | −0.400 | 0.462 | −1.875 |
| FOX (W) | 42.5 | 44.4 | 33.2 | 37.1 | 39.6 | −0.200 | 0.806 | −1.163 |
| LNZ (R) | 48.9 | 45.5 | 17.1 | 20.5 | 38.3 | −0.400 | 0.462 | −3.025 |
| TCY (A) | 36.4 | 44.2 | 48.7 | 30 | 28.8 | −0.400 | 0.462 | −2.017 |
| SXT (A) | 73.4 | 67.1 | 58.7 | 54.3 | 42.5 | −1.000 | 0.027 * | −7.538 |
| NIT (A) | 20.8 | 7.8 | 14.3 | 3.8 | 6.5 | −0.600 | 0.221 | −3.413 |
| Enterococcus sp. (n = 2844) | 237 | 504 | 600 | 614 | 889 | |||
| AMP (A) | 44.6 | 40.1 | 48.5 | 32.9 | 32.7 | −0.600 | 0.221 | −3.288 |
| PEN (A) | 42.5 | 47.5 | 34.1 | 54.4 | 51.9 | 0.400 | 0.462 | 2.900 |
| CIP (W) | 62.9 | 67.9 | 74.8 | 58.5 | 64.8 | 0.000 | 1.000 | −0.279 |
| LNZ (R) | 31.0 | 33.1 | 19.6 | 6.4 | 27.3 | −0.400 | 0.462 | −3.817 |
| TCY (A) | 61.7 | 79.0 | 86.0 | 71.7 | 77.0 | 0.200 | 0.806 | 3.579 |
| VAN (W) | 45.6 | 50.0 | 59.0 | 55.9 | 46.0 | 0.200 | 0.806 | 1.525 |
| NIT (A) | 45.5 | 18.3 | 28.1 | 34.1 | 26.1 | −0.200 | 0.806 | −2.400 |
| Key Finding | Policy Implication | Practice Recommendation |
|---|---|---|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chizimu, J.Y.; Kalungia, A.C.; Mudenda, S.; Muhimba, Z.; Lukwesa, C.; Mufwambi, W.; Siame, A.; Mwangilwa, K.; Gardner, P.; Hangoma, J.; et al. Diagnostic Stewardship Trends and Antimicrobial Resistance Profiles of Bacteria Isolated in Zambia: A Five-Year Retrospective Study (2020–2024). Antibiotics 2025, 14, 1136. https://doi.org/10.3390/antibiotics14111136
Chizimu JY, Kalungia AC, Mudenda S, Muhimba Z, Lukwesa C, Mufwambi W, Siame A, Mwangilwa K, Gardner P, Hangoma J, et al. Diagnostic Stewardship Trends and Antimicrobial Resistance Profiles of Bacteria Isolated in Zambia: A Five-Year Retrospective Study (2020–2024). Antibiotics. 2025; 14(11):1136. https://doi.org/10.3390/antibiotics14111136
Chicago/Turabian StyleChizimu, Joseph Yamweka, Aubrey Chichonyi Kalungia, Steward Mudenda, Zoran Muhimba, Chileshe Lukwesa, Webrod Mufwambi, Amon Siame, Kelvin Mwangilwa, Priscilla Gardner, Jimmy Hangoma, and et al. 2025. "Diagnostic Stewardship Trends and Antimicrobial Resistance Profiles of Bacteria Isolated in Zambia: A Five-Year Retrospective Study (2020–2024)" Antibiotics 14, no. 11: 1136. https://doi.org/10.3390/antibiotics14111136
APA StyleChizimu, J. Y., Kalungia, A. C., Mudenda, S., Muhimba, Z., Lukwesa, C., Mufwambi, W., Siame, A., Mwangilwa, K., Gardner, P., Hangoma, J., Daka, V., Chileshe, C., Mudenda, N. B., Kasanga, M., Shawa, M., Chambaro, H., Chanda, D., Sinyawa, T., Chibwe, B., ... Chilengi, R. (2025). Diagnostic Stewardship Trends and Antimicrobial Resistance Profiles of Bacteria Isolated in Zambia: A Five-Year Retrospective Study (2020–2024). Antibiotics, 14(11), 1136. https://doi.org/10.3390/antibiotics14111136









