A Predicted Helix-Turn-Helix Core Is Critical for Bacteriophage Kil Peptide to Disrupt Escherichia coli Cell Division
Abstract
1. Introduction
2. Results
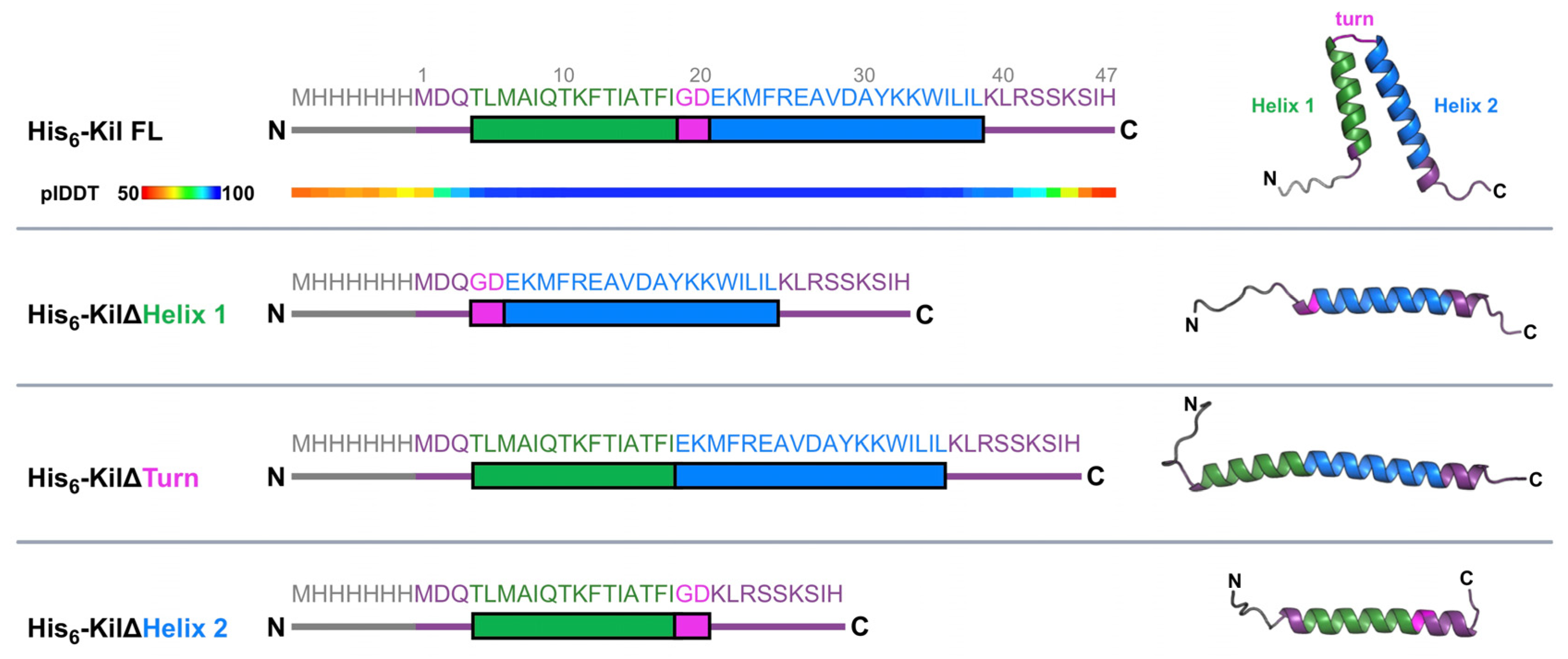
2.1. Kil Requires a Predicted Helix-Turn-Helix Structural Element for Its Anti-FtsZ Activity

2.2. Identifying the Minimal Region of Kil That Inhibits Cell Division

2.3. Kil Peptide from Enterobacteria Phage HK629 Shares a Common Mechanism to Inhibit Cytokinesis in E. coli
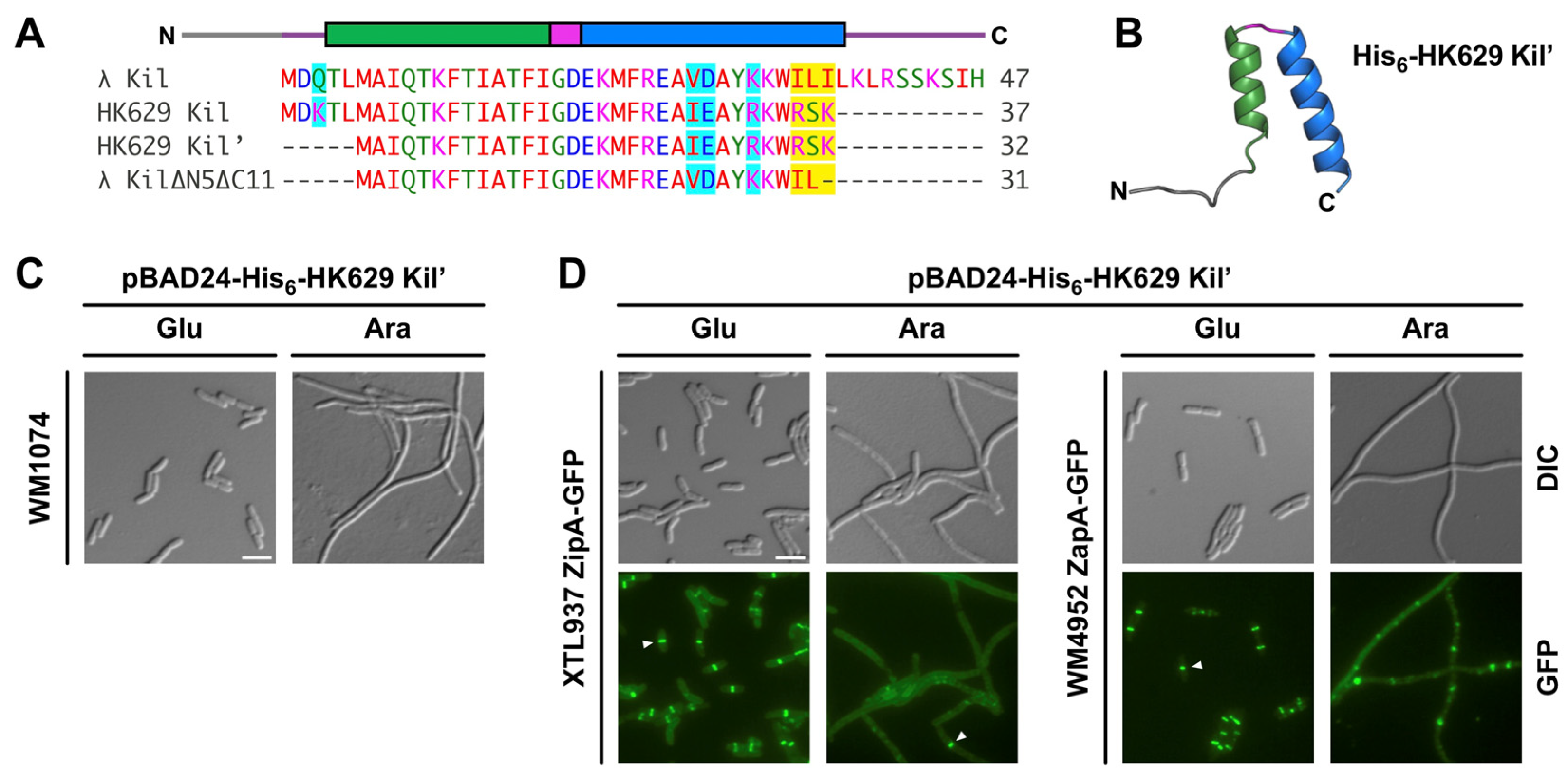

2.4. Kil Inhibits Cytokinesis of a Pathogenic E. coli Strain

3. Discussion
4. Materials and Methods
4.1. Bacterial Strains and Growth Conditions
4.2. Plasmid Construction
4.3. Microscopy
4.4. Immunoblot Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Errington, J.; Daniel, R.A.; Scheffers, D.J. Cytokinesis in bacteria. Microbiol. Mol. Biol. Rev. 2003, 67, 52–65. [Google Scholar] [CrossRef]
- Du, S.; Lutkenhaus, J. At the heart of bacterial cytokinesis: The Z ring. Trends Microbiol. 2019, 27, 781–791. [Google Scholar] [CrossRef]
- Bi, E.; Lutkenhaus, J. FtsZ ring structure associated with division in Escherichia coli. Nature 1991, 354, 161–164. [Google Scholar] [CrossRef] [PubMed]
- Goehring, N.W.; Beckwith, J. Diverse paths to midcell: Assembly of the bacterial cell division machinery. Curr. Biol. 2005, 15, 514–526. [Google Scholar] [CrossRef]
- Cameron, T.A.; Margolin, W. Insights into the assembly and regulation of the bacterial divisome. Nat. Rev. Microbiol. 2024, 22, 33–45. [Google Scholar] [CrossRef] [PubMed]
- Naha, A.; Haeusser, D.P.; Margolin, W. Anchors: A way for FtsZ filaments to stay membrane bound. Mol. Microbiol. 2023, 120, 525–538. [Google Scholar] [CrossRef] [PubMed]
- Duman, R.; Ishikawa, S.; Celik, I.; Strahl, H.; Ogasawara, N.; Troc, P.; Lowe, J.; Hamoen, L.W. Structural and genetic analyses reveal the protein SepF as a new membrane anchor for the Z ring. Proc. Natl. Acad. Sci. USA 2013, 110, E4601–E4610. [Google Scholar] [CrossRef] [PubMed]
- Wenzel, M.; Celik Gulsoy, I.N.; Gao, Y.; Teng, Z.; Willemse, J.; Middelkamp, M.; van Rosmalen, M.G.M.; Larsen, P.W.B.; van der Wel, N.N.; Wuite, G.J.L.; et al. Control of septum thickness by the curvature of SepF polymers. Proc. Natl. Acad. Sci. USA 2021, 118, e2002635118. [Google Scholar] [CrossRef] [PubMed]
- Schoenemann, K.M.; Krupka, M.; Rowlett, V.W.; Distelhorst, S.L.; Hu, B.; Margolin, W. Gain-of-function variants of FtsA form diverse oligomeric structures on lipids and enhance FtsZ protofilament bundling. Mol. Microbiol. 2018, 109, 676–693. [Google Scholar] [CrossRef]
- Nierhaus, T.; McLaughlin, S.H.; Bürmann, F.; Kureisaite-Ciziene, D.; Maslen, S.L.; Skehel, J.M.; Yu, C.W.H.; Freund, S.M.V.; Funke, L.F.H.; Chin, J.W.; et al. Bacterial divisome protein FtsA forms curved antiparallel double filaments when binding to FtsN. Nat. Microbiol. 2022, 7, 1686–1701. [Google Scholar] [CrossRef] [PubMed]
- Du, S.; Park, K.-T.; Lutkenhaus, J. Oligomerization of FtsZ converts the FtsZ Tail Motif (CCTP) into a multivalent ligand with high avidity for partners ZipA and SlmA. Mol. Microbiol. 2014, 95, 173–188. [Google Scholar] [CrossRef] [PubMed]
- Hernandez-Rocamora, V.M.; Garcia-Montanes, C.; Rivas, G.; Llorca, O. Reconstitution of the Escherichia coli cell division ZipA-FtsZ complexes in nanodiscs as revealed by electron microscopy. J. Struct. Biol. 2012, 180, 531–538. [Google Scholar] [CrossRef] [PubMed]
- Hale, C.A.; Rhee, A.C.; de Boer, P.A. ZipA-induced bundling of FtsZ polymers mediated by an interaction between C-terminal domains. J. Bacteriol. 2000, 182, 5153–5166. [Google Scholar] [CrossRef] [PubMed]
- Haeusser, D.P.; Schwartz, R.L.; Smith, A.M.; Oates, M.E.; Levin, P.A. EzrA prevents aberrant cell division by modulating assembly of the cytoskeletal protein FtsZ. Mol. Microbiol. 2004, 52, 801–814. [Google Scholar] [CrossRef] [PubMed]
- Pichoff, S.; Lutkenhaus, J. Unique and overlapping roles for ZipA and FtsA in septal ring assembly in Escherichia coli. EMBO J. 2002, 21, 685–693. [Google Scholar] [CrossRef] [PubMed]
- Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 2022, 399, 629–655. [CrossRef]
- Sass, P.; Brotz-Oesterhelt, H. Bacterial cell division as a target for new antibiotics. Curr. Opin. Microbiol. 2013, 16, 522–530. [Google Scholar] [CrossRef] [PubMed]
- Battaje, R.R.; Piyush, R.; Pratap, V.; Panda, D. Models versus pathogens: How conserved is the FtsZ in bacteria? Biosci. Rep. 2023, 43, BSR20221664. [Google Scholar] [CrossRef]
- Wang, Y.-T.; Liu, L.-T.; Hou, B.; Yao, C.-M.; Wang, X.-F.; Lu, B. Recent advances in studies on FtsZ inhibitors. Biochem. Pharmacol. 2024, 230, 116551. [Google Scholar] [CrossRef] [PubMed]
- Han, H.; Wang, Z.; Li, T.; Teng, D.; Mao, R.; Hao, Y.; Yang, N.; Wang, X.; Wang, J. Recent progress of bacterial FtsZ inhibitors with a focus on peptides. FEBS J. 2021, 288, 1091–1106. [Google Scholar] [CrossRef] [PubMed]
- Hirota, Y.; Ryter, A.; Jacob, F. Thermosensitive mutants of Escherichia coli affected in the process of dna synthesis and cellular division. Cold Spring Harb. Symp. Quant. Biol. 1968, 33, 677–694. [Google Scholar] [CrossRef] [PubMed]
- Georjon, H.; Bernheim, A. The highly diverse antiphage defence systems of bacteria. Nat. Rev. Microbiol. 2023, 21, 686–700. [Google Scholar] [CrossRef]
- Conter, A.; Bouché, J.P.; Dassain, M. Identification of a new inhibitor of essential division gene ftsZ as the kil gene of defective prophage Rac. J. Bacteriol. 1996, 178, 5100–5104. [Google Scholar] [CrossRef] [PubMed]
- Kiro, R.; Molshanski-Mor, S.; Yosef, I.; Milam, S.L.; Erickson, H.P.; Qimron, U. Gene product 0.4 increases bacteriophage T7 competitiveness by inhibiting host cell division. Proc. Natl. Acad. Sci. USA 2013, 110, 19549–19554. [Google Scholar] [CrossRef]
- Haeusser, D.P.; Hoashi, M.; Weaver, A.; Brown, N.; Pan, J.; Sawitzke, J.A.; Thomason, L.C.; Court, D.L.; Margolin, W. The Kil peptide of bacteriophage lambda blocks Escherichia coli cytokinesis via ZipA-dependent inhibition of FtsZ assembly. PLoS Genet. 2014, 10, e1004217. [Google Scholar] [CrossRef]
- Ragunathan, P.T.; Vanderpool, C.K. Cryptic-prophage-encoded small protein DicB protects Escherichia coli from phage infection by inhibiting inner membrane receptor proteins. J. Bacteriol. 2019, 201, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Hernandez-Rocamora, V.M.; Alfonso, C.; Margolin, W.; Zorrilla, S.; Rivas, G. Evidence that bacteriophage lambda Kil peptide inhibits bacterial cell division by disrupting FtsZ protofilaments and sequestering protein subunits. J. Biol. Chem. 2015, 290, 20325–20335. [Google Scholar] [CrossRef]
- Abramson, J.; Adler, J.; Dunger, J.; Evans, R.; Green, T.; Pritzel, A.; Ronneberger, O.; Willmore, L.; Ballard, A.J.; Bambrick, J.; et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature 2024, 630, 493–500. [Google Scholar] [CrossRef]
- Low, H.H.; Moncrieffe, M.C.; Löwe, J. The crystal structure of ZapA and its modulation of FtsZ polymerisation. J. Mol. Biol. 2004, 341, 839–852. [Google Scholar] [CrossRef] [PubMed]
- Galli, E.; Gerdes, K. Spatial resolution of two bacterial cell division proteins: ZapA recruits ZapB to the inner face of the Z-ring. Mol. Microbiol. 2010, 76, 1514–1526. [Google Scholar] [CrossRef] [PubMed]
- Geissler, B.; Margolin, W. Evidence for functional overlap among multiple bacterial cell division proteins: Compensating for the loss of FtsK. Mol. Microbiol. 2005, 58, 596–612. [Google Scholar] [CrossRef] [PubMed]
- Flores-Mireles, A.L.; Walker, J.N.; Caparon, M.; Hultgren, S.J. Urinary tract infections: Epidemiology, mechanisms of infection and treatment options. Nat. Rev. Microbiol. 2015, 13, 269–284. [Google Scholar] [CrossRef]
- Heller, D.M.; Tavag, M.; Hochschild, A. CbtA toxin of Escherichia coli inhibits cell division and cell elongation via direct and independent interactions with FtsZ and MreB. PLoS Genet. 2017, 13, e1007007. [Google Scholar] [CrossRef]
- Dhanoa, G.; Kushnir, I.; Qimron, U.; Roper, D.I.; Sagona, A.P. Investigating the effect of bacteriophages on bacterial FtsZ localisation. Front. Cell. Infect. Microbiol. 2022, 12, 863712. [Google Scholar] [CrossRef] [PubMed]
- Geissler, B.; Elraheb, D.; Margolin, W. A gain of function mutation in ftsA bypasses the requirement for the essential cell division gene zipA in Escherichia coli. Proc. Natl. Acad. Sci. USA 2003, 100, 4197–4202. [Google Scholar] [CrossRef] [PubMed]
- Tsang, M.J.; Bernhardt, T.G. A role for the FtsQLB complex in cytokinetic ring activation revealed by an ftsL allele that accelerates division. Mol. Microbiol. 2015, 95, 925–944. [Google Scholar] [CrossRef] [PubMed]
- Guzman, L.M.; Belin, D.; Carson, M.J.; Beckwith, J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose pBAD promoter. J. Bacteriol. 1995, 177, 4121–4130. [Google Scholar] [CrossRef] [PubMed]
| Strain/Plasmid | Genotype and/or Phenotype | Reference/Source |
|---|---|---|
| UTI89 | Uropathogenic E. coli (UPEC) | [32] |
| XL1-Blue | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac [F’ proAB lacIqZ∆M15 Tn10 (Tetr)] | Cloning host, lab collection |
| XTL937 | zipA-gfp at the native locus | Xintian Li |
| WM1074 | MG1655: ilvG rpb-50 rph-1 ΔlacU169 | Lab collection |
| WM1657 | WM1074 ftsAR286W ΔzipA::aph (ΔzipA::kan) | [35] |
| WM1659 | WM1074 ftsAR286W (ftsA*) | [35] |
| WM4952 | MT78 (leuA::Tn10 ftsA°, zapA-gfp, pSC101(ts)-PR-ftsA | [36] |
| WM5017 | BL21(DE3) ftsAR286W (ftsA*) sac284-Tn10 [pBS58] ΔzipA::aph | [25] |
| ANWM1 | WM1074 pANWM1 | This study |
| ANWM2 | WM1074 pANWM2 | This study |
| ANWM3 | WM1074 pANWM3 | This study |
| ANWM4 | WM1074 pANWM4 | This study |
| ANWM5 | WM1074 pANWM5 | This study |
| ANWM6 | WM1074 pANWM6 | This study |
| ANWM7 | WM1074 pANWM7 | This study |
| ANWM8 | WM1074 pANWM8 | This study |
| ANWM9 | WM1074 pANWM9 | This study |
| ANWM10 | WM4952 pANWM1 | This study |
| ANWM11 | WM4952 pANWM2 | This study |
| ANWM12 | WM4952 pANWM3 | This study |
| ANWM15 | WM4952 pANWM4 | This study |
| ANWM16 | WM4952 pANWM5 | This study |
| ANWM17 | WM4952 pANWM7 | This study |
| ANWM18 | WM4952 pANWM8 | This study |
| ANWM19 | XTL937 pANWM1 | This study |
| ANWM20 | XTL937 pANWM5 | This study |
| ANWM21 | XTL937 pANWM7 | This study |
| ANWM22 | XTL937 pANWM8 | This study |
| ANWM23 | WM1074 pANWM9 | This study |
| ANWM24 | XTL937 pANWM9 | This study |
| ANWM25 | WM4952 pANWM9 | This study |
| ANWM26 | WM1657 pANWM9 | This study |
| ANWM27 | WM1659 pANWM9 | This study |
| ANWM28 | WM1657 pANWM1 | This study |
| ANWM29 | WM1659 pANWM1 | This study |
| ANWM30 | UTI89 pBAD24 | This study |
| ANWM31 | UTI89 pANWM1 | This study |
| ANWM32 | UTI89 pANWM9 | This study |
| WM5018 | WM5017 pDH149 | [25] |
| ANWM33 | WM5017 pANWM10 | This study |
| ANWM34 | WM5017 pANWM11 | This study |
| pBAD24 | Arabinose inducible expression vector | [37] |
| pANWM1 | pBAD24-His6-Kil (Full length Kil; 47 aa) | This study |
| pANWM2 | pBAD24-His6-KilΔHelix1 (aa 4–18 deleted) | This study |
| pANWM3 | pBAD24-His6-KilΔTurn (aa G19D20 deleted) | This study |
| pANWM4 | pBAD24-His6-KilΔHelix2 (aa 21–38 deleted) | This study |
| pANWM5 | pBAD24-His6-KilΔN5 (Kil6–47, aa 1–5 deleted) | This study |
| pANWM6 | pBAD24-His6-KilΔC5 (Kil1–42, aa 43–47 deleted) | This study |
| pANWM7 | pBAD24-His6-KilΔC11 (Kil1–36, aa 37–47 deleted) | This study |
| pANWM8 | pBAD24-His6-Kil ΔN5ΔC11 (Kil6–36, aa 1–5, 37–47 deleted) | This study |
| pANWM9 | pBAD24-His6-HK629 Kil’ (aa 6–32) | This study |
| pDH149 | pET15b- His6-FLAG-Kil | [25] |
| pANWM10 | pET15b- His6-FLAG-Kil ΔHelix1 | This study |
| pANWM11 | pET15b- His6-FLAG-Kil ΔHelix2 | This study |
| Oligonucleotide | Sequence 5′-3′ | Template | Notes |
|---|---|---|---|
| WM2688 | AGCCGGAATTCCGGATGCATCATCATCATCATCAC | pRR48-Kil, HK629Kil’ oligo | 5′ EcoRI site, Used as forward primer (Fp) for pBAD24-his6-kil and pBAD24-his6-hk629kil’ cloning |
| WM2689 | GGAACAAGCTTTTAGTGAATGCTTTTGCTTGATC | pRR48-Kil | 3′ HindIII site, used as a reverse primer (Rp) for pBAD24-his6-kil cloning |
| WM2759 | GGCGATGAAAAGATGTTTC | pANWM1, pDH149 | KilΔHelix1 SDM Fp |
| WM2760 | TTGATCCATGTGATGATG | pANWM1 | His6-KilΔHelix1 SDM Rp |
| WM2778 | TTGATCCATACGCTGGATTTTG | pDH149 | His6 -FLAG-KilΔHelix1 SDM Rp |
| WM2757 | GAAAAGATGTTTCGTGAAG | pANWM1 | KilΔTurn SDM Fp |
| WM2758 | AATAAAAGTGGCGATAGTG | pANWM1 | KilΔTurn SDM Rp |
| WM2761 | AAACTGAGATCAAGCAAAAG | pANWM1, pDH149 | KilΔHelix2 SDM Fp |
| WM2762 | ATCGCCAATAAAAGTGGC | pANWM1, pDH149 | KilΔHelix2 SDM Rp |
| WM2753 | TAAAAGCTTGGCTGTTTTG | pANWM1 | KilΔC SDM Fp |
| WM2709 | TGATCTCAGTTTCAGTATTAATATC | pANWM1 | KilΔC5 SDM Rp |
| WM2655 | TAATATCCATTTTTTATAAGCGTC | pANWM1 | KilΔC11 SDM Rp |
| WM2748 | ATGGCTATCCAGACTAAATTC | pANWM1 | His6-KilΔN5 SDM Fp |
| WM2749 | GTGATGATGATGATGATGC | pANWM1 | His6-KilΔN5 SDM Rp |
| WM2813 | GGAACAAGCTTTCATTTTGACCTCCA | HK629Kil’ oligo | 3′ HindIII site, Used along with WM2688 for pBAD24-his6-HK629kil’ cloning as Rp |
| HK629Kil’ Oligo | ggGAATTCcggATGCATCATCATCATCATCACATGGC TATCCAGACTAAATTCACTATCGCCACTTTTATTGG CGATGAAAAGATGTTTCGTGAGGCCATCGAAGCCT ACAGAAAATGGAGGTCAAAATGAaAGCTTgg | His-tagged hk629kil’ oligo synthesized, Azenta Life Sciences | |
| pBAD Forward | ATGCCATAGCATTTTTATCC | pBAD24 derived clones | Sequencing primers from Azenta Life Sciences |
| pBAD Reverse | GATTTAATCTGTATCAGG | ||
| T7 Term | GCTAGTTATTGCTCAGCGG | pET15b derived clones | Sequencing primers from Azenta Life Sciences |
| T7 | TAATACGACTCACTATAGGG |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Naha, A.; Cameron, T.A.; Margolin, W. A Predicted Helix-Turn-Helix Core Is Critical for Bacteriophage Kil Peptide to Disrupt Escherichia coli Cell Division. Antibiotics 2025, 14, 52. https://doi.org/10.3390/antibiotics14010052
Naha A, Cameron TA, Margolin W. A Predicted Helix-Turn-Helix Core Is Critical for Bacteriophage Kil Peptide to Disrupt Escherichia coli Cell Division. Antibiotics. 2025; 14(1):52. https://doi.org/10.3390/antibiotics14010052
Chicago/Turabian StyleNaha, Arindam, Todd A. Cameron, and William Margolin. 2025. "A Predicted Helix-Turn-Helix Core Is Critical for Bacteriophage Kil Peptide to Disrupt Escherichia coli Cell Division" Antibiotics 14, no. 1: 52. https://doi.org/10.3390/antibiotics14010052
APA StyleNaha, A., Cameron, T. A., & Margolin, W. (2025). A Predicted Helix-Turn-Helix Core Is Critical for Bacteriophage Kil Peptide to Disrupt Escherichia coli Cell Division. Antibiotics, 14(1), 52. https://doi.org/10.3390/antibiotics14010052







