Abstract
Sequence-type 5 (ST5) of methicillin-resistant Staphylococcus aureus (MRSA), harboring the staphylococcal chromosomal cassette mec type IV (SCCmecIV), was first detected in Portugal. It emerged as a significant cause of healthcare-associated (HA) infection in pediatric units and was hence named the pediatric clone. Another ST5 lineage, which carries SCCmecII, also prevailed in the USA and Japan for multiple years. More recently, another MRSA lineage, ST105-SCCmecII, part of the evolution of clonal complex 5 (CC5) MRSA, has emerged as the cause of hospital-acquired bloodstream infection outbreaks in countries including Portugal, the USA, and Brazil. This article reviews studies on the epidemiology and evolution of these newly emerging pathogens. To this end, a search of PUBMED from inception to 2024 was performed to find articles reporting the occurrence of ST105 MRSA in epidemiologic studies. A second search was performed to find studies on MRSA, CC5, ST5, and SCCmecII. A search of PUBMED from 1999 to 2024 was also performed to identify studies on the genomics and evolution of ST5, CC5, and ST105 MRSA. Further studies were identified by analyzing the references of the previously selected articles from PUBMED. Most articles on ST105 MRSA were included in this review. Only articles written in English were included. Furthermore, only studies that used a reliable genotyping method (e.g., whole genome sequencing, or MLST) to classify the CC5 lineages were selected. The quality and selection of articles were based on the consensus assessment of the three authors in independent evaluations. In conclusion, ST105-SCCmecII is an emerging MRSA in several countries, being the second/third most important CC5 lineage, with a relatively high frequency in bloodstream infections. Of concern is the increased mortality from BSI in patients older than 15 years and the higher prevalence of ST105-SCCmecII in the blood of patients older than 60 years reported in some studies.
1. Introduction
Methicillin-resistant Staphylococcus aureus (MRSA) poses a significant public health challenge due to its high morbidity and mortality rates, extended hospital stays, healthcare expenses, and limited therapeutic options for serious and disseminated infections [1]. The death toll attributed to MRSA infections is alarming. In 2019, a study spanning 204 countries and territories estimated that MRSA caused 100,000 deaths and 3.5 million DALYs (disability-adjusted life years) [2]. MRSA is a burden even in wealthier nations like the United States. According to the 2019 Antibiotic Resistance Threats Report from the Centers for Disease Control and Prevention [3], the estimated number of hospitalized MRSA patients reached 323,700 in 2017. This accounted for approximately 10,600 deaths and an estimated USD 1.7 billion in healthcare costs that year. Although the estimated number of cases has decreased from 2012 (401,000) to 2017, it is still very high [3]. Furthermore, the rate (cases per 100,000 US population) of invasive MRSA infection for 2020 is similar to the rate for 2017 (20.1 and 20.7, respectively) [4,5]. In Latin American countries, the percentage of MRSA in bloodstream infections (BSIs) was roughly between 40 and 60% of total S. aureus isolates [6,7].
A limited number of MRSA strains are responsible for both healthcare-associated (HA) and community-associated (CA) infections. The molecular epidemiology studies of MRSA typically rely on defining lineages or clones using multilocus sequence typing (MLST), which groups strains into sequence types (STs) and clonal complexes (CCs) (https://pubmlst.org/, accessed on 23 August 2024), and staphylococcal cassette chromosome typing (SCCmec), which groups strains based on variations in the SCCmec (https://cge.food.dtu.dk/services/SCCmecFinder/, accessed on 23 August 2024). MRSA strains within the same lineage (ST-SCCmec) often share many more features than just SCCmec type and ST type separately. These lineages may also share a preferred environment to cause infection [e.g., community (ST8(CC8)-SCCmecIV related to USA300) and hospital settings [ST239(CC8)-SCCmecIII/related to Brazilian clone)] or a specific host [e.g., ST398(CC398)-SCCmecIV, associated with livestock animals (LA)]. Also, it is worth noting that MRSA lineages may share a similar antimicrobial resistance profile. For example, most ST239-SCCmecIII strains display high-level multidrug resistance, while most ST8-SCCmecIV strains are more susceptible to non-ß-lactam antibiotics [8,9,10,11]. However, these barriers can be overcome. For example, CA-MRSA can cause HA infections [8], and livestock MRSA (LA-MRSA) can infect humans [12]. Bacteremia is often observed in HA infections, frequently caused by typical HA-MRSA strains, while CA-MRSA is commonly associated with skin/soft tissue infections (SSTI) and other non-bloodstream infections [11,13]. However, severe infections can occasionally be associated with CA-MRSA strains, including necrotizing pneumonia and necrotizing fasciitis caused by Panton–Valentine-producing MRSA [14].
MRSA was initially classified into clones based on PFGE (pulsed-field gel electrophoresis) patterns, named according to the geographic region of isolation such as the Brazilian clone (ST239-SCCmecIII), New York/Japan clone (ST5(CC5)-SCCmecII), and Southwest Pacific clone (ST30(CC30)-SCCmecIV), or the locale they frequently caused infections, like the pediatric clone (ST5-SCCmecIV) [15,16,17,18]. However, the recent use of whole-genome sequencing (WGS) strategies and phylogenomic analysis has shown that MRSA isolates within the same MLST, SCCmec type, and even those sharing the same spa type (http://spaserver.ridom.de/, accessed on 23 August 2024) can cluster into unique phylogenetic clades [7,9,17,18]. Therefore, these should be regarded as separate clones. For instance, the CA-MRSA clone USA400 [ST1(CC1)-SCCmecIV] disseminated in the USA exhibits notable genetic and epidemiological differences compared to ST1-SCCmecIV MRSA strains observed in Brazil. ST1 from Brazil (ST1-BR) is associated with HA-MRSA infections and lacks lukSF genes for Panton–Valentine leucocidin (PVL), while USA400 is linked with CA-MRSA infections and carries lukSF genes. The genomes of USA400 and ST1-BR have been categorized into distinct phylogenetic clades, implying no direct relation [19]. Similarly, the CA-MRSA USA300 prevalent in North America (USA300-NA) belongs to a different phylogenetic clone than the USA300 variant (USA300-LA) disseminated in Colombia and other Latin American countries. These variants cluster in two dominant clades that vary by geographical region [20]. Hence, recent genomic epidemiology studies of MRSA suggest that WGS is the most precise technique for identifying MRSA-specific clones within the same lineage [9,11,19,20,21].
ST5-SCCmecII strains have spread beyond the borders of the USA and Japan, becoming more prevalent in various regions globally. The most common CC5 lineages are ST5-SCCmecII, associated with the New York/Japan clone, and ST5-SCCmecIV, linked to the pediatric clone [11,22,23,24]. The relatively recent diversification of ST105(CC5)-SCCmecII MRSA [21], its occurrence in different countries, its prevalence in bloodstream outbreaks in hospitals of industrialized countries, its extensive presence in BSI in the metropolitan area of Rio de Janeiro, Brazil [11], and the fact that its clinical importance may be underestimated due to the relatively expensive and time-consuming methods for MRSA genotyping [11,21] prompted us to write this review. In this article, we illustrate the global distribution of ST105-SCCmecII strains, highlight significant milestones in this MRSA strain’s genomic evolution, and discuss the possible threat this lineage presents to human health. Table 1 summarizes the most common MRSA lineages/clones, their molecular classification, and main geographical spread locations.

Table 1.
Methicillin-resistant Staphylococcus aureus lineages/clones common names and their most frequent geographical spread location.
2. The Clonal Complex 5
The clonal complex 5 (CC5) has an interesting historical trajectory. A study investigating the molecular chronology of CC5 strains in the Western Hemisphere estimated that the rise of this clonal complex began in the early 1960s/1970s, with the subsequent spread of ST5-SCCmecI in South America, particularly the Chilean-Cordobes clone. Conversely, ST5-SCCmecII spread throughout Central and North America [21]. ST5-SCCmecII MRSA, also known as the New York/Japan clone and USA100 clone, was first identified in the New York area in the late 1990s [18,25,26]. Later, a study examining 12 hospitals in seven US states revealed that the USA100 clone had spread beyond just New York [27]. In the early 2000s, the ST5-SCCmecII MRSA strains were also observed to be widespread in hospitals in Japan [28]. As a result, the clone initially named New York was renamed New York/Japan. Additionally, a systematic review of bacterial resistance within infections occurring in intensive care units (ICUs) highlighted the dominance of a few bacterial clones with multidrug-resistant phenotypes, with CC5-SCCmecII (USA100) still frequently circulating in US hospitals [29].
Since it was first detected in a Lisbon-based pediatric hospital in Portugal in 1992, strains of ST5-SCCmecIV (associated with the pediatric clone and USA800 clone) have circulated across various countries and continents [11,18,30,31,32]. This MRSA lineage is often found in young children, leading to its label as the pediatric clone [11,30,33,34]. Despite its capacity to provoke disseminated infections in adults, it is more repeatedly identified as a causative agent in infections in regions beyond the bloodstream, according to a study performed in Rio de Janeiro, Brazil [11]. However, in Spain, adult BSI was primarily attributed to CC5-SCCmecIV isolates [35].
In summary, these findings affirm that MRSA isolates in the same lineage can exhibit different epidemiological characteristics, despite having numerous shared genomic features. This implies that minor genomic alterations could potentially lead to differences in the epidemiological landscape.
In addition to the most commonly seen SCCmec types (II and IV), ST5 strains may also carry other SCCmec types, such as I and V [21]. For instance, a novel epidemic MRSA strain (ST5-SCCmecI) surfaced in Argentinean hospitals in 1999, replacing the prevailing Brazilian epidemic clone [BEC; ST239(CC8)-SCCmecIII] [36]. In 2014, an outbreak of Staphylococcal Scalded Skin Syndrome in Italy was attributed to a rare MRSA clone from the ST5-SCCmecV lineage that produced the exfoliative toxin A (ETA), as stated by Lamanna et al. (2017) [37]. The occurrence of ST5 strains carrying diverse SCCmec types and subtypes strongly suggests the multiple SCCmec acquisitions by ST5 strains [38,39].
Interestingly, the first vancomycin-resistant S. aureus (VRSA) strain was found in Michigan, USA, in 2002 [40,41]. Since then, approximately 52 VRSA strains carrying van genes have been reported. Many of these isolates belong to CC5 [42,43,44]. In the USA, at least 13 of the 16 reported VRSA belong to this clonal complex. Although the prevalence of the CC5 background among VRSA strains is well documented, it is not yet understood why this is the case [21]. VRSA isolates are not solely confined to CC5; a VRSA strain from CC30 has also been detected in the USA [45].
Currently, CC5 MRSA is one of the most prevalent MRSA clonal complexes worldwide. A study executed in various South American countries showed CC5-SCCmecIV isolates as the second most common MRSA isolates from BSI, as stated by Di Gregorio et al. (2023) [46]. In a tertiary hospital in Kuwait, the leading clonal complexes were CC5 (31.6%) and CC6 (15.0%). Among the CC5, the most widespread were those containing SCCmecV, accounting for 52.6% [47]. Moreover, CC5 MRSA was the dominant clonal complex causing BSI in children at a hospital in Mozambique, Africa [48]. The ST764-SCCmecII lineage, a single-nucleotide variant of ST5, has been documented as a prevalent nosocomial pathogen in Asian countries, including China [49], Japan [50], and Thailand [51].
As previously mentioned, MRSA CC5 strains are typically associated with healthcare-related infections, frequently causing serious conditions in both adults and children. However, most of these strains fall under ST5-SCCmecII and ST5-SCCmecIV. The PubMed search using the terms “ST5 MRSA” revealed that the number of publications on this topic increased from 2003 to 2013 and then plateaued until 2023. This may reflect the stable presence of these MRSA as important hospital pathogens worldwide over the years.
While other STs have been detected among CC5, ST105 is particularly noteworthy, also being linked to serious disseminated infections [11,21,52,53,54,55,56]. In the MLST scheme, ST105 is a single nucleotide variant of ST5(CC5) characterized by a single mutation in the yqiL gene [17,57]. CC5 MRSA from different lineages may be involved in quite different epidemiological scenarios. ST5-SCCmecIV has a tropism for pediatric infections, whereas ST5-SCCmecII and ST105-SCCmecII are more involved in adult diseases. Indeed, it has also been found that CC5-SCCmecII strains, mainly ST105-SCCmecII, are more involved in BSI compared to ST5-SCCmecIV in some hospitals [11,30,31]. Another interesting set of data is related to the differences in antimicrobial resistance patterns. ST5-SCCmecII and ST105-SCCmecII isolates are often multidrug-resistant bacteria, whereas ST5-SCCmecIV are more susceptible to non-β-lactam antibiotics [11]. However, the molecular basis of these differences is not fully understood, and some aspects that have driven the evolutionary path of these CC5 lineages are presented in the Section 7.
3. Epidemiological History of ST105-SCCmecII
To the best of our knowledge, ST105-SCCmecII was initially reported in North America and Europe in the late 1990s [52,58]. From January to June 1998, 17 isolates of this lineage were recovered from various infectious samples, such as blood, urine, wounds, and the respiratory tract, in two different Miami hospitals. In these institutions, ST105-SCCmecII isolates ranked third in terms of frequency among MRSA lineages, trailing ST8(CC8)-SCCmecIV (associated with USA300) and ST36(CC30)-SCCmecII (associated with EMRSA-16). In Western Switzerland, ST105 isolates triggered MRSA outbreaks from 1999 to 2004. During this period, MRSA isolates (one isolate per patient) with an identical PFGE profile to that of ST105 isolates (termed clone B) were the predominant MRSA in the region, constituting 32% of all MRSA isolates. The initial outbreak induced by clone B occurred in a tertiary hospital that had enacted strict infection control measures. In 2001, another outbreak attributed to this clone was reported, affecting several healthcare centers in the region [58].
At the Milton S. Hershey Medical Center in Philadelphia, USA, 62 MRSA isolates were obtained from nasal swabs collected from patients admitted between 2008 and 2009. Of these, 22.4% were identified as ST105-SCCmecII, comprising the second most common clone identified in the hospital. The most frequent lineage found was ST5-SCCmecII/IV, accounting for 38.8% [59]. In a separate study conducted at Penn State Hershey Medical Center, a rural hospital in Pennsylvania, between August 2009 and March 2010, a total of 94 MRSA isolates were recovered from 151 case patients (62.25%). Of these isolates, 11 were typed as ST105-SCCmecII (11.70%), making it the third most prevalent lineage, following ST5 (34 isolates) and ST8 (26 isolates) [60]. Due to the small number of hospitals and MRSA isolates analyzed in Philadelphia, it is not possible to determine whether the observed difference in ST105 rate represents a true decrease in the ST105 incidence.
David and colleagues analyzed clinical MRSA isolates from various sources, including patients’ blood from different regions of Alaska. These isolates were collected between 2000 and 2006 at the Alaska Native Medical Center in Anchorage. Out of their collection of 224 MRSA isolates, 12 were classified as ST105, accounting for 5.4% of the total isolates [53]. ST105 is not only present in Alaska and Pennsylvania, but it is also found in other regions of the USA. ST105-SCCmecII has also been reported to cause severe and invasive outbreaks at Mont Sinai Hospital in New York [61].
Also, in a study involving seven hospitals in Brooklyn, New York, Iregui et al. (2019) [62] found that 14 out of 348 MRSA analyzed were ST105 (4.0%). In another study conducted in Iowa, USA, Fischer et al. (2020) [63] investigated the prevalence of various STs of S. aureus among patients with cystic fibrosis. Out of the 98 MRSA collected from 75 patients, ST5 was the most common MLST type (n = 54; 55.1%), followed by ST105 (n = 14; 14.3%). Similar results were obtained in a subsequent study by the same authors in 2021 with cystic fibrosis patients in Iowa [64].
ST105 MRSA has also been reported on the Pacific Coast of the USA. These isolates were acquired from clinical samples, such as blood, taken from inpatients at 30 different hospitals in Orange County, California, between October 2008 and April 2010. The ST105-SCCmecII lineage accounted for 4% of the 284 isolates that were selected for MLST typing; this made it the third most common, falling behind ST5 (45%) related to USA100 and ST8 (CC8) related to USA300 (38%) [65].
ST105-SCCmecII has also been identified in other European countries. In 2012, a study conducted across hospitals in Northern, Central, and Southern Italy analyzed 102 MRSA isolates. The results revealed that the USA100-like clone (ST5/105-SCCmecII) ranked as the fourth most prevalent MRSA strain; ST5 comprised 13 cases and ST105 accounted for just two cases [66]. ST105-SCCmecII was found at a low frequency in a collection of 83 MRSA isolates from BSI in Athens, Greece, from 2000 to 2015. During this period, the dominant lineages were ST239-SCCmecIII and ST5-SCCmecII/IV, with only three (3.6%) ST105-SCCmecII isolates being recovered [67].
However, an increased prevalence of ST105 was observed in other European regions. Espadinha et al. analyzed the variations in the sample profile of MRSA obtained during two time periods, 1993 and 2010, and detected a clonal shift at a major tertiary teaching hospital in Portugal. By 2010, ST105 was found to be the second most frequent clone in the hospital (18.0%), trailing behind the dominant ST22-SCCmecIVh (72.0%) [68]. Furthermore, Faria et al. (2013) found that ST105-SCCmecII was the second most common MRSA in Portuguese hospitals, accounting for 19.0% [69]. Figure 1 provides a timeline summary of the major emergence events associated with ST105 MRSA strains.
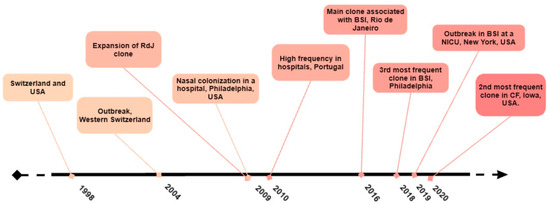
Figure 1.
Timeline showing the major emergence events of strains of the ST105-SCCmecII lineage.
A large cross-sectional study was conducted in Portugal to evaluate the carriage of S. aureus and MRSA between April 2010 and December 2012. This study involved a total of 3361 adults aged over 60 and examined both nasopharyngeal and oropharyngeal carriage of S. aureus. The results indicated the prevalence of ST105 MRSA. Most of the MRSA detected (82.3%) were related to three common HA-MRSA clones in Portugal: ST105-SCCmecII (43.5%), ST5-SCCmecIV (19.4%), and ST22-SCCmecIV (EMRSA-15 clone; 19.4%) [70]. The ST105 strain was also discovered in 2014 in Portugal, associated with SSTI, and was the main clone in this study (n = 7/34; 20.6%), followed by ST22 (n = 5/34; 14.7%) [71].
In 2010, a single ST105 MRSA isolate was reported in São Tome and Principe, Africa, from a colonization sample collected at Dr. Ayres Menezes Hospital. It has been suggested this isolate may originate from Portugal, a country where this clone is prevalent and maintains a demographic relationship with São Tome and Principe through tourism and patient transfers [72]. Evidence of international spread of isolates of the ST105 lineage has also been reported in genomic studies, where directly related strains from the USA and Brazil were grouped in the same phylogenetic clade [11].
A study conducted in Kuwait, Western Asia, analyzed 400 MRSA isolates from clinical samples collected between 1992 and 2010 from 13 hospitals. The study found the ST105-SCCmecII-t002 MRSA was first detected in 2010 at a low frequency of 0.5% [73]. Among a HA-MRSA group analyzed between 2012 and 2017 in China, only three isolates of ST105-SCCmecII-t688 (representing 2.5%) were found. The majority of the identified MRSA was ST239-SCCmecIII (at 61.7%) [74]. These data suggest that ST105 is still uncommon in some continents, including Asia.
ST105 MRSA has been detected in both Central and South America. Studies analyzing 386 MRSA genomes from the American continent, collected from BSI samples from 2011 to 2018, revealed that ST105 isolates represented 17.9% of those samples [75]. In a separate prospective study that was carried out in nine Latin American countries between January 2011 and July 2014 and included S. aureus strains, it was discovered that the ST5-SCCmecII strain was widespread (>80%) in Mexico, Guatemala, and Brazil. However, in Colombia and Ecuador, over 70% of MRSA strains were classified as USA300-LV/LAE, demonstrating a diversity of strains present in the region. Out of the 96 isolates that were subjected to WGS, ST105 accounted for approximately 10% and was found in Brazil, Chile, and Colombia [6].
In a study conducted on S. aureus isolates collected between 2005 and 2013 from various clinical settings in Quito, Ecuador, it was reported that two out of 62 MRSA isolates were ST105 [76]. Martínez and colleagues used WGS to analyze 469 MRSA isolates obtained from Chilean hospitals between 1999 and 2018. They found the most widespread clone was ST5-SCCmecI, accounting for 70.1%, followed by ST105-SCCmecII at 14.9% [55].
While ST105-SCCmecII is not commonly found in certain Latin American countries, it was present in a study conducted in a São Paulo hospital from August 2010 to January 2012. The study identified nine clones of MRSA from nasal and groin swabs taken from 190 pre- and post-liver transplant patients. Out of the 69 MRSA isolates identified, 25, equivalent to 36.2%, belonged to the predominant clone, ST105-SCCmecII [77].
It is noteworthy that USA300 and EMRSA-15 isolates, which were found at higher frequencies in hospitals in the USA and Portugal, respectively, compared to ST105 strains, were also common causes of community-acquired infections, whereas ST105 strains are typically HA-MRSA. In addition, both USA300 and EMRSA-15 are generally more susceptible to non-beta-lactam antibiotics than ST105 [11,20,21,53,70,71]. This suggests that resistance characteristics alone cannot explain the prevalence of some MRSA lineages. However, the molecular mechanisms involved in the prevalence of specific MRSA lineages remain to be elucidated [19]. Figure 2 illustrates the prevalence of ST105 MRSA in different countries.
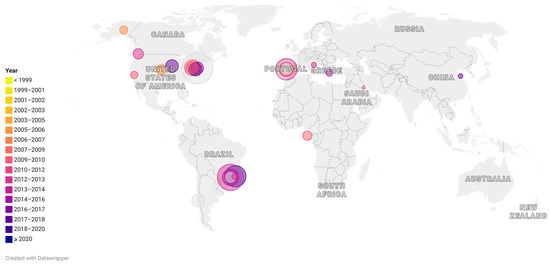
Figure 2.
Representative map showing the occurrence of Sequence-type (ST)105 around the world. Each circle represents a scientific article in which ST105 was reported. The size of the circles indicates the percentage of ST105 reported among the collection included in each study. The colors indicate the date (in years) of the sample collection. For studies performed in a large period of years, the last year of the reported period was considered.
4. ST105-SCCmecII in Bloodstream Infections
A 2019 study estimated that among 33 bacterial pathogens, just five leading pathogens were responsible for 56.2% of all sepsis-related deaths. S. aureus was the leading cause of death in 135 countries. The most deadly infectious syndrome and pathogen were associated with patient region and age, and the majority of deaths occurred in patients older than 15 years. In 2019, more than 6 million deaths were caused by three types of bacterial infections, with BSI and lower respiratory tract infections dominating with more than 2 million deaths each, followed by perineal and intra-abdominal infections, which caused more than 1 million deaths [78]. In another global study, S. aureus was second only to Escherichia coli among the top six antimicrobial-resistant pathogens, each causing approximately 50,000 to 100,000 deaths [2]. These data underscore the importance of MRSA in serious and life-threatening infections such as BSI.
Among MRSA, CC5 isolates, mainly ST5-SCCmecII, have been described in high frequency in BSI in different hospitals worldwide, and it was reported with a higher global frequency of 68% in a large medical center in Minnesota, USA [46,79,80]. As in this Minnesota study, the emerging ST105-SCCmecII has been reported as a cause of BSI with variable frequency in different European and American hospitals, generally being the second most common MRSA lineage after ST5-SCCmecII [6,11,54,55,61,69,75,81,82]. Read and colleagues analyzed 105 MRSA isolates from bloodstream infection cases in two hospitals in Philadelphia, Pennsylvania, between July 2018 and June 2019, using WGS. Of the 105 isolates, 16 belonged to ST105 (15.84%). The majority of the MRSA isolates were classified as CC8 (n = 55), with USA300 (n = 49) being the dominant strain, followed by CC5 (n = 40). ST105 was the second most common CC5 lineage [54].
Berbel Caban and collaborators (2020) traced an “under-the-radar” outbreak caused by ST105-SCCmecII-t002 involving 16 patients with bacteremia and two hospitals in New York [83]. Another survey of 132 MRSA genomes from blood isolates showed that a multi-month outbreak in a neonatal intensive care unit (NICU) at Mont Sinai Hospital in New York was caused by ST105-SCCmecII isolates. Hospital-wide genomic surveillance data traced the origin of the outbreak to 3 patients admitted to adult wards 4 months before the NICU outbreak. Genomic studies show that the ST105 strain responsible for the outbreak had unique mutations and genetic elements that affected genes related to metabolism, resistance, and persistence. Transcriptome sequencing (RNA-Seq) profiling revealed that epigenetic changes in the outbreak clone repressed agr gene expression and upregulated genes associated with stress response and biofilm formation [61]. Furthermore, Faria et al. (2013), focusing exclusively on isolates from bloodstream infections in Portuguese hospitals, identified two predominant MRSA clones: EMRSA-15, representing 75.0% of MRSA isolates, and ST105-SCCmecII, accounting for 19.0% [69].
Studies analyzing 386 MRSA genomes from the American continent, collected from BSI samples between 2011 and 2018, found that ST105 isolates represented 17.9% of these samples [75]. A large epidemiological study of 600 MRSA isolates from 51 hospitals in the Rio de Janeiro metropolitan area showed that ST105-SCCmecII was the most prevalent among BSI isolates. Of the 245 MRSA isolates from BSI, 115 (46.9%) were CC5-SCCmecII, and 73.2% of these were ST105-SCCmecII. Moreover, ST105-SCCmecII was shown to have a greater ability to evade phagocytosis compared to other CC5-SCCmecII/IV clones. As suggested by the authors, this may explain its higher prevalence in BSI [11]. Additionally, an independent study conducted at a university hospital in Rio de Janeiro documented that the majority of blood isolates (43.2%) were ST105-SCCmecII [81]. Viana et al. found a higher frequency of this lineage in patients older than 60 years compared to other MRSA lineages, such as ST5-SCCmecIV and ST30-SCCmecIV [11]. Also in Portugal, Almeida and coworkers found an increased isolation of ST105-SCCmecII in BSI in patients older than 60 years [70].
5. ST105-SCCmecII and Its Dissemination among Animals and the Environment
MRSA has been identified in domestic animals. A study conducted in 2006 explored the diversity of MRSA isolates within veterinary clinics in the Midwest and Northeastern areas of the USA. From 24 isolates, approximately half were found to match the PFGE profile USA100, correlated with clones ST5 and ST105. The study also reported the existence of other human MRSA clones, thus suggesting potential transmission from humans to animals [84].
Couto et al., in Portugal, found four strains that belonged to CC5 among animal isolates, one of which was of canine origin and belonged to ST105-SCCmecII [85]. The ST105-SCCmecII lineage has been identified as a nasal colonizer of MRSA in individuals in Portugal who are in regular contact with animals. Moreover, the study also detected the presence of the primary human healthcare clones in Portugal, including ST22-SCCmecIV and ST105-SCCmecII, along with LA-MRSA ST398-SCCmecIV [86].
In 2014, it was discovered that two urban Norway rats in Vancouver’s Downtown Eastside neighborhood were colonized by ST105 MRSA. In addition to ST105, these rats were also colonized by other clones, such as ST97 and ST398, which are frequently found in both human and livestock populations [87]. More recently, an ST105-SCCmecII isolate was identified among 16 MRSA samples taken from purulent lesions in rabbits at a Portuguese slaughterhouse [88].
Hospital wastewater often contains antibiotic-resistant bacteria, including MRSA. A study conducted with waste samples from three hospitals in northern Portugal detected MRSA isolates belonging to ST22-SCCmecIV, ST8-SCCmecIV, and ST105-SCCmecII. These findings highlight the capacity of these lineages to survive in untreated hospital effluents [89]. The environmental discharge of MRSA, including ST105-SCCmecII, brings about its detection in rats. Given the promiscuity of gene transfer in bacteria, the presence of multidrug-resistant MRSA such as ST105 isolates in the environment should be considered an ecological concern. Furthermore, the environmental spread of ST105-SCCmecII may reflect a higher frequency of this lineage in humans than has actually been reported. Table 2 shows the occurrence of ST105 MRSA in humans and animals from different countries.

Table 2.
Articles reporting the occurrence methicillin-resistant Staphylococcus aureus belonging to the sequence-type (ST) 105 from human and animal sources.
6. Antibiotic-Resistance Pattern of ST105-SCCmecII
In 2007, Mwangi et al. conducted a study exploring the mechanisms of vancomycin resistance. Isogenic S. aureus isolates were obtained from a patient’s bloodstream both before the commencement of vancomycin treatment and after the failure of the said treatment. The initial isolate, identified as JH1 [GenBank Accession number (Acc): NC_009632], showed susceptibility to vancomycin, while the final isolate, JH9 (Acc: GCA_000016805.1), had developed resistance to it. Both isolates were categorized as ST105-SCCmecII and displayed a parallel 100-fold decrease in susceptibility to daptomycin [95]. Daptomycin, a cationic antimicrobial agent, coupled with vancomycin, is considered a last resort for treating MRSA BSI [96].
There have also been reports of daptomycin-resistant ST105 isolates due to mutations in various genes related to phospholipid syntheses and metabolisms, such as mprF (multiple peptide resistance factors), cls (cardiolipin synthesis), and pgsA (phospholipid metabolism). Further, alterations in regulatory genes yycF/yycG and vraSR, related to cell membrane stress and permeability, have been implicated too. Additionally, the transcriptional regulator dltABCD’s upregulation, which boosts the surface positive charge through teichoic acid D-alanination as seen with mprF mutations, has been linked to nonsusceptibility to daptomycin [97]. However, the overall impact of this resistance profile among ST105 isolates worldwide remains undetermined.
According to a study by Melo-Cristino et al. (2013), the first case of vancomycin-resistant S. aureus (VRSA) identified in Portugal is linked to the clonal background ST105-SCCmecII [98]. Notably, the strain FCFHV36 (Acc: CP011147), reported in 2015 as an MRSA strain heterogeneously intermediate to vancomycin, was recovered from a vertebral biopsy of an osteomyelitis patient at a hospital in Santa Catarina, Brazil, and belongs to the ST105-SCCmecII lineage [99]. These studies suggest that ST105 can acquire not only the vanA gene but also increase the minimum inhibitory concentrations (MICs) of vancomycin.
Silva et al. (2020) examined the antimicrobial resistance profiles of 16 bloodstream infection (BSI) isolates from Portugal. Among these, ST105-SCCmecII isolates exhibited resistance to quinolones and an inducible macrolide-lincosamide-streptogramin B (iMLSB) phenotype [94].
Iregui et al. (2020) investigated delafloxacin-resistant S. aureus isolates in seven hospitals in Brooklyn, NY (USA) [62]. They found that 14 of the 16 delafloxacin-resistant isolates selected for MLST were ST105-MRSA, and one was ST105-MSSA (MSSA: methicillin-susceptible Staphylococcus aureus). Analysis of these isolates revealed that all were found in patients with extensive healthcare system contact, and the resistance was due to an accumulation of multiple mutations in gyrase and topoisomerase IV genes. Viana et al. (2022) showed through genomic research involving 82 ST105 isolates from 2014–2016 in Rio de Janeiro, that 100% of ST105-SCCmecII isolates were aminoglycoside-resistant [mediated by the genes aad, aph(3′), or ant(9)]; 98.7% (n = 81/82) to macrolides [erm(A), erm(C), msr(A), or mph(C)] and 50% (n = 41/82) were resistant to chloramphenicol [cat(pC221) gene] [100]. It is concerning that all tigecycline-resistant isolates collected from ICU patients in Brazil over 6 months were identified as ST105 MRSA (n = 10/36; 27.8%) [101].
ST105-SCCmecII isolates from Rio de Janeiro have been discovered to have low-level triclosan resistance, which is facilitated by the TnSha1 transposon [100,102]. Although resistance to low-dose triclosan is unlikely to affect the biocide’s effectiveness because it is typically used in high concentrations, it could potentially lead to complete resistance and the emergence of antimicrobial persisters [103].
7. Evolution of ST105-SCCmecII-t002
Challagundla et al. (2018) analyzed the genomic sequence of 598 CC5 isolates from various countries in the Western Hemisphere [21]. According to their studies, the most basal group, termed CC5-Basal, was predominantly represented by ST5-SCCmecIV and ST5 MSSA. This group also includes the genome of strain N315, which was previously reported to be related to the USA100/New York Japan clone, suggesting that strains related to N315 are not directly related with other NY/Japan-related genomes, which cluster in a different phylogenetic group. In fact, it was even proposed that the NY/Japan clone consists of two independent clones of the ST5-SCCmecII lineage [21]. The second group identified by these authors, the CC5-SCCmecI (CC5-I), likely emerged in the early 1970s. Its earliest branches represent isolates from Europe, with its expansion in South America occurring in the mid-1990s. CC5-I groups Chilean/Cordobes (ST5-SCCmecI) and South German (ST228-SCCmecI) clones, representing one of the three presumed acquisitions of SCCmecI by CC5 strains (the other two instances occurred in the CC5-Basal and CC5-IIB groups [21].
Strains from the CC5-II group predominantly hail from Central and North America, with some from South American countries like Brazil, Venezuela, and Ecuador. The CC5-II bifurcates into two subgroups: the paraphyletic basal CC5-IIA and the monophyletic terminal CC5-IIB, both of which encompass CC5-SCCmecII-carrying isolates procured at the onset of clade formation. Both subgroups contain USA100-related reference genomes (associated with the NY clone). This suggests that MRSA isolates from the NY clone are likely polyphyletic and have therefore accumulated mutations in their independent evolution [21]. This finding further bolsters the capacity of CC5 to adapt and evolve into multiple successful clones [11,21,54,55,75]. The CC5-IIA represents the early branches of ST5-II genomes from the USA and boasts a notable subclade with the ST228-II derivative of ST5 isolated in Mexico. The CC5-IIB terminal subclades signify the expansion of ST5-SCCmecII and the emergence of ST105-SCCmecII, ST225-SCCmecII, ST231-SCCmecII, and ST496-SCCmecII, isolated in both South America (specifically Chile, Brazil, and Ecuador) and North America (USA and Canada), with subsequent expansion in the USA. A distinct feature of the CC5-IIB clade is the loss of the sep gene encoding the staphylococcal enterotoxin P [21]. According to molecular chronology studies, it is estimated that ST105-SCCmecII originated in the early 1980s, several years after the specialization of ST5-SCCmecII [11,21]. Table 3 describes the major events in the evolution of clonal complex 5.

Table 3.
Evolutionary events and acquisition of virulence and antimicrobial resistance traits along the evolution of clonal complex 5.
Viana et al. (2021) reported a significant spread of ST105-SCCmecII in hospitals throughout Rio de Janeiro [11]. These MRSA isolates, grouped into the terminal subclade of the CC5-IIB clade, became known as the Rio de Janeiro (RdJ) clone. In addition, they discovered that during the evolution of CC5-SCCmecII, ST105 isolates not only lost sep but also lacked the splD gene encoding for serine protease D. This omission persisted during the ST105-SCCmecII expansion, implying an evolutionary benefit for this lineage. Notably, the ST105 isolates from Rio de Janeiro demonstrated greater evasion of phagocytosis by THP-1 monocytes compared to the ST5-SCCmecII and ST5-SCCmecIV isolates, possibly explaining their higher prevalence in BSI [11]. Concurrent studies by these researchers suggest the loss of splD might have affected ST105 phagocytosis [105]. Pangenomic studies identified a specific mutation in the aur gene, which encodes the protease aureolysin, only in the RdJ clone. This protease contributes significantly to bacterial virulence, or ability to cause disease, by cleaving host factors of the innate immune system. The nonsynonymous mutation A1106G found in RdJ isolates was located in the recognition site of aur. However, the role played by this mutation in RdJ virulence remains to be elucidated [104].
Arias et al. (2017) detected the presence of ST105-SCCmecII in Brazil and Chile as well [6]. However, the Chilean ST105-SCCmecII strains described by Arias lack aph(39)-III, msrA, mphC (genes responsible for antimicrobial resistance), and the chp gene (from the immune evasion cluster; IEC). This is different from the recently described RdJ clone and other Brazilian ST5 strains, which carried these genes [6,11,55]. This observation suggests that the Chilean and RdJ clones present different genomic traits, despite belonging to the same lineage, ST105-SCCmecII. Once again, this demonstrates the great plasticity of these MRSA. In fact, pangenomic studies of CC30, CC5, and CC8 revealed that CC5 has the highest genetic variability. In addition, multiple virulence and resistance genes were identified, highlighting the complex virulence profiles of these MRSA strains [106].
Sullivan and colleagues (2019) analyzed the genome of ST105 strains involved in a BSI outbreak in a neonatal ICU in New York City [61]. They found that the outbreak strains had mutations in genes associated with bacterial metabolism, resistance, and persistence. Furthermore, they discovered a recombination present in the DNA recognition domain of the hsdS gene of the type I restriction-modification (R-M) system that had altered the DNA methylation. The study’s findings revealed that the transcriptome sequencing profile exhibited epigenetic alterations in the outbreak clone, an attenuation of agr expression, and an upregulation of genes governing stress response and biofilm development. It has been previously suggested that agr attenuation reduces bacterial cytotoxicity and is thus related to the bacterium’s ability to cause persistent bloodstream infections and disseminate within the hospital setting [107,108,109].
Altogether, these studies imply that ST105 strains not only have a high propensity to acquire resistance genes but can also adapt effectively to the human host to modulate their virulence and elude the immune system. These evolutionary adaptations may be attributable to mechanisms that can augment biofilm accumulation and stress response, for example, and reduce bacterial cytotoxicity. This allows the bacteria to evade the immune system and escape phagocytosis through mechanisms requiring further elucidation [11,21,61]. Moreover, the observation that ST105 is prevalent in blood infections also mandates further exploration, with the need to investigate the mechanisms implicated in this prevalence.
8. Conclusions
ST105-SCCmecII is an emerging MRSA lineage in European and American hospitals. Among CC5 isolates, it is often the second most important MRSA lineage, with a relatively high frequency in bloodstream infections in some countries. Moreover, the increased mortality from BSI in patients older than 15 years and the high prevalence of ST105-SCCmecII in the blood of patients older than 60 years should be of great concern.
9. Note of Authors
With the coronavirus disease 2019 (COVID-19) pandemic, the ST105-SCCmecII in Rio de Janeiro has drastically dropped in bloodstream infections, with the rise of both North and Latin American USA300 variants, although RdJ are still frequent in BSI affecting mainly old people [110].
Author Contributions
A.S.V.: data curation, formal analysis, investigation, and writing—original draft; L.P.d.V.T.: data curation, formal analysis, investigation, and writing—original draft; A.M.S.F.: funding acquisition, writing—review and editing, conceptualization, project administration, supervision, and validation. All authors have read and agreed to the published version of the manuscript.
Funding
This work was partially funded by the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) (443804/2018-4 and 307672/2019-0 to AMSF), the Fundação Carlos Chagas Filho de Amparo à Pesquisa do Estado do Rio de Janeiro (FAPERJ) (grant numbers E-26/010.001280/2016, E-26.202.803/2017 and E-26/010.002435/2019 to AMSF), the Fundação Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) (grant number 001) and also by the Bill & Melinda Gates Foundation (grant number INV-007641). ASV is a postdoctoral fellow of FAPERJ (grant number E-26/200.56/2024).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Mahjabeen, F.; Saha, U.; Mostafa, M.N.; Siddique, F.; Ahsan, E.; Fathma, S.; Tasnim, A.; Rahman, T.; Faruq, R.; Sakibuzzaman, M.; et al. An Update on Treatment Options for Methicillin-Resistant Staphylococcus aureus (MRSA) Bacteremia: A Systematic Review. Cureus 2022, 14, e31486. [Google Scholar] [CrossRef] [PubMed]
- Murray, C.J.L.; Ikuta, K.S.; Sharara, F.; Swetschinski, L.; Robles Aguilar, G.; Gray, A.; Han, C.; Bisignano, C.; Rao, P.; Wool, E.; et al. Global Burden of Bacterial Antimicrobial Resistance in 2019: A Systematic Analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention. Active Bacterial Core Surveillance (ABCs) Report Emerging Infections Program Network Methicillin-Resistant Staphylococcus aureus. 2014. Available online: https://stacks.cdc.gov/view/cdc/39907/cdc_39907_DS1.pdf (accessed on 23 August 2024).
- Centers for Disease Control and Prevention. Emerging Infections Program Healthcare-Associated Infections Community Interface Report Invasive Staphylococcus aureus, 2020; U.S. Department of Health and Human Services, CDC: Atlanta, GA, USA, 2022. Available online: https://www.cdc.gov/healthcare-associated-infections/media/pdfs/2020-MRSA-Report-508.pdf (accessed on 23 August 2024).
- Centers for Disease Control and Prevention. Healthcare-Associated Infections-Community Interface (HAIC): Emerging Infections Program (EIP) Network Report Invasive Staphylococcus aureus, 2017; U.S. Department of Health and Human Services, CDC: Atlanta, GA, USA, 2024; Available online: https://www.cdc.gov/healthcare-associated-infections/media/pdfs/2017-mrsa-report-508.pdf (accessed on 23 August 2024).
- Arias, C.A.; Reyes, J.; Carvajal, L.P.; Rincon, S.; Diaz, L.; Panesso, D.; Ibarra, G.; Rios, R.; Munita, J.M.; Salles, M.J.; et al. A Prospective Cohort Multicenter Study of Molecular Epidemiology and Phylogenomics of Staphylococcus aureus Bacteremia in Nine Latin American Countries. Antimicrob. Agents Chemother. 2017, 61, AAC.00816-17. [Google Scholar] [CrossRef]
- Seas, C.; Garcia, C.; Salles, M.J.; Labarca, J.; Luna, C.; Alvarez-Moreno, C.; Mejía-Villatoro, C.; Zurita, J.; Guzmán-Blanco, M.; Rodríguez-Noriega, E.; et al. Staphylococcus aureus Bloodstream Infections in Latin America: Results of a Multinational Prospective Cohort Study. J. Antimicrob. Chemother. 2018, 73, 212–222. [Google Scholar] [CrossRef] [PubMed]
- Figueiredo, A.M.S.; Ferreira, F.A. The Multifaceted Resources and Microevolution of the Successful Human and Animal Pathogen Methicillin-Resistant Staphylococcus aureus. Memórias Do Inst. Oswaldo Cruz 2014, 109, 265–278. [Google Scholar] [CrossRef][Green Version]
- Botelho, A.M.N.; Cerqueira E Costa, M.O.; Moustafa, A.M.; Beltrame, C.O.; Ferreira, F.A.; Côrtes, M.F.; Costa, B.S.S.; Silva, D.N.S.; Bandeira, P.T.; Lima, N.C.B.; et al. Local Diversification of Methicillin-Resistant Staphylococcus aureus ST239 in South America after Its Rapid Worldwide Dissemination. Front. Microbiol. 2019, 10, 82. [Google Scholar] [CrossRef] [PubMed]
- Schnitt, A.; Lienen, T.; Wichmann-Schauer, H.; Cuny, C.; Tenhagen, B.-A. The Occurrence and Distribution of Livestock-Associated Methicillin-Resistant Staphylococcus aureus ST398 on German Dairy Farms. J. Dairy Sci. 2020, 103, 11806–11819. [Google Scholar] [CrossRef]
- Viana, A.S.; Botelho, A.M.N.; Moustafa, A.M.; Boge, C.L.K.; Ferreira, A.L.P.; da Silva Carvalho, M.C.; Guimarães, M.A.; de Souza Scramignon Costa, B.; de Mattos, M.C.; Maciel, S.P.; et al. Multidrug-Resistant Methicillin- Resistant Staphylococcus aureus Associated with Bacteremia and Monocyte Evasion, Rio de Janeiro, Brazil. Emerg. Infect. Dis. 2021, 27, 2825–2835. [Google Scholar] [CrossRef]
- Chen, C.; Wu, F. Livestock-Associated Methicillin-Resistant Staphylococcus aureus (LA-MRSA) Colonisation and Infection among Livestock Workers and Veterinarians: A Systematic Review and Meta-Analysis. Occup Environ. Med 2021, 78, 530–540. [Google Scholar] [CrossRef]
- Breyre, A.; Frazee, B.W. Skin and Soft Tissue Infections in the Emergency Department. Emerg. Med. Clin. N. Am. 2018, 36, 723–750. [Google Scholar] [CrossRef]
- Hoppe, P.-A.; Holzhauer, S.; Lala, B.; Bührer, C.; Gratopp, A.; Hanitsch, L.G.; Humme, D.; Kieslich, M.; Kallinich, T.; Lau, S.; et al. Severe Infections of Panton-Valentine Leukocidin Positive Staphylococcus aureus in Children. Medicine 2019, 98, e17185. [Google Scholar] [CrossRef] [PubMed]
- Teixeira, L.A.; Resende, C.A.; Ormonde, L.R.; Rosenbaum, R.; Figueiredo, A.M.; de Lencastre, H.; Tomasz, A. Geographic Spread of Epidemic Multiresistant Staphylococcus aureus Clone in Brazil. J. Clin. Microbiol. 1995, 33, 2400–2404. [Google Scholar] [CrossRef] [PubMed]
- Melo, M.C.N.; Silva-Carvalho, M.C.; Ferreira, R.L.; Coelho, L.R.; Souza, R.R.; Gobbi, C.N.; Rozenbaum, R.; Solari, C.A.; Ferreira-Carvalho, B.T.; Figueiredo, A.M.S. Detection and Molecular Characterization of a Gentamicin-Susceptible, Methicillin-Resistant Staphylococcus aureus (MRSA) Clone in Rio de Janeiro That Resembles the New York/Japanese Clone. J. Hosp. Infect. 2004, 58, 276–285. [Google Scholar] [CrossRef]
- Enright, M.C.; Robinson, D.A.; Randle, G.; Feil, E.J.; Grundmann, H.; Spratt, B.G. The Evolutionary History of Methicillin-Resistant Staphylococcus aureus (MRSA). Proc. Natl. Acad. Sci. USA 2002, 99, 7687–7692. [Google Scholar] [CrossRef]
- Roberts, R.B.; Chung, M.; de Lencastre, H.; Hargrave, J.; Tomasz, A.; Nicolau, D.P.; John, J.F.; Korzeniowski, O.; Group, T.-S.M.C.S. Distribution of Methicillin-Resistant Staphylococcus aureus Clones among Health Care Facilities in Connecticut, New Jersey, and Pennsylvania. Microb. Drug Resist. 2000, 6, 245–251. [Google Scholar] [CrossRef] [PubMed]
- Côrtes, M.F.; Botelho, A.M.N.; Bandeira, P.T.; Mouton, W.; Badiou, C.; Bes, M.; Lima, N.C.B.; Soares, A.E.R.; Souza, R.C.; Almeida, L.G.P.; et al. Reductive Evolution of Virulence Repertoire to Drive the Divergence between Community- and Hospital-Associated Methicillin-Resistant Staphylococcus aureus of the ST1 Lineage. Virulence 2021, 12, 951–967. [Google Scholar] [CrossRef]
- Planet, P.J.; Diaz, L.; Kolokotronis, S.-O.; Narechania, A.; Reyes, J.; Xing, G.; Rincon, S.; Smith, H.; Panesso, D.; Ryan, C.; et al. Parallel Epidemics of Community-Associated Methicillin-Resistant Staphylococcus aureus USA300 Infection in North and South America. J. Infect. Dis. 2015, 212, 1874–1882. [Google Scholar] [CrossRef]
- Challagundla, L.; Reyes, J.; Rafiqullah, I.; Sordelli, D.O.; Echaniz-Aviles, G.; Velazquez-Meza, M.E.; Castillo-Ramírez, S.; Fittipaldi, N.; Feldgarden, M.; Chapman, S.B.; et al. Phylogenomic Classification and the Evolution of Clonal Complex 5 Methicillin-Resistant Staphylococcus aureus in the Western Hemisphere. Front. Microbiol. 2018, 9, 1901. [Google Scholar] [CrossRef]
- Gu, F.; He, W.; Xiao, S.; Wang, S.; Li, X.; Zeng, Q.; Ni, Y.; Han, L. Antimicrobial Resistance and Molecular Epidemiology of Staphylococcus aureus Causing Bloodstream Infections at Ruijin Hospital in Shanghai from 2013 to 2018. Sci. Rep. 2020, 10, 6019. [Google Scholar] [CrossRef]
- Tkadlec, J.; Capek, V.; Brajerova, M.; Smelikova, E.; Melter, O.; Bergerova, T.; Polivkova, S.; Balejova, M.; Hanslianova, M.; Fackova, D.; et al. The Molecular Epidemiology of Methicillin-Resistant Staphylococcus aureus (MRSA) in the Czech Republic. J. Antimicrob. Chemother. 2021, 76, 55–64. [Google Scholar] [CrossRef]
- Shoaib, M.; Aqib, A.I.; Muzammil, I.; Majeed, N.; Bhutta, Z.A.; Kulyar, M.F.-A.; Fatima, M.; Zaheer, C.-N.F.; Muneer, A.; Murtaza, M.; et al. MRSA Compendium of Epidemiology, Transmission, Pathophysiology, Treatment, and Prevention within One Health Framework. Front. Microbiol. 2023, 13, 1067284. [Google Scholar] [CrossRef] [PubMed]
- de Lencastre, H.; de Lencastre, A.; Tomasz, A. Methicillin-Resistant Staphylococcus aureus Isolates Recovered from a New York City Hospital: Analysis by Molecular Fingerprinting Techniques. J. Clin. Microbiol 1996, 34, 2121–2124. [Google Scholar] [CrossRef] [PubMed]
- Roberts, R.B.; de Lencastre, A.; Eisner, W.; Severina, E.P.; Shopsin, B.; Kreiswirth, B.N.; Tomasz, A. Molecular Epidemiology of Methicillin-Resistant Staphylococcus aureus in 12 New York Hospitals. MRSA Collaborative Study Group. J. Infect. Dis. 1998, 178, 164–171. [Google Scholar] [CrossRef]
- da Silva Coimbra, M.V.; Silva-Carvalho, M.C.; Wisplinghoff, H.; Hall, G.O.; Tallent, S.; Wallace, S.; Edmond, M.B.; Figueiredo, A.M.S.; Wenzel, R.P. Clonal Spread of Methicillin-Resistant Staphylococcus aureus in a Large Geographic Area of the United States. J. Hosp. Infect. 2003, 53, 103–110. [Google Scholar] [CrossRef]
- Ohkura, T.; Yamada, K.; Okamoto, A.; Baba, H.; Ike, Y.; Arakawa, Y.; Hasegawa, T.; Ohta, M. Nationwide Epidemiological Study Revealed the Dissemination of Meticillin-Resistant Staphylococcus aureus Carrying a Specific Set of Virulence-Associated Genes in Japanese Hospitals. J. Med. Microbiol. 2009, 58, 1329–1336. [Google Scholar] [CrossRef][Green Version]
- Loaiza, W.M.; Ruiz, A.K.R.; Patiño, C.C.O.; Vivas, M.C. Bacterial Resistance in Hospital-Acquired Infections Acquired in the Intensive Care Unit: A Systematic Review. Acta Med. 2023, 66, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Sá-Leão, R.; Santos Sanches, I.; Dias, D.; Peres, I.; Barros, R.M.; de Lencastre, H. Detection of an Archaic Clone of Staphylococcus aureus with Low-Level Resistance to Methicillin in a Pediatric Hospital in Portugal and in International Samples: Relics of a Formerly Widely Disseminated Strain? J. Clin. Microbiol. 1999, 37, 1913–1920. [Google Scholar] [CrossRef] [PubMed]
- de Miranda, O.P.; Silva-Carvalho, M.C.; Ribeiro, A.; Portela, F.; Cordeiro, R.P.; Caetano, N.; Vidal, C.F.L.; Figueiredo, A.M.S. Emergence in Brazil of Methicillin-Resistant Staphylococcus aureus Isolates Carrying SCCmecIV That Are Related Genetically to the USA800 Clone. Clin. Microbiol. Infect. 2007, 13, 1166–1172. [Google Scholar] [CrossRef]
- De Sousa-Junior, F.C.; Silva-Carvalho, M.C.; Fernandes, M.J.B.C.; Vieira, M.F.P.; Pellegrino, F.L.P.C.; Figueiredo, A.M.S.; de Melo, M.C.N.; Milan, E.P.; de Sousa-Junior, F.C.; Silva-Carvalho, M.C.; et al. Genotyping of Methicillin-Resistant Staphylococcus aureus Isolates Obtained in the Northeast Region of Brazil. Braz. J. Med. Biol. Res. = Rev. Bras. Pesqui. Medicas E Biol. 2009, 42, 877–881. [Google Scholar] [CrossRef]
- Pereira, V.C.; Riboli, D.F.M.; Da Cunha, M.D.L.R.D.S. Characterization of the Clonal Profile of MRSA Isolated in Neonatal and Pediatric Intensive Care Units of a University Hospital. Ann. Clin. Microbiol. Antimicrob. 2014, 13, 50. [Google Scholar] [CrossRef]
- Rokney, A.; Baum, M.; Ben-Shimol, S.; Sagi, O.; Anuka, E.; Agmon, V.; Greenberg, D.; Valinsky, L.; Danino, D. Dissemination of the Methicillin-Resistant Staphylococcus aureus Pediatric Clone (ST5-T002-IV-PVL+) as a Major Cause of Community-Associated Staphylococcal Infections in Bedouin Children, Southern Israel. Pediatr. Infect. Dis. J. 2019, 38, 230–235. [Google Scholar] [CrossRef]
- Vázquez-Sánchez, D.A.; Grillo, S.; Carrera-Salinas, A.; González-Díaz, A.; Cuervo, G.; Grau, I.; Camoez, M.; Martí, S.; Berbel, D.; Tubau, F.; et al. Molecular Epidemiology, Antimicrobial Susceptibility, and Clinical Features of Methicillin-Resistant Staphylococcus aureus Bloodstream Infections over 30 Years in Barcelona, Spain (1990–2019). Microorganisms 2022, 10, 2401. [Google Scholar] [CrossRef] [PubMed]
- Sola, C.; Gribaudo, G.; Vindel, A.; Patrito, L.; Bocco, J.L. Identification of a Novel Methicillin-Resistant Staphylococcus aureus Epidemic Clone in Córdoba, Argentina, Involved in Nosocomial Infections. J. Clin. Microbiol. 2002, 40, 1427–1435. [Google Scholar] [CrossRef] [PubMed]
- Lamanna, O.; Bongiorno, D.; Bertoncello, L.; Grandesso, S.; Mazzucato, S.; Pozzan, G.B.; Cutrone, M.; Chirico, M.; Baesso, F.; Brugnaro, P.; et al. Rapid Containment of Nosocomial Transmission of a Rare Community-Acquired Methicillin-Resistant Staphylococcus aureus (CA-MRSA) Clone, Responsible for the Staphylococcal Scalded Skin Syndrome (SSSS). Ital. J. Pediatr. 2017, 43, 5. [Google Scholar] [CrossRef] [PubMed]
- Lina, G.; Durand, G.; Berchich, C.; Short, B.; Meugnier, H.; Vandenesch, F.; Etienne, J.; Enright, M.C. Staphylococcal Chromosome Cassette Evolution in Staphylococcus aureus Inferred from Ccr Gene Complex Sequence Typing Analysis. Clin. Microbiol. Infect. 2006, 12, 1175–1184. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Sola, C.; Paganini, H.; Egea, A.L.; Moyano, A.J.; Garnero, A.; Kevric, I.; Culasso, C.; Vindel, A.; Lopardo, H.; Bocco, J.L.; et al. Spread of Epidemic MRSA-ST5-IV Clone Encoding PVL as a Major Cause of Community Onset Staphylococcal Infections in Argentinean Children. PLoS ONE 2012, 7, e30487. [Google Scholar] [CrossRef] [PubMed]
- Chang, S.; Sievert, D.M.; Hageman, J.C.; Boulton, M.L.; Tenover, F.C.; Downes, F.P.; Shah, S.; Rudrik, J.T.; Pupp, G.R.; Brown, W.J.; et al. Infection with Vancomycin-Resistant Staphylococcus aureus Containing the vanA Resistance Gene. N. Engl. J. Med. 2003, 348, 1342–1347. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention. Staphylococcus aureus Resistant to Vancomycin—United States, 2002. MMWR Morb. Mortal. Wkly. Rep. 2002, 51, 565–567. [Google Scholar]
- Kos, V.N.; Desjardins, C.A.; Griggs, A.; Cerqueira, G.; Van Tonder, A.; Holden, M.T.G.; Godfrey, P.; Palmer, K.L.; Bodi, K.; Mongodin, E.F.; et al. Comparative Genomics of Vancomycin-Resistant Staphylococcus aureus Strains and Their Positions within the Clade Most Commonly Associated with Methicillin-Resistant S. aureus Hospital-Acquired Infection in the United States. mBio 2012, 3, e00112-12. [Google Scholar] [CrossRef]
- Cong, Y.; Yang, S.; Rao, X. Vancomycin Resistant Staphylococcus aureus Infections: A Review of Case Updating and Clinical Features. J. Adv. Res. 2020, 21, 169–176. [Google Scholar] [CrossRef]
- Haas, W.; Singh, N.; Lainhart, W.; Mingle, L.; Nazarian, E.; Mitchell, K.; Nattanmai, G.; Kohlerschmidt, D.; Dickinson, M.C.; Kacica, M.; et al. Genomic Analysis of Vancomycin-Resistant Staphylococcus aureus Isolates from the 3rd Case Identified in the United States Reveals Chromosomal Integration of the vanA Locus. Microbiol. Spectr. 2023, 11, e04317-22. [Google Scholar] [CrossRef] [PubMed]
- Limbago, B.M.; Kallen, A.J.; Zhu, W.; Eggers, P.; McDougal, L.K.; Albrecht, V.S. Report of the 13th Vancomycin-Resistant Staphylococcus aureus Isolate from the United States. J. Clin. Microbiol. 2014, 52, 998–1002. [Google Scholar] [CrossRef]
- Di Gregorio, S.; Haim, M.S.; Famiglietti, Á.M.R.; Di Conza, J.; Mollerach, M. Comparative Genomics Identifies Novel Genetic Changes Associated with Oxacillin, Vancomycin and Daptomycin Susceptibility in ST100 Methicillin-Resistant Staphylococcus aureus. Antibiotics 2023, 12, 372. [Google Scholar] [CrossRef]
- Alfouzan, W.A.; Boswihi, S.S.; Udo, E.E. Methicillin-Resistant Staphylococcus aureus (MRSA) in a Tertiary Care Hospital in Kuwait: A Molecular and Genetic Analysis. Microorganisms 2023, 12, 17. [Google Scholar] [CrossRef]
- Garrine, M.; Costa, S.S.; Messa, A.; Massora, S.; Vubil, D.; Ácacio, S.; Nhampossa, T.; Bassat, Q.; Mandomando, I.; Couto, I. Antimicrobial Resistance and Clonality of Staphylococcus aureus Causing Bacteraemia in Children Admitted to the Manhiça District Hospital, Mozambique, over Two Decades. Front. Microbiol. 2023, 14, 1208131. [Google Scholar] [CrossRef]
- Zhang, H.; Tian, L.; Chen, T.; Chen, W.; Ge, Y.; Bi, J.; Fang, Z.; Chen, M. Prevalence and WGS-Based Characteristics of MRSA Isolates in Hospitals in Shanghai, China. Front. Microbiol. 2022, 13, 1002691. [Google Scholar] [CrossRef] [PubMed]
- Kaku, N.; Sasaki, D.; Ota, K.; Miyazaki, T.; Yanagihara, K. Changing Molecular Epidemiology and Characteristics of MRSA Isolated from Bloodstream Infections: Nationwide Surveillance in Japan in 2019. J. Antimicrob. Chemother. 2022, 77, 2130–2141. [Google Scholar] [CrossRef] [PubMed]
- Kondo, S.; Phokhaphan, P.; Tongsima, S.; Ngamphiw, C.; Phornsiricharoenphant, W.; Ruangchai, W.; Disratthakit, A.; Tingpej, P.; Mahasirimongkol, S.; Lulitanond, A.; et al. Molecular Characterization of Methicillin-Resistant Staphylococcus aureus Genotype ST764-SCCmec Type II in Thailand. Sci. Rep. 2022, 12, 2085. [Google Scholar] [CrossRef]
- Chung, M.; Dickinson, G.; de Lencastre, H.; Tomasz, A. International Clones of Methicillin-Resistant Staphylococcus aureus in Two Hospitals in Miami, Florida. J. Clin. Microbiol. 2004, 42, 542–547. [Google Scholar] [CrossRef][Green Version]
- David, M.Z.; Rudolph, K.M.; Hennessy, T.W.; Zychowski, D.L.; Asthi, K.; Boyle-Vavra, S.; Daum, R.S. MRSA USA300 at Alaska Native Medical Center, Anchorage, Alaska, USA, 2000–2006. Emerg. Infect. Dis. 2012, 18, 105–108. [Google Scholar] [CrossRef]
- Read, T.D.; Jacko, N.F.; Petit, R.A.; Pegues, D.A.; David, M.Z. 852. Genomic Clusters of Methicillin-Resistant Staphylococcus aureus (MRSA) Causing Bloodstream Infections (BSIs) in Hospitalized Adults, 2018-19. Open Forum Infect. Dis. 2020, 7, S466–S467. [Google Scholar] [CrossRef]
- Martínez, J.R.; Planet, P.J.; Maria, S.-S.; Lina, R.; Lorena, D.; Ana, Q.-V.; Roberto, R.-N.; Manuel, A.-R.; Blake, H.; Carvajal, L.P.; et al. Dynamics of the MRSA Population in A Chilean Hospital: A Phylogenomic Analysis (2000–2016). Microbiol. Spectr. 2023, 11, e05351-22. [Google Scholar] [CrossRef] [PubMed]
- Bouiller, K.; Jacko, N.F.; Shumaker, M.J.; Talbot, B.M.; Read, T.D.; David, M.Z. Factors Associated with Foreign Body Infection in Methicillin-Resistant Staphylococcus aureus Bacteremia. Front. Immunol. 2024, 15, 1335867. [Google Scholar] [CrossRef]
- Jolley, K.A.; Bray, J.E.; Maiden, M.C.J. Open-Access Bacterial Population Genomics: BIGSdb Software, the PubMLST.Org Website and Their Applications. Wellcome Open Res. 2018, 3, 124. [Google Scholar] [CrossRef]
- Blanc, D.S.; Petignat, C.; Wenger, A.; Kuhn, G.; Vallet, Y.; Fracheboud, D.; Trachsel, S.; Reymond, M.; Troillet, N.; Siegrist, H.H.; et al. Changing Molecular Epidemiology of Methicillin-Resistant Staphylococcus aureus in a Small Geographic Area over an Eight-Year Period. J. Clin. Microbiol. 2007, 45, 3729–3736. [Google Scholar] [CrossRef]
- Verghese, B.; Schwalm, N.D.; Dudley, E.G.; Knabel, S.J. A Combined Multi-Virulence-Locus Sequence Typing and Staphylococcal Cassette Chromosome Mec Typing Scheme Possesses Enhanced Discriminatory Power for Genotyping MRSA. Infect. Genet. Evol. 2012, 12, 1816–1821. [Google Scholar] [CrossRef]
- Peterson, A.E.; Davis, M.F.; Julian, K.G.; Awantang, G.; Greene, W.H.; Price, L.B.; Waters, A.; Doppalapudi, A.; Krain, L.J.; Nelson, K.; et al. Molecular and Phenotypic Characteristics of Healthcare- and Community-Associated Methicillin-Resistant Staphylococcus aureus at a Rural Hospital. PLoS ONE 2012, 7, e38354. [Google Scholar] [CrossRef]
- Sullivan, M.J.; Altman, D.R.; Chacko, K.I.; Ciferri, B.; Webster, E.; Pak, T.R.; Deikus, G.; Lewis-Sandari, M.; Khan, Z.; Beckford, C.; et al. A Complete Genome Screening Program of Clinical Methicillin-Resistant Staphylococcus aureus Isolates Identifies the Origin and Progression of a Neonatal Intensive Care Unit Outbreak. J. Clin. Microbiol. 2019, 57, e01261-19. [Google Scholar] [CrossRef] [PubMed]
- Iregui, A.; Khan, Z.; Malik, S.; Landman, D.; Quale, J. Emergence of Delafloxacin-Resistant Staphylococcus aureus in Brooklyn, New York. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2020, 70, 1758–1760. [Google Scholar] [CrossRef]
- Fischer, A.J.; Porterfield, H.S.; LaMarche, M.M.; Hansen, A.R.; Pitcher, N.; Thurman, A.L.; Reeb, V.L. Staphylococcus aureus Genome Sequencing Reveals Strains Associated with Persistence and Poor Outcome in CF. Am. J. Respir. Crit. Care Med. 2020, 201, A7457. [Google Scholar]
- Porterfield, H.S.; Maakestad, L.J.; LaMarche, M.M.; Thurman, A.L.; Kienenberger, Z.E.; Pitcher, N.J.; Hansen, A.R.; Zirbes, C.F.; Boyken, L.; Muyskens, B.L.; et al. MRSA Strains with Distinct Accessory Genes Predominate at Different Ages in Cystic Fibrosis. Pediatr. Pulmonol. 2021, 56, 2868–2878. [Google Scholar] [CrossRef] [PubMed]
- Hudson, L.O.; Murphy, C.R.; Spratt, B.G.; Enright, M.C.; Elkins, K.; Nguyen, C.; Terpstra, L.; Gombosev, A.; Kim, D.; Hannah, P.; et al. Diversity of Methicillin-Resistant Staphylococcus aureus (MRSA) Strains Isolated from Inpatients of 30 Hospitals in Orange County, California. PLoS ONE 2013, 8, e62117. [Google Scholar] [CrossRef] [PubMed]
- Campanile, F.; Bongiorno, D.; Perez, M.; Mongelli, G.; Sessa, L.; Benvenuto, S.; Gona, F.; Varaldo, P.E.; Stefani, S. Epidemiology of Staphylococcus aureus in Italy: First Nationwide Survey, 2012. J. Glob. Antimicrob. Resist. 2015, 3, 247–254. [Google Scholar] [CrossRef]
- Nikolaras, G.P.; Papaparaskevas, J.; Samarkos, M.; Tzouvelekis, L.S.; Psychogiou, M.; Pavlopoulou, I.; Goukos, D.; Polonyfi, K.; Pantazatou, A.; Deliolanis, I.; et al. Changes in the Rates and Population Structure of MRSA from Bloodstream Infections. A Single Center Experience (2000–2015). J. Glob. Antimicrob. Resist. 2019, 17, 117–122. [Google Scholar] [CrossRef]
- Espadinha, D.; Faria, N.A.; Miragaia, M.; Lito, L.M.; Melo-Cristino, J.; de Lencastre, H. Extensive Dissemination of Methicillin-Resistant Staphylococcus aureus (MRSA) between the Hospital and the Community in a Country with a High Prevalence of Nosocomial MRSA. PLoS ONE 2013, 8, e59960. [Google Scholar] [CrossRef][Green Version]
- Faria, N.A.; Miragaia, M.; de Lencastre, H.; Multi Laboratory Project Collaborators. Massive Dissemination of Methicillin Resistant Staphylococcus aureus in Bloodstream Infections in a High MRSA Prevalence Country: Establishment and Diversification of EMRSA-15. Microb. Drug Resist. 2013, 19, 483–490. [Google Scholar] [CrossRef]
- Almeida, S.T.; Nunes, S.; Paulo, A.C.S.; Faria, N.A.; de Lencastre, H.; Sá-Leão, R. Prevalence, Risk Factors, and Epidemiology of Methicillin-resistantStaphylococcus aureus Carried by Adults over 60 Years of Age. Eur. J. Clin. Microbiol. Infect. Dis. 2015, 34, 593–600. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, F.A.; Helmersen, K.; Visnovska, T.; Jørgensen, S.B.; Aamot, H.V. Rapid Nanopore-Based DNA Sequencing Protocol of Antibiotic-Resistant Bacteria for Use in Surveillance and Outbreak Investigation. Microb. Genom. 2021, 7, 000557. [Google Scholar] [CrossRef] [PubMed]
- Conceição, T.; Santos Silva, I.; de Lencastre, H.; Aires-de-Sousa, M. Staphylococcus aureus Nasal Carriage Among Patients and Health Care Workers in São Tomé and Príncipe. Microb. Drug Resist. 2014, 20, 57–66. [Google Scholar] [CrossRef]
- Boswihi, S.S.; Udo, E.E.; Al-Sweih, N. Shifts in the Clonal Distribution of Methicillin-Resistant Staphylococcus aureus in Kuwait Hospitals: 1992–2010. PLoS ONE 2016, 11, e0162744. [Google Scholar] [CrossRef]
- Peng, H.; Liu, D.; Ma, Y.; Gao, W. Comparison of Community- and Healthcare-Associated Methicillin-Resistant Staphylococcus aureus Isolates at a Chinese Tertiary Hospital, 2012–2017. Sci. Rep. 2018, 8, 17916. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.T.; Eckhardt, E.M.; Hansel, N.B.; Rahmani Eliato, T.; Martin, I.W.; Andam, C.P. Genome Evolution of Invasive Methicillin-Resistant Staphylococcus aureus in the Americas. Microbiol. Spectr. 2022, 10, e0020122. [Google Scholar] [CrossRef] [PubMed]
- Zurita, J.; Barba, P.; Ortega-Paredes, D.; Mora, M.; Rivadeneira, S. Local Circulating Clones of Staphylococcus aureus in Ecuador. Braz. J. Infect. Dis. 2016, 20, 525–533. [Google Scholar] [CrossRef]
- van der Heijden, I.M.; de Oliveira, L.M.; Brito, G.C.; Abdala, E.; Freire, M.P.; Rossi, F.; D’Albuquerque, L.A.C.; Levin, A.S.S.; Costa, S.F. Virulence and Resistance Profiles of MRSA Isolates in Pre- and Post-Liver Transplantation Patients Using Microarray. J. Med. Microbiol. 2016, 65, 1060–1073. [Google Scholar] [CrossRef]
- Ikuta, K.S.; Swetschinski, L.R.; Robles Aguilar, G.; Sharara, F.; Mestrovic, T.; Gray, A.P.; Davis Weaver, N.; Wool, E.E.; Han, C.; Gershberg Hayoon, A.; et al. Global Mortality Associated with 33 Bacterial Pathogens in 2019: A Systematic Analysis for the Global Burden of Disease Study 2019. Lancet 2022, 400, 2221–2248. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.W.; Bae, S.; Yang, E.; Chung, H.; Kim, E.; Jung, J.; Kim, M.J.; Chong, Y.P.; Kim, S.-H.; Choi, S.-H.; et al. Clinical and Microbiological Characteristics of Hospital-Acquired Methicillin-Resistant Staphylococcus aureus Bacteremia Caused by a Community-Associated PVL-Negative Strain. Open Forum Infect. Dis. 2021, 8, ofab424. [Google Scholar] [CrossRef] [PubMed]
- Park, K.-H.; Greenwood-Quaintance, K.E.; Uhl, J.R.; Cunningham, S.A.; Chia, N.; Jeraldo, P.R.; Sampathkumar, P.; Nelson, H.; Patel, R. Molecular Epidemiology of Staphylococcus aureus Bacteremia in a Single Large Minnesota Medical Center in 2015 as Assessed Using MLST, Core Genome MLST and Spa Typing. PLoS ONE 2017, 12, e0179003. [Google Scholar] [CrossRef]
- Augusto, M.F.; Da Silva Fernandes, D.C.; De Oliveira, T.L.R.; Cavalcante, F.S.; Chamon, R.C.; Ferreira, A.L.P.; Nouér, S.A.; Infection Control Group HUCFF/UFRJ; Rangel, A.P.; Castiñeiras, A.C.; et al. Pandemic Clone USA300 in a Brazilian Hospital: Detection of an Emergent Lineage among Methicillin-Resistant Staphylococcus aureus Isolates from Bloodstream Infections. Antimicrob. Resist. Infect. Control 2022, 11, 114. [Google Scholar] [CrossRef]
- Silva, V.; Hermenegildo, S.; Ferreira, C.; Manaia, C.M.; Capita, R.; Alonso-Calleja, C.; Carvalho, I.; Pereira, J.E.; Maltez, L.; Capelo, J.L.; et al. Genetic Characterization of Methicillin-Resistant Staphylococcus aureus Isolates from Human Bloodstream Infections: Detection of MLSB Resistance. Antibiotics 2020, 9, 375. [Google Scholar] [CrossRef]
- Berbel Caban, A.; Pak, T.R.; Obla, A.; Dupper, A.C.; Chacko, K.I.; Fox, L.; Mills, A.; Ciferri, B.; Oussenko, I.; Beckford, C.; et al. PathoSPOT Genomic Epidemiology Reveals Under-the-Radar Nosocomial Outbreaks. Genome Med. 2020, 12, 96. [Google Scholar] [CrossRef]
- Lin, Y.; Barker, E.; Kislow, J.; Kaldhone, P.; Stemper, M.E.; Pantrangi, M.; Moore, F.M.; Hall, M.; Fritsche, T.R.; Novicki, T.; et al. Evidence of Multiple Virulence Subtypes in Nosocomial and Community-Associated MRSA Genotypes in Companion Animals from the Upper Midwestern and Northeastern United States. Clin. Med. Res. 2011, 9, 7–16. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Couto, N.; Belas, A.; Kadlec, K.; Schwarz, S.; Pomba, C. Clonal Diversity, Virulence Patterns and Antimicrobial and Biocide Susceptibility among Human, Animal and Environmental MRSA in Portugal. J. Antimicrob. Chemother. 2015, 70, 2483–2487. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, A.C.; Belas, A.; Marques, C.; Cruz, L.; Gama, L.T.; Pomba, C. Risk Factors for Nasal Colonization by Methicillin-Resistant Staphylococci in Healthy Humans in Professional Daily Contact with Companion Animals in Portugal. Microb. Drug Resist. 2017, 24, 434–446. [Google Scholar] [CrossRef] [PubMed]
- Himsworth, C.G.; Miller, R.R.; Montoya, V.; Hoang, L.; Romney, M.G.; Al-Rawahi, G.N.; Kerr, T.; Jardine, C.M.; Patrick, D.M.; Tang, P.; et al. Carriage of Methicillin-Resistant Staphylococcus aureus by Wild Urban Norway Rats (Rattus Norvegicus). PLoS ONE 2014, 9, e87983. [Google Scholar] [CrossRef] [PubMed]
- Silva, V.; de Sousa, T.; Gómez, P.; Sabença, C.; Vieira-Pinto, M.; Capita, R.; Alonso-Calleja, C.; Torres, C.; Capelo, J.L.; Igrejas, G.; et al. Livestock-Associated Methicillin-Resistant Staphylococcus Aureus (MRSA) in Purulent Subcutaneous Lesions of Farm Rabbits. Foods 2020, 9, 439. [Google Scholar] [CrossRef]
- Silva, V.; Ribeiro, J.; Rocha, J.; Manaia, C.M.; Silva, A.; Pereira, J.E.; Maltez, L.; Capelo, J.L.; Igrejas, G.; Poeta, P. High Frequency of the EMRSA-15 Clone (ST22-MRSA-IV) in Hospital Wastewater. Microorganisms 2022, 10, 147. [Google Scholar] [CrossRef]
- Zuma, A.V.P.; Lima, D.F.; Assef, A.P.D.C.D.C.; Marques, E.A.; Leão, R.S. Molecular Characterization of Methicillin-Resistant Staphylococcus aureus Isolated from Blood in Rio de Janeiro Displaying Susceptibility Profiles to Non-β-Lactam Antibiotics. Braz. J. Microbiol. 2017, 48, 237–241. [Google Scholar] [CrossRef]
- Okado, J.B.; Bogni, S.C.; Reinato, L.A.F.; Martinez, R.; Gir, E.; Camargo, I.L.B.D.C. Molecular Analysis of Methicillin-Resistant Staphylococcus aureus Dissemination among Healthcare Professionals and/or HIV Patients from a Tertiary Hospital. Rev. Soc. Bras. Med. Trop. 2016, 49, 51–56. [Google Scholar] [CrossRef][Green Version]
- Caiaffa-Filho, H.H.; Trindade, P.A.; Gabriela da Cunha, P.; Alencar, C.S.; Prado, G.V.B.B.; Rossi, F.; Levin, A.S. Methicillin-Resistant Staphylococcus aureus Carrying SCCmec Type II Was More Frequent than the Brazilian Endemic Clone as a Cause of Nosocomial Bacteremia. Diagn. Microbiol. Infect. Dis. 2013, 76, 518–520. [Google Scholar] [CrossRef]
- Ferreira, C.; Costa, S.S.; Serrano, M.; Oliveira, K.; Trigueiro, G.; Pomba, C.; Couto, I. Clonal Lineages, Antimicrobial Resistance, and PVL Carriage of Staphylococcus aureus Associated to Skin and Soft-Tissue Infections from Ambulatory Patients in Portugal. Antibiotics 2021, 10, 345. [Google Scholar] [CrossRef]
- Salgueiro, V.; Manageiro, V.; Bandarra, N.M.; Ferreira, E.; Clemente, L.; Caniça, M. Genetic Relatedness and Diversity of Staphylococcus aureus from Different Reservoirs: Humans and Animals of Livestock, Poultry, Zoo, and Aquaculture. Microorganisms 2020, 8, 1345. [Google Scholar] [CrossRef] [PubMed]
- Mwangi, M.M.; Wu, S.W.; Zhou, Y.; Sieradzki, K.; de Lencastre, H.; Richardson, P.; Bruce, D.; Rubin, E.; Myers, E.; Siggia, E.D.; et al. Tracking the In Vivo Evolution of Multidrug Resistance in Staphylococcus aureus by Whole-Genome Sequencing. Proc. Natl. Acad. Sci. USA 2007, 104, 9451–9456. [Google Scholar] [CrossRef] [PubMed]
- Adamu, Y.; Puig-Asensio, M.; Dabo, B.; Schweizer, M.L. Comparative Effectiveness of Daptomycin versus Vancomycin among Patients with Methicillin-Resistant Staphylococcus aureus (MRSA) Bloodstream Infections: A Systematic Literature Review and Meta-Analysis. PLoS ONE 2024, 19, e0293423. [Google Scholar] [CrossRef] [PubMed]
- Mlynarczyk-Bonikowska, B.; Kowalewski, C.; Krolak-Ulinska, A.; Marusza, W. Molecular Mechanisms of Drug Resistance in Staphylococcus aureus. Int. J. Mol. Sci. 2022, 23, 8088. [Google Scholar] [CrossRef]
- Melo-Cristino, J.; Resina, C.; Manuel, V.; Lito, L.; Ramirez, M. First Case of Infection with Vancomycin-Resistant Staphylococcus aureus in Europe. Lancet 2013, 382, 205. [Google Scholar] [CrossRef]
- McCulloch, J.A.; de, O. Silveira, A.C.; da C. Lima Moraes, A.; Pérez-Chaparro, P.J.; Ferreira Silva, M.; Almeida, L.M.; D’Azevedo, P.A.; Mamizuka, E.M. Complete Genome Sequence of Staphylococcus aureus FCFHV36, a Methicillin-Resistant Strain Heterogeneously Resistant to Vancomycin. Genome Announc. 2015, 3, e00893-15. [Google Scholar] [CrossRef] [PubMed]
- Viana, A.S.; Botelho, A.M.N.; Feder, A.; Moustafa, A.M.; Santos Silva, D.N.; Martini, C.L.; Ferreira, A.L.P.; Silva-Carvalho, M.C.; Ferreira-Carvalho, B.T.; Planet, P.J.; et al. High Frequency of Increased Triclosan MIC among CC5 MRSA and Risk of Misclassification of the SCC Mec into Types. J. Antimicrob. Chemother. 2022, 77, 3340–3348. [Google Scholar] [CrossRef]
- Dabul, A.N.G.; Camargo, I.L.B.C. Molecular Characterization of Methicillin-Resistant Staphylococcus aureus Resistant to Tigecycline and Daptomycin Isolated in a Hospital in Brazil. Epidemiol. Infect. 2018, 142, 479–483. [Google Scholar] [CrossRef]
- Furi, L.; Haigh, R.; Al Jabri, Z.J.H.; Morrissey, I.; Ou, H.Y.; León-Sampedro, R.; Martinez, J.L.; Coque, T.M.; Oggioni, M.R. Dissemination of Novel Antimicrobial Resistance Mechanisms through the Insertion Sequence Mediated Spread of Metabolic Genes. Front. Microbiol. 2016, 7, 1008. [Google Scholar] [CrossRef]
- Vulin, C.; Leimer, N.; Huemer, M.; Ackermann, M.; Zinkernagel, A.S. Prolonged Bacterial Lag Time Results in Small Colony Variants That Represent a Sub-Population of Persisters. Nat. Commun. 2018, 9, 4074. [Google Scholar] [CrossRef]
- Esteves, M.A.C.; Viana, A.S.; Viçosa, G.N.; Botelho, A.M.N.; Moustafa, A.M.; Mansoldo, F.R.P.; Ferreira, A.L.P.; Vermelho, A.B.; Ferreira-Carvalho, B.T.; Planet, P.J.; et al. RdJ Detection Tests to Identify a Unique MRSA Clone of ST105-SCCmecII Lineage and Its Variants Disseminated in the Metropolitan Region of Rio de Janeiro. Front. Microbiol. 2023, 14, 1275918. [Google Scholar] [CrossRef] [PubMed]
- Bandeira, P.T.; Viana, A.S.; Côrtes, A.F.; Botelho, A.M.N.; Tasse, J.; Guimarães, M.A.; Abreu, U.d.S.; Lima, J.E.; Laurent, F.; Planet, P.J.; et al. Spl Proteases Modulate Important Virulence Attributes of Staphylococcus aureus. Virulence, 2024; Submitted manuscript. [Google Scholar]
- Guillén, R.; Salinas, C.; Mendoza-Álvarez, A.; Rubio Rodríguez, L.A.; Díaz-de Usera, A.; Lorenzo-Salazar, J.M.; González-Montelongo, R.; Flores, C.; Rodríguez, F. Genomic Epidemiology of the Primary Methicillin-Resistant Staphylococcus aureus Clones Causing Invasive Infections in Paraguayan Children. Microbiol. Spectr. 2024, 12, e03012-23. [Google Scholar] [CrossRef] [PubMed]
- Park, S.-Y.; Chong, Y.P.; Park, H.J.; Park, K.-H.; Moon, S.M.; Jeong, J.-Y.; Kim, M.-N.; Kim, S.-H.; Lee, S.-O.; Choi, S.-H.; et al. Agr Dysfunction and Persistent Methicillin-Resistant Staphylococcus aureus Bacteremia in Patients with Removed Eradicable Foci. Infection 2013, 41, 111–119. [Google Scholar] [CrossRef] [PubMed]
- Smeltzer, M.S. Staphylococcus aureus Pathogenesis: The Importance of Reduced Cytotoxicity. Trends Microbiol. 2016, 24, 681–682. [Google Scholar] [CrossRef]
- Lee, S.O.; Lee, S.; Lee, J.E.; Song, K.-H.; Kang, C.K.; Wi, Y.M.; San-Juan, R.; López-Cortés, L.E.; Lacoma, A.; Prat, C.; et al. Dysfunctional Accessory Gene Regulator (Agr) as a Prognostic Factor in Invasive Staphylococcus aureus Infection: A Systematic Review and Meta-Analysis. Sci. Rep. 2020, 10, 20697. [Google Scholar] [CrossRef]
- Figueiredo, A.M. Molecular Epidemiology of Methicillin-Resistant Staphylococcus aureus in Hospitals of Metropolitan Region of Rio de Janeiro, Brazil; Universidade Federal do Rio de Janeiro: Rio de Janeiro, RJ, Brazil, 2024; Manuscript in preparation. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).