Persistence and Culturability of Escherichia coli under Induced Toxin Expression
Abstract
1. Introduction
2. Results
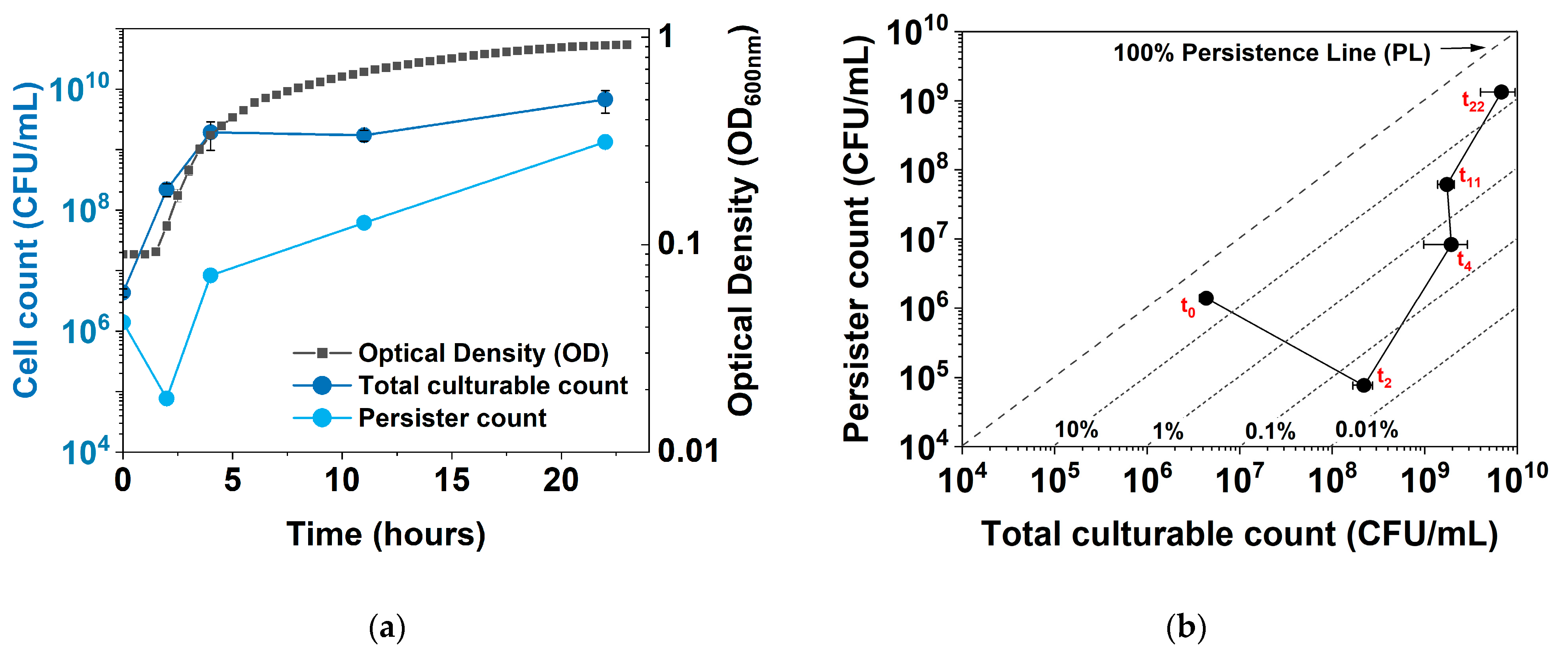
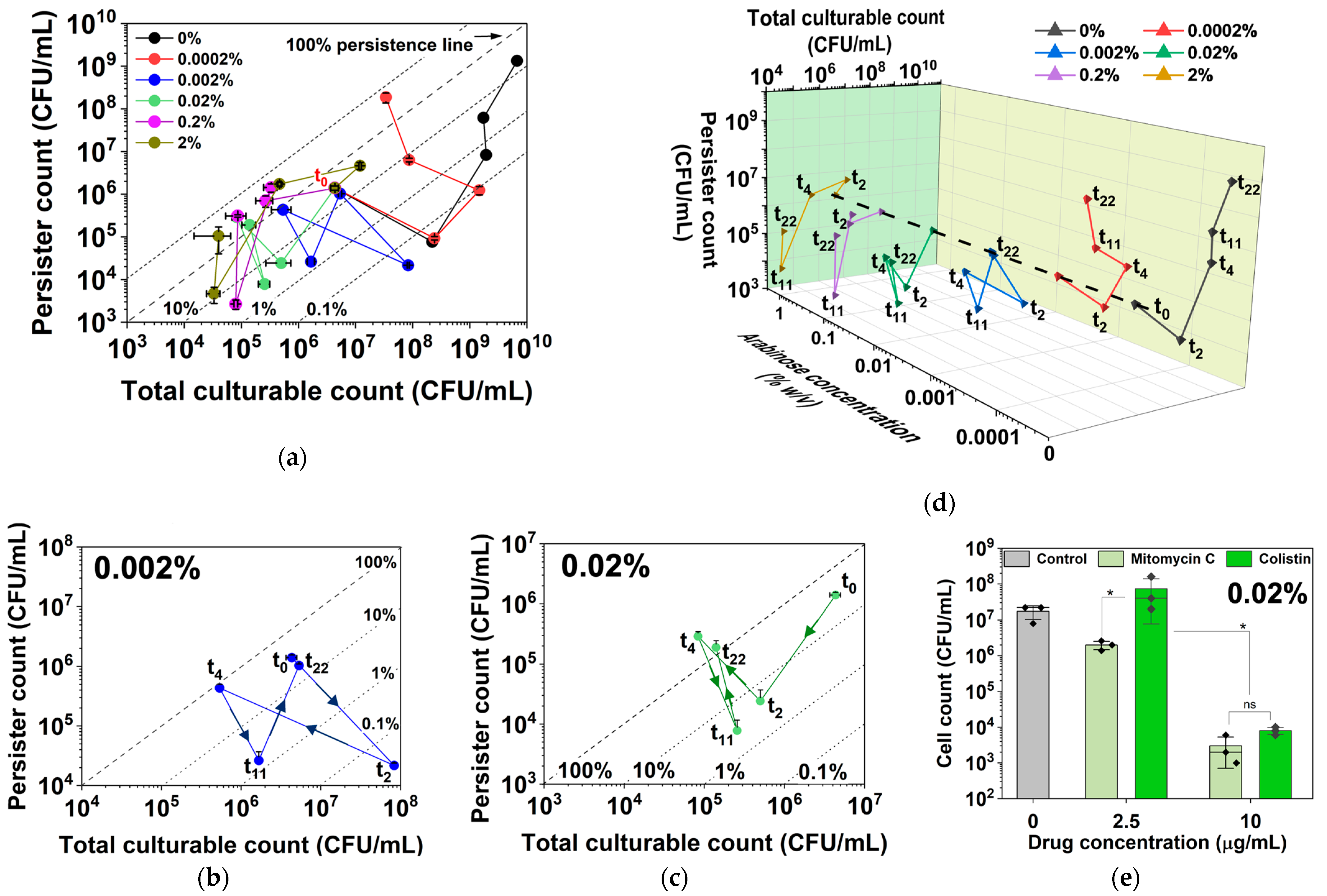
2.1. Dynamic Changes in Persistence and Culturability under hipA Expression
2.2. Dynamic Changes in Culturability and Dormancy under hipA Induction
2.3. Dynamic Changes in Cell Counts Revealed More Information about Dormancy
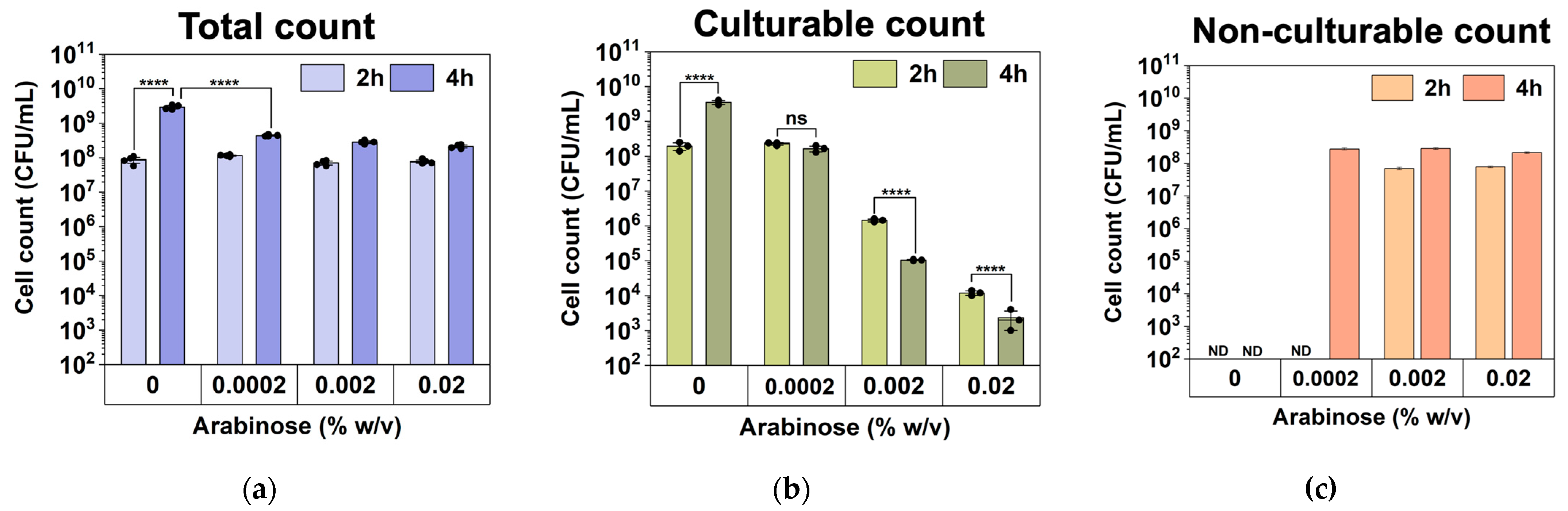
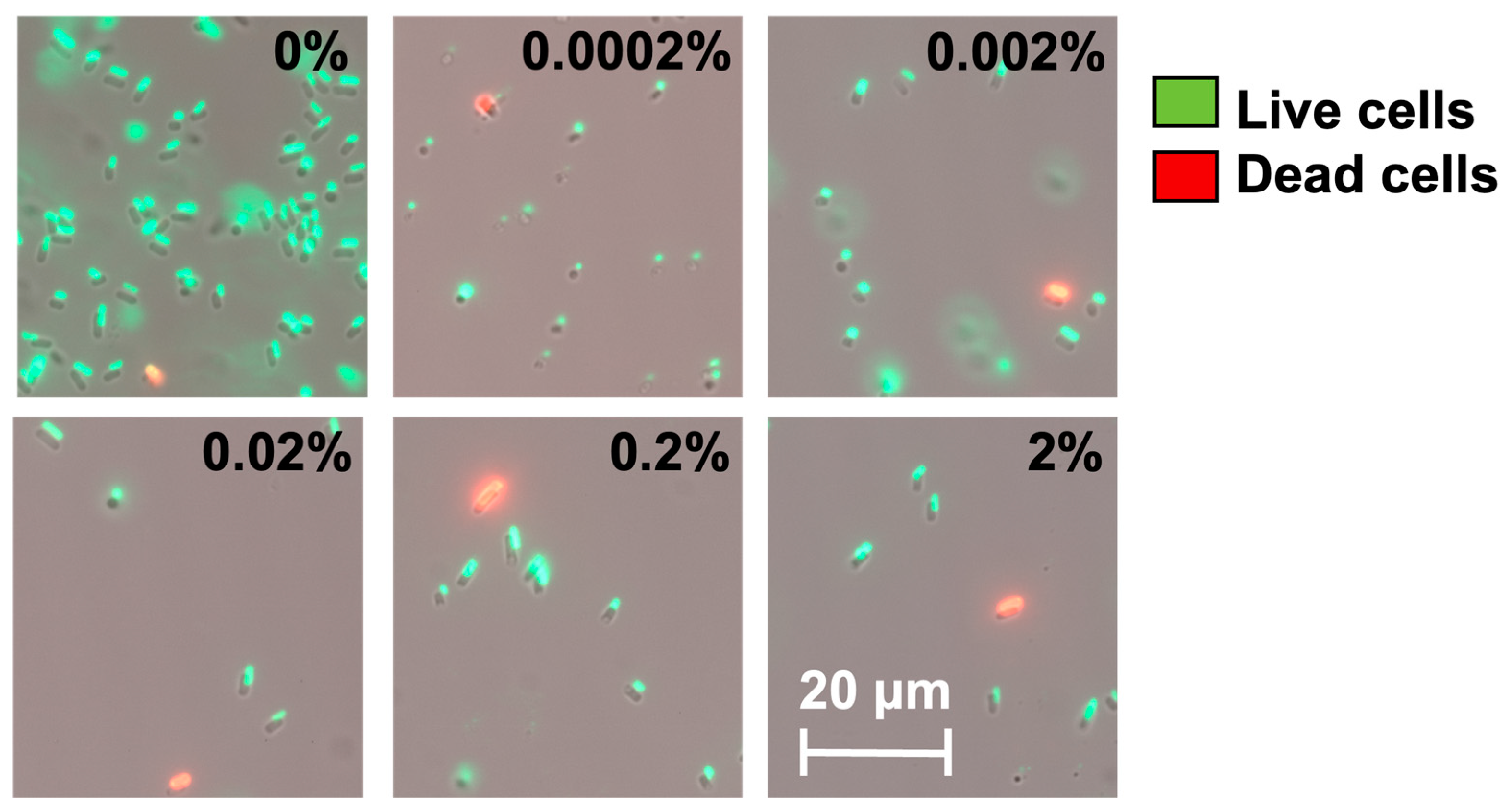
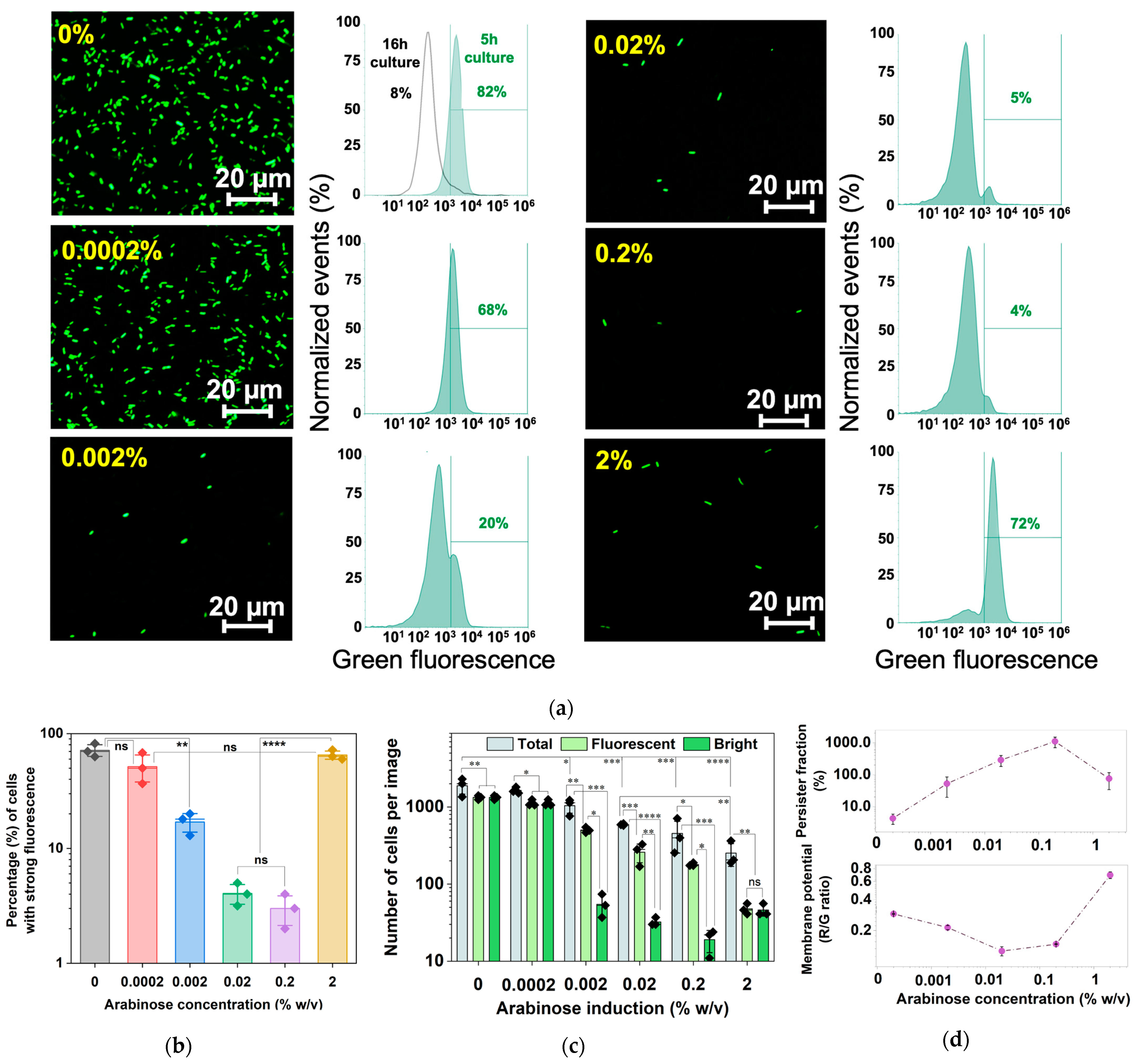
2.4. Arabinose-Induced Expression of hipA
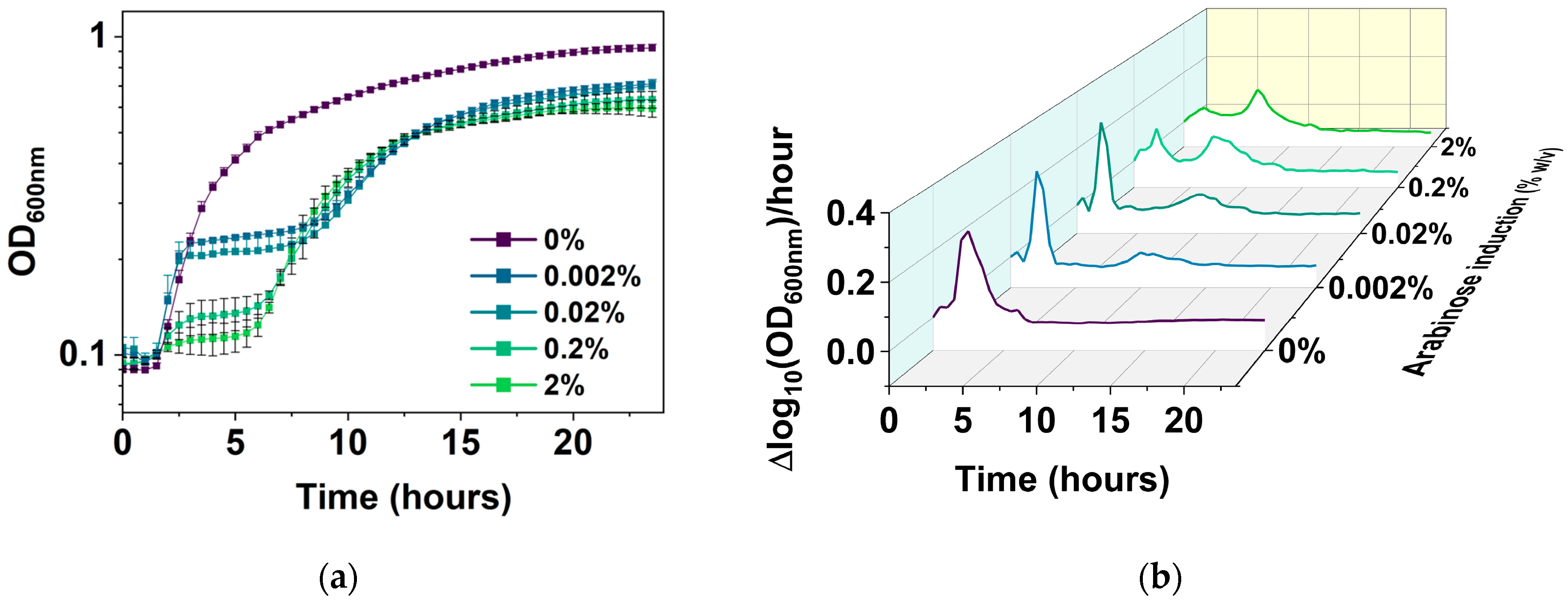
2.5. Corroborating the Results Using a Growth Reporter Strain
3. Discussion
4. Materials and Methods
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Keren, I.; Kaldalu, N.; Spoering, A.; Wang, Y.; Lewis, K. Persister cells and tolerance to antimicrobials. FEMS Microbiol. Lett. 2004, 230, 13–18. [Google Scholar] [CrossRef]
- Gollan, B.; Grabe, G.; Michaux, C.; Helaine, S. Bacterial Persisters and Infection: Past, Present, and Progressing. Annu. Rev. Microbiol. 2019, 73, 359–385. [Google Scholar] [CrossRef] [PubMed]
- Windels, E.M.; Michiels, J.E.; Fauvart, M.; Wenseleers, T.; Van den Bergh, B.; Michiels, J. Bacterial persistence promotes the evolution of antibiotic resistance by increasing survival and mutation rates. ISME J. 2019, 13, 1239–1251. [Google Scholar] [CrossRef] [PubMed]
- Salam, M.A.; Al-Amin, M.Y.; Salam, M.T.; Pawar, J.S.; Akhter, N.; Rabaan, A.A.; Alqumber, M.A.A. Antimicrobial Resistance: A Growing Serious Threat for Global Public Health. Healthcare 2023, 11, 1946. [Google Scholar] [CrossRef]
- Levin-Reisman, I.; Brauner, A.; Ronin, I.; Balaban, N.Q. Epistasis between antibiotic tolerance, persistence, and resistance mutations. Proc. Natl. Acad. Sci. USA 2019, 116, 14734–14739. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Liao, H.; Pei, L.; Pu, Y. Combatting persister cells: The daunting task in post-antibiotics era. Cell Insight 2023, 2, 100104. [Google Scholar] [CrossRef] [PubMed]
- Niu, H.; Gu, J.; Zhang, Y. Bacterial persisters: Molecular mechanisms and therapeutic development. Signal Transduct. Target. Ther. 2024, 9, 174. [Google Scholar] [CrossRef]
- Pan, H.; Ren, Q. Wake Up! Resuscitation of Viable but Nonculturable Bacteria: Mechanism and Potential Application. Foods 2022, 12, 82. [Google Scholar] [CrossRef]
- Zhang, X.H.; Ahmad, W.; Zhu, X.Y.; Chen, J.; Austin, B. Viable but nonculturable bacteria and their resuscitation: Implications for cultivating uncultured marine microorganisms. Mar. Life Sci. Technol. 2021, 3, 189–203. [Google Scholar] [CrossRef]
- Song, S.; Wood, T.K. ‘Viable but non-culturable cells’ are dead. Environ. Microbiol. 2021, 23, 2335–2338. [Google Scholar] [CrossRef]
- Mu, D.S.; Du, Z.J.; Chen, J.; Austin, B.; Zhang, X.H. What do we mean by viability in terms of ‘viable but non-culturable’ cells? Environ. Microbiol. Rep. 2021, 13, 248–252. [Google Scholar] [CrossRef] [PubMed]
- Ayrapetyan, M.; Williams, T.; Oliver, J.D. Relationship between the Viable but Nonculturable State and Antibiotic Persister Cells. J. Bacteriol. 2018, 200, e00249-18. [Google Scholar] [CrossRef] [PubMed]
- Qi, Z.; Liu, C. Metabolic characteristics and markers in viable but nonculturable state of Pseudomonas aeruginosa induced by chlorine stress. Environ. Res. 2022, 214, 114111. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Huang, T.Y.; Mao, Y.; Chen, Y.; Shi, F.; Peng, R.; Chen, J.; Yuan, L.; Bai, C.; Chen, L.; et al. Study on the Viable but Non-culturable (VBNC) State Formation of. Front. Microbiol. 2020, 11, 599739. [Google Scholar] [CrossRef]
- Liu, J.; Yang, L.; Kjellerup, B.V.; Xu, Z. Viable but nonculturable (VBNC) state, an underestimated and controversial microbial survival strategy. Trends Microbiol. 2023, 31, 1013–1023. [Google Scholar] [CrossRef] [PubMed]
- Balaban, N.Q.; Merrin, J.; Chait, R.; Kowalik, L.; Leibler, S. Bacterial persistence as a phenotypic switch. Science 2004, 305, 1622–1625. [Google Scholar] [CrossRef] [PubMed]
- Kojic, M.; Milisavljevic, M. When disaster strikes: Reconstitution of population density by expansion of survivors. Mol. Ecol. 2020, 29, 4757–4764. [Google Scholar] [CrossRef]
- Wilmaerts, D.; Windels, E.M.; Verstraeten, N.; Michiels, J. General Mechanisms Leading to Persister Formation and Awakening. Trends Genet. 2019, 35, 401–411. [Google Scholar] [CrossRef]
- Hong, S.H.; Wang, X.; O’Connor, H.F.; Benedik, M.J.; Wood, T.K. Bacterial persistence increases as environmental fitness decreases. Microb. Biotechnol. 2012, 5, 509–522. [Google Scholar] [CrossRef]
- Barrett, M.P.; Kyle, D.E.; Sibley, L.D.; Radke, J.B.; Tarleton, R.L. Protozoan persister-like cells and drug treatment failure. Nat. Rev. Microbiol. 2019, 17, 607–620. [Google Scholar] [CrossRef]
- Fisher, R.A.; Gollan, B.; Helaine, S. Persistent bacterial infections and persister cells. Nat. Rev. Microbiol. 2017, 15, 453–464. [Google Scholar] [CrossRef] [PubMed]
- Arastehfar, A.; Daneshnia, F.; Cabrera, N.; Penalva-Lopez, S.; Sarathy, J.; Zimmerman, M.; Shor, E.; Perlin, D.S. Macrophage internalization creates a multidrug-tolerant fungal persister reservoir and facilitates the emergence of drug resistance. Nat. Commun. 2023, 14, 1183. [Google Scholar] [CrossRef] [PubMed]
- Hastings, C.J.; Himmler, G.E.; Patel, A.; Marques, C.N.H. Immune Response Modulation by Pseudomonas aeruginosa Persister Cells. mBio 2023, 14, e0005623. [Google Scholar] [CrossRef]
- Urbaniec, J.; Xu, Y.; Hu, Y.; Hingley-Wilson, S.; McFadden, J. Phenotypic heterogeneity in persisters: A novel ‘hunker’ theory of persistence. FEMS Microbiol. Rev. 2022, 46, fuab042. [Google Scholar] [CrossRef] [PubMed]
- Berkvens, A.; Chauhan, P.; Bruggeman, F.J. Integrative biology of persister cell formation: Molecular circuitry, phenotypic diversification and fitness effects. J. R. Soc. Interface 2022, 19, 20220129. [Google Scholar] [CrossRef] [PubMed]
- Moyed, H.S.; Bertrand, K.P. hipA, a newly recognized gene of Escherichia coli K-12 that affects frequency of persistence after inhibition of murein synthesis. J. Bacteriol. 1983, 155, 768–775. [Google Scholar] [CrossRef]
- Hansen, S.; Vulić, M.; Min, J.; Yen, T.J.; Schumacher, M.A.; Brennan, R.G.; Lewis, K. Regulation of the Escherichia coli HipBA toxin-antitoxin system by proteolysis. PLoS ONE 2012, 7, e39185. [Google Scholar] [CrossRef]
- Germain, E.; Castro-Roa, D.; Zenkin, N.; Gerdes, K. Molecular mechanism of bacterial persistence by HipA. Mol. Cell 2013, 52, 248–254. [Google Scholar] [CrossRef]
- Black, D.S.; Kelly, A.J.; Mardis, M.J.; Moyed, H.S. Structure and organization of hip, an operon that affects lethality due to inhibition of peptidoglycan or DNA synthesis. J. Bacteriol. 1991, 173, 5732–5739. [Google Scholar] [CrossRef]
- Semanjski, M.; Germain, E.; Bratl, K.; Kiessling, A.; Gerdes, K.; Macek, B. The kinases HipA and HipA7 phosphorylate different substrate pools in Escherichia coli to promote multidrug tolerance. Sci. Signal 2018, 11, eaat5750. [Google Scholar] [CrossRef]
- Korch, S.B.; Hill, T.M. Ectopic overexpression of wild-type and mutant hipA genes in Escherichia coli: Effects on macromolecular synthesis and persister formation. J. Bacteriol. 2006, 188, 3826–3836. [Google Scholar] [CrossRef]
- Kaspy, I.; Rotem, E.; Weiss, N.; Ronin, I.; Balaban, N.Q.; Glaser, G. HipA-mediated antibiotic persistence via phosphorylation of the glutamyl-tRNA-synthetase. Nat. Commun. 2013, 4, 3001. [Google Scholar] [CrossRef]
- Correia, F.F.; D’Onofrio, A.; Rejtar, T.; Li, L.; Karger, B.L.; Makarova, K.; Koonin, E.V.; Lewis, K. Kinase activity of overexpressed HipA is required for growth arrest and multidrug tolerance in Escherichia coli. J. Bacteriol. 2006, 188, 8360–8367. [Google Scholar] [CrossRef]
- Lin, C.Y.; Hamini, S.; Tupa, P.R.; Masuda, H. Cellular Memory of HipA-Induced Growth Arrest: The Length of Cell Growth Arrest Becomes Shorter for Each Successive Induction. Microorganisms 2021, 9, 2594. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Yin, N.; Liu, H.; Pei, J.; Lai, L. Novel Inhibitors of Toxin HipA Reduce Multidrug Tolerant Persisters. ACS Med. Chem. Lett. 2016, 7, 449–453. [Google Scholar] [CrossRef] [PubMed]
- Rotem, E.; Loinger, A.; Ronin, I.; Levin-Reisman, I.; Gabay, C.; Shoresh, N.; Biham, O.; Balaban, N.Q. Regulation of phenotypic variability by a threshold-based mechanism underlies bacterial persistence. Proc. Natl. Acad. Sci. USA 2010, 107, 12541–12546. [Google Scholar] [CrossRef] [PubMed]
- Arvizu, I.S.; Murray, S.R. A simple, quantitative assay for the detection of viable but non-culturable (VBNC) bacteria. STAR Protoc. 2021, 2, 100738. [Google Scholar] [CrossRef]
- Schumacher, M.A.; Min, J.; Link, T.M.; Guan, Z.; Xu, W.; Ahn, Y.H.; Soderblom, E.J.; Kurie, J.M.; Evdokimov, A.; Moseley, M.A.; et al. Role of unusual P loop ejection and autophosphorylation in HipA-mediated persistence and multidrug tolerance. Cell Rep. 2012, 2, 518–525. [Google Scholar] [CrossRef]
- Zheng, E.J.; Stokes, J.M.; Collins, J.J. Eradicating Bacterial Persisters with Combinations of Strongly and Weakly Metabolism-Dependent Antibiotics. Cell Chem. Biol. 2020, 27, 1544–1552.e3. [Google Scholar] [CrossRef]
- Andersen, J.B.; Sternberg, C.; Poulsen, L.K.; Bjorn, S.P.; Givskov, M.; Molin, S. New unstable variants of green fluorescent protein for studies of transient gene expression in bacteria. Appl. Environ. Microbiol. 1998, 64, 2240–2246. [Google Scholar] [CrossRef]
- Barer, M.R.; Harwood, C.R. Bacterial viability and culturability. Adv. Microb. Physiol. 1999, 41, 93–137. [Google Scholar] [CrossRef] [PubMed]
- Kell, D.B.; Kaprelyants, A.S.; Weichart, D.H.; Harwood, C.R.; Barer, M.R. Viability and activity in readily culturable bacteria: A review and discussion of the practical issues. Antonie Van Leeuwenhoek 1998, 73, 169–187. [Google Scholar] [CrossRef]
- Zhao, S.; Lu, X.; Zhang, J.; Kan, B. Absolute Quantification of Viable but Nonculturable Vibrio cholerae Using Droplet Digital PCR with Oil-Enveloped Bacterial Cells. Microbiol. Spectr. 2022, 10, e0070422. [Google Scholar] [CrossRef]
- Kim, J.S.; Chowdhury, N.; Yamasaki, R.; Wood, T.K. Viable but non-culturable and persistence describe the same bacterial stress state. Environ. Microbiol. 2018, 20, 2038–2048. [Google Scholar] [CrossRef] [PubMed]
- Korch, S.B.; Henderson, T.A.; Hill, T.M. Characterization of the hipA7 allele of Escherichia coli and evidence that high persistence is governed by (p)ppGpp synthesis. Mol. Microbiol. 2003, 50, 1199–1213. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.S.; Wood, T.K. Persistent Persister Misperceptions. Front. Microbiol. 2016, 7, 2134. [Google Scholar] [CrossRef]
- Spanka, D.T.; Konzer, A.; Edelmann, D.; Berghoff, B.A. High-Throughput Proteomics Identifies Proteins with Importance to Postantibiotic Recovery in Depolarized Persister Cells. Front. Microbiol. 2019, 10, 378. [Google Scholar] [CrossRef]
- Lewis, K. Persister Cells. Annu. Rev. Microbiol. 2010, 64, 357–372. [Google Scholar] [CrossRef]
- Ayrapetyan, M.; Williams, T.C.; Oliver, J.D. Bridging the gap between viable but non-culturable and antibiotic persistent bacteria. Trends Microbiol. 2015, 23, 7–13. [Google Scholar] [CrossRef]
- Kirschner, A.K.T.; Vierheilig, J.; Flemming, H.C.; Wingender, J.; Farnleitner, A.H. How dead is dead? Viable but non-culturable versus persister cells. Environ. Microbiol. Rep. 2021, 13, 243–245. [Google Scholar] [CrossRef]
- Asakura, H.; Ishiwa, A.; Arakawa, E.; Makino, S.; Okada, Y.; Yamamoto, S.; Igimi, S. Gene expression profile of Vibrio cholerae in the cold stress-induced viable but non-culturable state. Environ. Microbiol. 2007, 9, 869–879. [Google Scholar] [CrossRef]
- Orman, M.A.; Brynildsen, M.P. Establishment of a method to rapidly assay bacterial persister metabolism. Antimicrob. Agents Chemother. 2013, 57, 4398–4409. [Google Scholar] [CrossRef] [PubMed]
- Datsenko, K.A.; Wanner, B.L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 2000, 97, 6640–6645. [Google Scholar] [CrossRef] [PubMed]
- Amato, S.M.; Brynildsen, M.P. Nutrient transitions are a source of persisters in Escherichia coli biofilms. PLoS ONE 2014, 9, e93110. [Google Scholar] [CrossRef]
- de Lorenzo, V.; Herrero, M.; Jakubzik, U.; Timmis, K.N. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J. Bacteriol. 1990, 172, 6568–6572. [Google Scholar] [CrossRef]
- Zhou, K.; Zhou, L.; Lim, Q.; Zou, R.; Stephanopoulos, G.; Too, H.P. Novel reference genes for quantifying transcriptional responses of Escherichia coli to protein overexpression by quantitative PCR. BMC Mol. Biol. 2011, 12, 18. [Google Scholar] [CrossRef] [PubMed]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef]
- Novo, D.; Perlmutter, N.G.; Hunt, R.H.; Shapiro, H.M. Accurate flow cytometric membrane potential measurement in bacteria using diethyloxacarbocyanine and a ratiometric technique. Cytometry 1999, 35, 55–63. [Google Scholar] [CrossRef]
- Baba, T.; Ara, T.; Hasegawa, M.; Takai, Y.; Okumura, Y.; Baba, M.; Datsenko, K.A.; Tomita, M.; Wanner, B.L.; Mori, H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: The Keio collection. Mol. Syst. Biol. 2006, 2, 2006.0008. [Google Scholar] [CrossRef]


| Gene | Primer Sequence |
|---|---|
| rrsA | FP: 5′-ctcttgccatcggatgtgccca-3′ RP: 5′-cagtgtggctggtcatcctctca-3′ |
| hipA | FP: 5′-ggcggtacgggaatacacat-3′ RP: 5′-ccgggaatctcagcaccttt-3′ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dhaouadi, Y.; Hashemi, M.J.; Ren, D. Persistence and Culturability of Escherichia coli under Induced Toxin Expression. Antibiotics 2024, 13, 863. https://doi.org/10.3390/antibiotics13090863
Dhaouadi Y, Hashemi MJ, Ren D. Persistence and Culturability of Escherichia coli under Induced Toxin Expression. Antibiotics. 2024; 13(9):863. https://doi.org/10.3390/antibiotics13090863
Chicago/Turabian StyleDhaouadi, Yousr, Mohamad Javad Hashemi, and Dacheng Ren. 2024. "Persistence and Culturability of Escherichia coli under Induced Toxin Expression" Antibiotics 13, no. 9: 863. https://doi.org/10.3390/antibiotics13090863
APA StyleDhaouadi, Y., Hashemi, M. J., & Ren, D. (2024). Persistence and Culturability of Escherichia coli under Induced Toxin Expression. Antibiotics, 13(9), 863. https://doi.org/10.3390/antibiotics13090863








