Rapid Phenotypic and Genotypic Antimicrobial Susceptibility Testing Approaches for Use in the Clinical Laboratory
Abstract
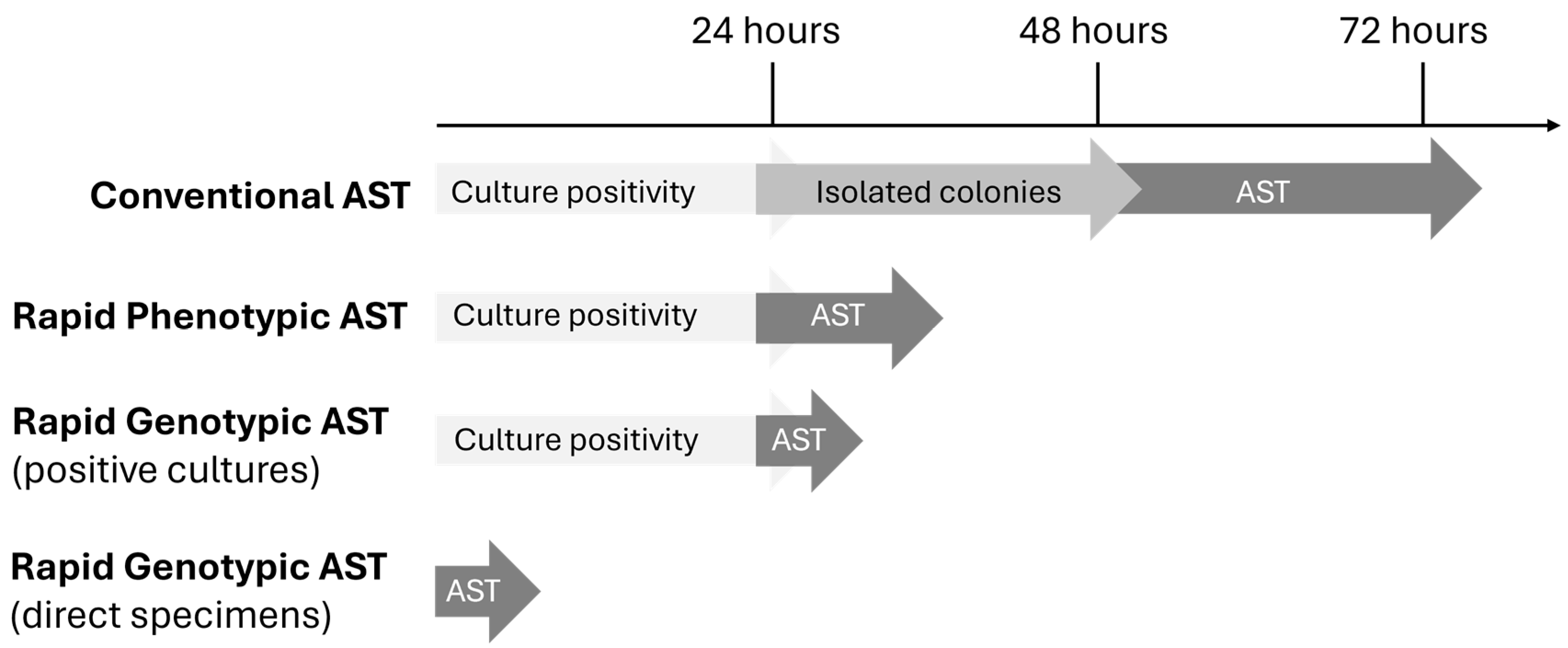
1. Introduction
2. Conventional Approaches to Antimicrobial Susceptibility Testing
3. Rapid Phenotypic AST Approaches
4. Rapid Genotypic AST Platforms
4.1. Genotypic Approaches with Limited On-Panel Targets
4.2. Genotypic Approaches for Syndromic, Multiplex Testing
5. Phenotypic versus Genotypic Approaches
6. Clinical Impact
7. Considerations for Implementation of Rapid AST Technologies in the Clinical Laboratory
8. The Future of Rapid AST
9. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Collaborators, A.R. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef]
- Boccabella, L.; Palma, E.G.; Abenavoli, L.; Scarlata, G.G.M.; Boni, M.; Ianiro, G.; Santori, P.; Tack, J.F.; Scarpellini, E. Post-Coronavirus Disease 2019 Pandemic Antimicrobial Resistance. Antibiotics 2024, 13, 233. [Google Scholar] [CrossRef]
- Larkin, H. Increasing Antimicrobial Resistance Poses Global Threat, WHO Says. JAMA 2023, 329, 200. [Google Scholar] [CrossRef]
- Balouiri, M.; Sadiki, M.; Ibnsouda, S.K. Methods for in vitro evaluating antimicrobial activity: A review. J. Pharm. Anal. 2016, 6, 71–79. [Google Scholar] [CrossRef]
- Ferrer, R.; Martin-Loeches, I.; Phillips, G.; Osborn, T.M.; Townsend, S.; Dellinger, R.P.; Artigas, A.; Schorr, C.; Levy, M.M. Empiric antibiotic treatment reduces mortality in severe sepsis and septic shock from the first hour: Results from a guideline-based performance improvement program. Crit. Care Med. 2014, 42, 1749–1755. [Google Scholar] [CrossRef]
- Brennan-Krohn, T.; Smith, K.P.; Kirby, J.E. The Poisoned Well: Enhancing the Predictive Value of Antimicrobial Susceptibility Testing in the Era of Multidrug Resistance. J. Clin. Microbiol. 2017, 55, 2304–2308. [Google Scholar] [CrossRef]
- Clinical Laboratory and Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing, CLSI Guideline M1000, 34th ed.; Clinical Laboratory and Standards Institute: Wayne, PA, USA, 2024. [Google Scholar]
- United States Food and Drug Administration. Antibacterial Susceptibility Test Interpretive Criteria. Available online: https://www.fda.gov/drugs/development-resources/antibacterial-susceptibility-test-interpretive-criteria (accessed on 15 July 2024).
- European Committee on Antimicrobial Susceptibility Testing. Clinical Breakpoints—Breakpoints and Guidance. Available online: https://www.eucast.org/clinical_breakpoints (accessed on 15 July 2024).
- Jorgensen, J.H.; Ferraro, M.J. Antimicrobial susceptibility testing: A review of general principles and contemporary practices. Clin. Infect. Dis. 2009, 49, 1749–1755. [Google Scholar] [CrossRef] [PubMed]
- Clark, R.B.; Lewinski, M.A.; Loeffelholz, M.J.; Tibbetts, R.J. Cumitech 31A: Verification and Validation of Procedures in the Clinical Microbiology Laboratory; ASM Press: Washington, DC, USA, 2009. [Google Scholar]
- Cenci, E.; Paggi, R.; Socio, G.V.; Bozza, S.; Camilloni, B.; Pietrella, D.; Mencacci, A. Accelerate Pheno™ blood culture detection system: A literature review. Future Microbiol. 2020, 15, 1595–1605. [Google Scholar] [CrossRef] [PubMed]
- Ullberg, M.; Özenci, V. Identification and antimicrobial susceptibility testing of Gram-positive and Gram-negative bacteria from positive blood cultures using the Accelerate Pheno™ system. Eur. J. Clin. Microbiol. Infect. Dis. 2020, 39, 139–149. [Google Scholar] [CrossRef]
- Marschal, M.; Bachmaier, J.; Autenrieth, I.; Oberhettinger, P.; Willmann, M.; Peter, S. Evaluation of the Accelerate Pheno System for Fast Identification and Antimicrobial Susceptibility Testing from Positive Blood Cultures in Bloodstream Infections Caused by Gram-Negative Pathogens. J. Clin. Microbiol. 2017, 55, 2116–2126. [Google Scholar] [CrossRef]
- Lutgring, J.D.; Bittencourt, C.; McElvania TeKippe, E.; Cavuoti, D.; Hollaway, R.; Burd, E.M. Evaluation of the Accelerate Pheno System: Results from Two Academic Medical Centers. J. Clin. Microbiol. 2018, 56, e01672-17. [Google Scholar] [CrossRef]
- Elliott, G.; Malczynski, M.; Barr, V.O.; Aljefri, D.; Martin, D.; Sutton, S.; Zembower, T.R.; Postelnick, M.; Qi, C. Evaluation of the impact of the Accelerate Pheno™ system on time to result for differing antimicrobial stewardship intervention models in patients with gram-negative bloodstream infections. BMC Infect. Dis. 2019, 19, 942. [Google Scholar] [CrossRef]
- Starr, K.F.; Robinson, D.C.; Hazen, K.C. Performance of the Accelerate Diagnostics Pheno(TM) system with resin-containing BacT/ALERT® Plus blood culture bottles. Diagn. Microbiol. Infect. Dis. 2019, 94, 122–128. [Google Scholar] [CrossRef]
- De Angelis, G.; Posteraro, B.; Menchinelli, G.; Liotti, F.M.; Spanu, T.; Sanguinetti, M. Antimicrobial susceptibility testing of pathogens isolated from blood culture: A performance comparison of Accelerate Pheno™ and VITEK® 2 systems with the broth microdilution method. J. Antimicrob. Chemother. 2019, 74, i24–i31. [Google Scholar] [CrossRef]
- Burg, T.P.; Godin, M.; Knudsen, S.M.; Shen, W.; Carlson, G.; Foster, J.S.; Babcock, K.; Manalis, S.R. Weighing of biomolecules, single cells and single nanoparticles in fluid. Nature 2007, 446, 1066–1069. [Google Scholar] [CrossRef]
- Montelongo-Jauregui, D.; Slechta, E.S.; Fisher, M. Evaluation of the LifeScale system for rapid phenotypic antimicrobial susceptibility testing from positive blood cultures. Am. J. Clin. Pathol. 2023, 160, S137–S138. [Google Scholar] [CrossRef]
- Banchini, I.; Borgatti, E.C.; Foschi, C.; Lazzarotto, T.; Ambretti, S. Evaluation of an automated rapid phenotypic antimicrobial susceptibility testing (ASTar, Q-linea AB) applied directly on blood cultures bottles positive for Gram-negative pathogens. New Microbiol. 2024, 47, 107–110. [Google Scholar]
- Göransson, J.; Sundqvist, M.; Ghaderi, E.; Lisby, J.G.; Molin, Y.; Eriksson, E.; Carlsson, S.; Cederlöf, A.; Ellis, L.; Melin, J. Performance of a System for Rapid Phenotypic Antimicrobial Susceptibility Testing of Gram-Negative Bacteria Directly from Positive Blood Culture Bottles. J. Clin. Microbiol. 2023, 61, e0152522. [Google Scholar] [CrossRef]
- Esse, J.; Träger, J.; Valenza, G.; Bogdan, C.; Held, J. Rapid phenotypic antimicrobial susceptibility testing of Gram-negative rods directly from positive blood cultures using the novel Q-linea ASTar system. J. Clin. Microbiol. 2023, 61, e0054923. [Google Scholar] [CrossRef]
- Tibbetts, R.; George, S.; Burwell, R.; Rajeev, L.; Rhodes, P.A.; Singh, P.; Samuel, L. Performance of the Reveal Rapid Antibiotic Susceptibility Testing System on Gram-Negative Blood Cultures at a Large Urban Hospital. J. Clin. Microbiol. 2022, 60, e0009822. [Google Scholar] [CrossRef]
- Flentie, K.; Spears, B.R.; Chen, F.; Purmort, N.B.; DaPonte, K.; Viveiros, E.; Phelan, N.; Krebill, C.; Flyer, A.N.; Hooper, D.C.; et al. Microplate-based surface area assay for rapid phenotypic antibiotic susceptibility testing. Sci. Rep. 2019, 9, 237. [Google Scholar] [CrossRef] [PubMed]
- Baker, K.R.; Flentie, K.; Spears, B.R.; Mozharov, S.; Roberts, K.; El Ganbour, A.; Somers, M.; Calkwood, J.; Liu, J.; DaPonte, K.; et al. Multicenter evaluation of the Selux Next-Generation Phenotyping antimicrobial susceptibility testing system. J. Clin. Microbiol. 2024, 62, e0054623. [Google Scholar] [CrossRef]
- Wistrand-Yuen, P.; Malmberg, C.; Fatsis-Kavalopoulos, N.; Lübke, M.; Tängdén, T.; Kreuger, J. A Multiplex Fluidic Chip for Rapid Phenotypic Antibiotic Susceptibility Testing. mBio 2020, 11, e03109-19. [Google Scholar] [CrossRef]
- Malmberg, C.; Torpner, J.; Fernberg, J.; Öhrn, H.; Ångström, J.; Johansson, C.; Tängdén, T.; Kreuger, J. Evaluation of the Speed, Accuracy and Precision of the QuickMIC Rapid Antibiotic Susceptibility Testing Assay with Gram-Negative Bacteria in a Clinical Setting. Front. Cell. Infect. Microbiol. 2022, 12, 758262. [Google Scholar] [CrossRef]
- Boland, L.; Streel, C.; De Wolf, H.; Rodriguez, H.; Verroken, A. Rapid antimicrobial susceptibility testing on positive blood cultures through an innovative light scattering technology: Performances and turnaround time evaluation. BMC Infect. Dis. 2019, 19, 989. [Google Scholar] [CrossRef]
- Cupaiolo, R.; Cherkaoui, S.; Serrano, G.; Dauby, N.; Georgala, A.; Blumental, S.; Maillart, E.; Hites, M.; Hallin, M.; Martiny, D. Antimicrobial susceptibility testing determined by Alfred 60/AST (Alifax®) in a multi-sites lab: Performance’s evaluation and optimization of workflow. J. Microbiol. Methods 2022, 194, 106433. [Google Scholar] [CrossRef]
- Anton-Vazquez, V.; Adjepong, S.; Suarez, C.; Planche, T. Evaluation of a new Rapid Antimicrobial Susceptibility system for Gram-negative and Gram-positive bloodstream infections: Speed and accuracy of Alfred 60AST. BMC Microbiol. 2019, 19, 268. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Carrillo, C.; Pescador, P.; Ricote, R.; Fuentes, J.; Losada, C.; Candela, A.; Cercenado, E. Evaluation of the Alfred AST® system for rapid antimicrobial susceptibility testing directly from positive blood cultures. Eur. J. Clin. Microbiol. Infect. Dis. 2019, 38, 1665–1670. [Google Scholar] [CrossRef] [PubMed]
- Giordano, C.; Piccoli, E.; Brucculeri, V.; Barnini, S. A Prospective Evaluation of Two Rapid Phenotypical Antimicrobial Susceptibility Technologies for the Diagnostic Stewardship of Sepsis. BioMed Res. Int. 2018, 2018, 6976923. [Google Scholar] [CrossRef]
- Choi, J.; Jeong, H.Y.; Lee, G.Y.; Han, S.; Han, S.; Jin, B.; Lim, T.; Kim, S.; Kim, D.Y.; Kim, H.C.; et al. Direct, rapid antimicrobial susceptibility test from positive blood cultures based on microscopic imaging analysis. Sci. Rep. 2017, 7, 1148. [Google Scholar] [CrossRef]
- Wong, A.Y.W.; Johnsson, A.T.A.; Özenci, V. Performance of dRAST on Prospective Clinical Blood Culture Samples in a Simulated Clinical Setting and on Multidrug-Resistant Bacteria. Microbiol. Spectr. 2022, 10, e0210721. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.Y.; Kang, M.; Shim, H.J.; Kang, O.K.; Huh, H.J.; Lee, N.Y. Evaluation of the QMAC-dRAST System Version 2.5 for Rapid Antimicrobial Susceptibility Testing of Gram-Negative Bacteria from Positive Blood Culture Broth and Subcultured Colony Isolates. J. Clin. Lab. Anal. 2024, 38, e25043. [Google Scholar] [CrossRef]
- Rosselin, M.; Prod’hom, G.; Greub, G.; Croxatto, A. Performance Evaluation of the Quantamatrix QMAC-dRAST System for Rapid Antibiotic Susceptibility Testing Directly from Blood Cultures. Microorganisms 2022, 10, 1212. [Google Scholar] [CrossRef] [PubMed]
- Pina-Vaz, C.; Silva-Dias, A.; Martins-Oliveira, I.; Gomes, R.; Perez-Viso, B.; Cruz, S.; Rodrigues, A.G.; Sarmento, A.; Cantón, R. A multisite validation of a two hours antibiotic susceptibility flow cytometry assay directly from positive blood cultures. BMC Microbiol. 2024, 24, 187. [Google Scholar] [CrossRef]
- Silva-Dias, A.; Pérez-Viso, B.; Martins-Oliveira, I.; Gomes, R.; Rodrigues, A.G.; Cantón, R.; Pina-Vaz, C. Evaluation of FASTinov Ultrarapid Flow Cytometry Antimicrobial Susceptibility Testing Directly from Positive Blood Cultures. J. Clin. Microbiol. 2021, 59, e0054421. [Google Scholar] [CrossRef]
- Fonseca, E.S.D.; Silva-Dias, A.; Gomes, R.; Martins-Oliveira, I.; Ramos, M.H.; Rodrigues, A.G.; Cantón, R.; Pina-Vaz, C. Evaluation of rapid colistin susceptibility directly from positive blood cultures using a flow cytometry assay. Int. J. Antimicrob. Agents 2019, 54, 820–823. [Google Scholar] [CrossRef] [PubMed]
- Fonseca, E.S.D.; Andrade, F.F.; Gomes, R.; Silva-Dias, A.; Martins-Oliveira, I.; Pérez-Viso, B.; Ramos, M.H.; Rodrigues, A.G.; Cantón, R.; Pina-Vaz, C. Ultra-rapid flow cytometry assay for colistin MIC determination in Enterobacterales, Pseudomonas aeruginosa and Acinetobacter baumannii. Clin. Microbiol. Infect. 2020, 26, 1559.e1–1559.e4. [Google Scholar] [CrossRef]
- Cruz, S.; Abreu, D.; Gomes, R.; Martins-Oliveira, I.; Silva-Dias, A.; Perez-Viso, B.; Cantón, R.; Pina-Vaz, C. An improved protocol for bacteria identification by MALDI-TOF MS directly from positive blood cultures. Eur. J. Clin. Microbiol. Infect. Dis. 2024, 43, 605–610. [Google Scholar] [CrossRef]
- Åkerlund, A.; Jonasson, E.; Matuschek, E.; Serrander, L.; Sundqvist, M.; Kahlmeter, G.; the RAST Study Group. EUCAST rapid antimicrobial susceptibility testing (RAST) in blood cultures: Validation in 55 European laboratories. J. Antimicrob. Chemother. 2020, 75, 3230–3238. [Google Scholar] [CrossRef]
- Jonasson, E.; Matuschek, E.; Kahlmeter, G. The EUCAST rapid disc diffusion method for antimicrobial susceptibility testing directly from positive blood culture bottles. J. Antimicrob. Chemother. 2020, 75, 968–978. [Google Scholar] [CrossRef]
- Humphries, R.; Bobenchik, A.M.; Hindler, J.A.; Schuetz, A.N. Overview of Changes to the Clinical and Laboratory Standards Institute Performance Standards for Antimicrobial Susceptibility Testing, M100, 31st Edition. J. Clin. Microbiol. 2021, 59, e0021321. [Google Scholar] [CrossRef] [PubMed]
- Savage, T.J.; Rao, S.; Joerger, J.; Ozonoff, A.; McAdam, A.J.; Sandora, T.J. Predictive Value of Direct Disk Diffusion Testing from Positive Blood Cultures in a Children’s Hospital and Its Utility in Antimicrobial Stewardship. J. Clin. Microbiol. 2021, 59. [Google Scholar] [CrossRef] [PubMed]
- Bard, J.D.; Lee, F. Why Can’t We Just Use PCR? The Role of Genotypic versus Phenotypic Testing for Antimicrobial Resistance Testing. Clin. Microbiol. Newsl. 2018, 40, 87–95. [Google Scholar] [CrossRef] [PubMed]
- Smith, K.P.; Kirby, J.E. Rapid Susceptibility Testing Methods. Clin. Lab. Med. 2019, 39, 333–344. [Google Scholar] [CrossRef]
- Simner, P.J.; Dien Bard, J.; Doern, C.; Kristie Johnson, J.; Westblade, L.; Yenokyan, G.; Patel, R.; Hanson, K.E. Reporting of Antimicrobial Resistance from Blood Cultures, an Antibacterial Resistance Leadership Group Survey Summary: Resistance Marker Reporting Practices from Positive Blood Cultures. Clin. Infect. Dis. 2023, 76, 1550–1558. [Google Scholar] [CrossRef]
- Becker, K.; Pagnier, I.; Schuhen, B.; Wenzelburger, F.; Friedrich, A.W.; Kipp, F.; Peters, G.; von Eiff, C. Does nasal cocolonization by methicillin-resistant coagulase-negative staphylococci and methicillin-susceptible Staphylococcus aureus strains occur frequently enough to represent a risk of false-positive methicillin-resistant S. aureus determinations by molecular methods? J. Clin. Microbiol. 2006, 44, 229–231. [Google Scholar] [CrossRef]
- Okuma, K.; Iwakawa, K.; Turnidge, J.D.; Grubb, W.B.; Bell, J.M.; O’Brien, F.G.; Coombs, G.W.; Pearman, J.W.; Tenover, F.C.; Kapi, M.; et al. Dissemination of new methicillin-resistant Staphylococcus aureus clones in the community. J. Clin. Microbiol. 2002, 40, 4289–4294. [Google Scholar] [CrossRef]
- Jennings, L.; Van Deerlin, V.M.; Gulley, M.L. Recommended principles and practices for validating clinical molecular pathology tests. Arch. Pathol. Lab. Med. 2009, 133, 743–755. [Google Scholar] [CrossRef]
- Burd, E.M. Validation of laboratory-developed molecular assays for infectious diseases. Clin. Microbiol. Rev. 2010, 23, 550–576. [Google Scholar] [CrossRef]
- Simner, P.J.; Musser, K.A.; Mitchell, K.; Wise, M.G.; Lewis, S.; Yee, R.; Bergman, Y.; Good, C.E.; Abdelhamed, A.M.; Li, H.; et al. Multicenter Evaluation of the Acuitas AMR Gene Panel for Detection of an Extended Panel of Antimicrobial Resistance Genes among Bacterial Isolates. J. Clin. Microbiol. 2022, 60, e0209821. [Google Scholar] [CrossRef]
- Piatek, A.S.; Van Cleeff, M.; Alexander, H.; Coggin, W.L.; Rehr, M.; Van Kampen, S.; Shinnick, T.M.; Mukadi, Y. GeneXpert for TB diagnosis: Planned and purposeful implementation. Glob. Health Sci. Pract. 2013, 1, 18–23. [Google Scholar] [CrossRef]
- Cattamanchi, A.; Berger, C.A.; Shete, P.B.; Turyahabwe, S.; Joloba, M.; Moore, D.A.; Davis, L.J.; Katamba, A. Implementation science to improve the quality of tuberculosis diagnostic services in Uganda. J. Clin. Tuberc. Other Mycobact. Dis. 2020, 18, 100136. [Google Scholar] [CrossRef]
- Brown, S.; Leavy, J.E.; Jancey, J. Implementation of GeneXpert for TB Testing in Low- and Middle-Income Countries: A Systematic Review. Glob. Health Sci. Pract. 2021, 9, 698–710. [Google Scholar] [CrossRef]
- Li, S.; Liu, B.; Peng, M.; Chen, M.; Yin, W.; Tang, H.; Luo, Y.; Hu, P.; Ren, H. Diagnostic accuracy of Xpert MTB/RIF for tuberculosis detection in different regions with different endemic burden: A systematic review and meta-analysis. PLoS ONE 2017, 12, e0180725. [Google Scholar] [CrossRef] [PubMed]
- Cuong, N.K.; Ngoc, N.B.; Hoa, N.B.; Dat, V.Q.; Nhung, N.V. GeneXpert on patients with human immunodeficiency virus and smear-negative pulmonary tuberculosis. PLoS ONE 2021, 16, e0253961. [Google Scholar] [CrossRef]
- Bouzouita, I.; Ghariani, A.; Dhaou, K.B.; Jemaeil, S.; Essaalah, L.; Bejaoui, S.; Draoui, H.; El Marzouk, N.; Mehiri, E.; Slim-Saidi, L. Usefulness of Xpert MTB/RIF Ultra for rapid diagnosis of extrapulmonary tuberculosis in Tunisia. Sci. Rep. 2024, 14, 2217. [Google Scholar] [CrossRef] [PubMed]
- Dorman, S.E.; Schumacher, S.G.; Alland, D.; Nabeta, P.; Armstrong, D.T.; King, B.; Hall, S.L.; Chakravorty, S.; Cirillo, D.M.; Tukvadze, N.; et al. Xpert MTB/RIF Ultra for detection of Mycobacterium tuberculosis and rifampicin resistance: A prospective multicentre diagnostic accuracy study. Lancet Infect. Dis. 2018, 18, 76–84. [Google Scholar] [CrossRef]
- Chakravorty, S.; Simmons, A.M.; Rowneki, M.; Parmar, H.; Cao, Y.; Ryan, J.; Banada, P.P.; Deshpande, S.; Shenai, S.; Gall, A.; et al. The New Xpert MTB/RIF Ultra: Improving Detection of Mycobacterium tuberculosis and Resistance to Rifampin in an Assay Suitable for Point-of-Care Testing. mBio 2017, 8, e00812-17. [Google Scholar] [CrossRef] [PubMed]
- Armstrong, D.T.; Fisher, S.; Totten, M.; Schwartz, M.; Gnanashanmugam, D.; Parrish, N. Clinical Validation of the Xpert MTB/RIF Test for Identification of the Mycobacterium tuberculosis Complex in Acid-Fast Bacillus Smear-Positive MGIT Broth Cultures. J. Clin. Microbiol. 2022, 60, e0216421. [Google Scholar] [CrossRef]
- Altun, O.; Almuhayawi, M.; Ullberg, M.; Ozenci, V. Clinical evaluation of the FilmArray blood culture identification panel in identification of bacteria and yeasts from positive blood culture bottles. J. Clin. Microbiol. 2013, 51, 4130–4136. [Google Scholar] [CrossRef]
- She, R.C.; Bender, J.M. Advances in Rapid Molecular Blood Culture Diagnostics: Healthcare Impact, Laboratory Implications, and Multiplex Technologies. J. Appl. Lab. Med. 2019, 3, 617–630. [Google Scholar] [CrossRef]
- Ginocchio, C.C.; Garcia-Mondragon, C.; Mauerhofer, B.; Rindlisbacher, C. Multinational evaluation of the BioFire® FilmArray® Pneumonia plus Panel as compared to standard of care testing. Eur. J. Clin. Microbiol. Infect. Dis. 2021, 40, 1609–1622. [Google Scholar] [CrossRef] [PubMed]
- Klein, M.; Bacher, J.; Barth, S.; Atrzadeh, F.; Siebenhaller, K.; Ferreira, I.; Beisken, S.; Posch, A.E.; Carroll, K.C.; Wunderink, R.G.; et al. Multicenter Evaluation of the Unyvero Platform for Testing Bronchoalveolar Lavage Fluid. J. Clin. Microbiol. 2021, 59. [Google Scholar] [CrossRef]
- Murdoch, D.R.; O’Brien, K.L.; Scott, J.A.; Karron, R.A.; Bhat, N.; Driscoll, A.J.; Knoll, M.D.; Levine, O.S. Breathing new life into pneumonia diagnostics. J. Clin. Microbiol. 2009, 47, 3405–3408. [Google Scholar] [CrossRef] [PubMed]
- Esteban, J.; Salar-Vidal, L.; Schmitt, B.H.; Waggoner, A.; Laurent, F.; Abad, L.; Bauer, T.W.; Mazariegos, I.; Balada-Llasat, J.-M.; Horn, J.; et al. Multicenter evaluation of the BIOFIRE Joint Infection Panel for the detection of bacteria, yeast, and AMR genes in synovial fluid samples. J. Clin. Microbiol. 2023, 61, e00357-00323. [Google Scholar] [CrossRef]
- Banerjee, R.; Patel, R. Molecular diagnostics for genotypic detection of antibiotic resistance: Current landscape and future directions. JAC-Antimicrob. Resist. 2023, 5, dlad018. [Google Scholar] [CrossRef]
- Yee, R.; Dien Bard, J.; Simner, P.J. The Genotype-to-Phenotype Dilemma: How Should Laboratories Approach Discordant Susceptibility Results? J. Clin. Microbiol. 2021, 59. [Google Scholar] [CrossRef]
- Tamma, P.D.; Sharara, S.L.; Pana, Z.D.; Amoah, J.; Fisher, S.L.; Tekle, T.; Doi, Y.; Simner, P.J. Molecular Epidemiology of Ceftriaxone Non-Susceptible Enterobacterales Isolates in an Academic Medical Center in the United States. Open Forum Infect. Dis. 2019, 6, ofz353. [Google Scholar] [CrossRef]
- Perez, K.K.; Olsen, R.J.; Musick, W.L.; Cernoch, P.L.; Davis, J.R.; Peterson, L.E.; Musser, J.M. Integrating rapid diagnostics and antimicrobial stewardship improves outcomes in patients with antibiotic-resistant Gram-negative bacteremia. J. Infect. 2014, 69, 216–225. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, R.; Komarow, L.; Virk, A.; Rajapakse, N.; Schuetz, A.N.; Dylla, B.; Earley, M.; Lok, J.; Kohner, P.; Ihde, S.; et al. Randomized Trial Evaluating Clinical Impact of RAPid IDentification and Susceptibility Testing for Gram-negative Bacteremia: RAPIDS-GN. Clin. Infect. Dis. 2021, 73, e39–e46. [Google Scholar] [CrossRef]
- Banerjee, R.; Teng, C.B.; Cunningham, S.A.; Ihde, S.M.; Steckelberg, J.M.; Moriarty, J.P.; Shah, N.D.; Mandrekar, J.N.; Patel, R. Randomized Trial of Rapid Multiplex Polymerase Chain Reaction-Based Blood Culture Identification and Susceptibility Testing. Clin. Infect. Dis. 2015, 61, 1071–1080. [Google Scholar] [CrossRef] [PubMed]
- Darie, A.M.; Khanna, N.; Jahn, K.; Osthoff, M.; Bassetti, S.; Osthoff, M.; Schumann, D.M.; Albrich, W.C.; Hirsch, H.; Brutsche, M.; et al. Fast multiplex bacterial PCR of bronchoalveolar lavage for antibiotic stewardship in hospitalised patients with pneumonia at risk of Gram-negative bacterial infection (Flagship II): A multicentre, randomised controlled trial. Lancet Respir. Med. 2022, 10, 877–887. [Google Scholar] [CrossRef] [PubMed]
- Miller, M.M.; Van Schooneveld, T.C.; Stohs, E.J.; Marcelin, J.R.; Alexander, B.T.; Watkins, A.B.; Creager, H.M.; Bergman, S.J. Implementation of a Rapid Multiplex Polymerase Chain Reaction Pneumonia Panel and Subsequent Antibiotic De-escalation. Open Forum Infect. Dis. 2023, 10, ofad382. [Google Scholar] [CrossRef] [PubMed]
- WHO. WHO Guidelines Approved by the Guidelines Review Committee. In Global Guidelines for the Prevention of Surgical Site Infection; World Health Organization: Geneva, Switzerland, 2018. [Google Scholar]
- Coia, J.E.; Wilson, J.A.; Bak, A.; Marsden, G.L.; Shimonovich, M.; Loveday, H.P.; Humphreys, H.; Wigglesworth, N.; Demirjian, A.; Brooks, J.; et al. Joint Healthcare Infection Society (HIS) and Infection Prevention Society (IPS) guidelines for the prevention and control of meticillin-resistant Staphylococcus aureus (MRSA) in healthcare facilities. J. Hosp. Infect. 2021, 118, S1–S39. [Google Scholar] [CrossRef]
- Birgand, G.; Ruimy, R.; Schwarzinger, M.; Lolom, I.; Bendjelloul, G.; Houhou, N.; Armand-Lefevre, L.; Andremont, A.; Yazdanpanah, Y.; Lucet, J.C. Rapid detection of glycopeptide-resistant enterococci: Impact on decision-making and costs. Antimicrob. Resist. Infect. Control 2013, 2, 30. [Google Scholar] [CrossRef]
- Zhou, M.; Kudinha, T.; Du, B.; Peng, J.; Ma, X.; Yang, Y.; Zhang, G.; Zhang, J.; Yang, Q.; Xu, Y.C. Active Surveillance of Carbapenemase-Producing Organisms (CPO) Colonization with Xpert Carba-R Assay Plus Positive Patient Isolation Proves to Be Effective in CPO Containment. Front. Cell. Infect. Microbiol. 2019, 9, 162. [Google Scholar] [CrossRef]
- Wolfe, K.H.; Pierce, V.M.; Humphries, R.M. How New Regulation of Laboratory-Developed Antimicrobial Susceptibility Tests Will Affect Infectious Diseases Clinical Practice. Clin. Infect. Dis. 2024, 78, 1140–1147. [Google Scholar] [CrossRef]
- Elnifro, E.M.; Ashshi, A.M.; Cooper, R.J.; Klapper, P.E. Multiplex PCR: Optimization and application in diagnostic virology. Clin. Microbiol. Rev. 2000, 13, 559–570. [Google Scholar] [CrossRef]
- Truong, T.T.; Mongkolrattanothai, K.; Flores, I.I.; Dien Bard, J. Evaluation of the Performance and Clinical Impact of a Rapid Phenotypic Susceptibility Testing Method Directly from Positive Blood Culture at a Pediatric Hospital. J. Clin. Microbiol. 2022, 60, e0012222. [Google Scholar] [CrossRef]
- Patel, Y.A.; Kirn, T.J.; Weinstein, M.P.; Uprety, P. Systematic Evaluation of the Accelerate Pheno System for Susceptibility Testing of Gram-Negative Bacteria Isolated from Blood Cultures. Microbiol. Spectr. 2021, 9, e0183621. [Google Scholar] [CrossRef]
- Patel, J.B.; Alby, K.; Humphries, R.; Weinstein, M.; Lutgring, J.D.; Naccache, S.N.; Simner, P.J. Updating breakpoints in the United States: A summary from the ASM Clinical Microbiology Open 2022. J. Clin. Microbiol. 2023, 61, e0115422. [Google Scholar] [CrossRef] [PubMed]
- Donner, L.M.; Campbell, W.S.; Lyden, E.; Van Schooneveld, T.C. Assessment of Rapid-Blood-Culture-Identification Result Interpretation and Antibiotic Prescribing Practices. J. Clin. Microbiol. 2017, 55, 1496–1507. [Google Scholar] [CrossRef] [PubMed]
- Gotham, D.; McKenna, L.; Deborggraeve, S.; Madoori, S.; Branigan, D. Public investments in the development of GeneXpert molecular diagnostic technology. PLoS ONE 2021, 16, e0256883. [Google Scholar] [CrossRef]
- Boutal, H.; Vogel, A.; Bernabeu, S.; Devilliers, K.; Creton, E.; Cotellon, G.; Plaisance, M.; Oueslati, S.; Dortet, L.; Jousset, A.; et al. A multiplex lateral flow immunoassay for the rapid identification of NDM-, KPC-, IMP- and VIM-type and OXA-48-like carbapenemase-producing Enterobacteriaceae. J. Antimicrob. Chemother. 2018, 73, 909–915. [Google Scholar] [CrossRef] [PubMed]
- Hopkins, K.L.; Meunier, D.; Naas, T.; Volland, H.; Woodford, N. Evaluation of the NG-Test CARBA 5 multiplex immunochromatographic assay for the detection of KPC, OXA-48-like, NDM, VIM and IMP carbapenemases. J. Antimicrob. Chemother. 2018, 73, 3523–3526. [Google Scholar] [CrossRef]
- Jenkins, S.; Ledeboer, N.A.; Westblade, L.F.; Burnham, C.A.; Faron, M.L.; Bergman, Y.; Yee, R.; Mesich, B.; Gerstbrein, D.; Wallace, M.A.; et al. Evaluation of NG-Test CARBA 5 for Rapid Phenotypic Detection and Differentiation of Five Common Carbapenemase Families: Results of a Multicenter Clinical Evaluation. J. Clin. Microbiol. 2020, 58. [Google Scholar] [CrossRef]
- Trienski, T.L.; Barrett, H.L.; Pasquale, T.R.; DiPersio, J.R.; File, T.M., Jr. Evaluation and use of a rapid Staphylococcus aureus assay by an antimicrobial stewardship program. Am. J. Health-Syst. Pharm. AJHP 2013, 70, 1908–1912. [Google Scholar] [CrossRef]
- Dupieux, C.; Trouillet-Assant, S.; Tasse, J.; Freydière, A.M.; Raulin, O.; Roure-Sobas, C.; Salord, H.; Tigaud, S.; Laurent, F. Evaluation of a commercial immunochromatographic assay for rapid routine identification of PBP2a-positive Staphylococcus aureus and coagulase-negative staphylococci. Diagn. Microbiol. Infect. Dis. 2016, 86, 262–264. [Google Scholar] [CrossRef]
- Munier, C.; Dupieux, C.; Kolenda, C.; Ranc, A.G.; Dauwalder, O.; Bes, M.; Vandenesch, F.; Tristan, A.; Laurent, F. Sensitivity of the PBP2a SA Culture Colony Test on shortly incubated subcultures of methicillin-resistant staphylococci from positive blood cultures. Diagn. Microbiol. Infect. Dis. 2023, 106, 115917. [Google Scholar] [CrossRef]
- van Belkum, A.; Burnham, C.D.; Rossen, J.W.A.; Mallard, F.; Rochas, O.; Dunne, W.M., Jr. Innovative and rapid antimicrobial susceptibility testing systems. Nat. Rev. Microbiol. 2020, 18, 299–311. [Google Scholar] [CrossRef]
- Choi, J.; Yoo, J.; Lee, M.; Kim, E.G.; Lee, J.S.; Lee, S.; Joo, S.; Song, S.H.; Kim, E.C.; Lee, J.C.; et al. A rapid antimicrobial susceptibility test based on single-cell morphological analysis. Sci. Transl. Med. 2014, 6, 267ra174. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Jing, W.; Iriya, R.; Yang, Y.; Syal, K.; Mo, M.; Grys, T.E.; Haydel, S.E.; Wang, S.; Tao, N. Phenotypic Antimicrobial Susceptibility Testing with Deep Learning Video Microscopy. Anal. Chem. 2018, 90, 6314–6322. [Google Scholar] [CrossRef] [PubMed]
- Frymier, P.D.; Ford, R.M.; Berg, H.C.; Cummings, P.T. Three-dimensional tracking of motile bacteria near a solid planar surface. Proc. Natl. Acad. Sci. USA 1995, 92, 6195–6199. [Google Scholar] [CrossRef] [PubMed]
| Test [Manufactuer] | Specimen | Functionality | Organisms | Technology | Run Time per Test | Performance | Regulatory Status | Comments |
|---|---|---|---|---|---|---|---|---|
| PhenoTest BC [Accelerate Diagnostics Inc., (Tucson, AZ, USA)] | Blood cultures | ID and AST | GP and GN | Morphokinetic cellular analysis and fluorescence in situ hybridization | Identification in 2 h, AST in 7 h |
| FDA, CE-IVD | |
| LifeScale [Affinity Biosensors (Santa, Barbara, CA, USA)] | Blood cultures | AST only | GN | Microfluidic sensor and resonant frequency to determine organism concentration and mass distribution (e.g., growth-independent) | 5 h |
| FDA, CE-IVD | |
| ASTar [Q-linea (Uppsala, Sweden)] | Blood cultures | AST only | GN | Time-lapse imaging of bacterial growth | 6 h |
| FDA, CE-IVD | |
| VITEK REVEAL [bioMerieux (Mountain view, CA, USA)] | Blood cultures | AST only | GN | Colorimetric sensors reacting to volatile organic compounds due to bacterial metabolism during growth | 5 h |
| FDA, CE-IVD | 1. Real-time monitoring of MICs |
| Selux Next-Generation Phenotyping (NGP) Test [SeluxDX (Boston, MA, USA)] | Blood cultures and bacterial colonies | AST only | GP (isolates) and GN (isolates and blood) | Fluorescent growth indictor using a viability and surface-binding assay | 6–7 h |
| FDA, CE-IVD | 1. Requires several instruments (e.g., Separator, Inoculator, and Analyzer) 2. Can be modified into high-throughput workflow in 384-well format 3. Multiple growth control wells |
| QuickMIC [Gradientech (Uppsala, Sweden)] | Blood cultures | AST only | GN | Microscopic analysis of a microfluidic device | 2–4 h |
| CE-IVD | 1. Determination of precise MIC values (not in doubling dilutions) |
| Alfred [Alifax (Padova, Italy)] | Blood cultures | AST only | GP and GN | Light scattering to detect bacterial growth | 4–7 h |
| CE-IVD | 1. MIC determination is not available |
| dRAST [QuantaMatrix (Seoul, South Korea)] | Blood cultures | AST only | GP and GN | Time-lapse microscopic imaging of bacterial cells | 4–7 h |
| CE-IVD | 1. Inclusion of ESBL detection |
| FASTinov [FASTinov (Porto, Portugal)] | Blood cultures | AST only | GP and GN | Flow cytometry using fluorescent dyes to reveal cell damage and metabolic changes (e.g., growth-independent) | 2 h |
| CE-IVD | 1. Requires flow cytometry instrumentation 2. Bacterial suspension for sample preparation can be repurposed for bacterial identification (off-label) 3. Inclusion of colistin testing 4. Inclusion of ESBL detection and AmpC plasmid screening |
| Test | Manufacturer | Technology | Run Time | Specimen Type | Organism | Resistance Markers | ||
|---|---|---|---|---|---|---|---|---|
| Methicillin | Vancomycin | Rifampin | ||||||
| Xpert MRSA/SA Blood Culture Assay | Cepheid (Sunnyvale, CA, USA) | PCR | 1 h | Blood cultures | Staphylococcus species/S. aureus | mecA, SCCmec/attB | n/a | n/a |
| mecA XpressFish | AdvanDx (Woburn, MA, USA) | Fluorescence in situ hybridization | 1 h | Blood cultures | Staphylococcus species/S. aureus | mecA | n/a | n/a |
| BD GeneOhM StaphSR Assay | Becton, Dickinson and Company (Sparks, MD, USA) | PCR | 2 h | Blood cultures | Staphylococcus species/S. aureus | SCCmec/orfX | n/a | n/a |
| Great Basin Staph ID/R Blood Culture Panel | Great Basin Scientific, Inc., (West Valley City, UT, USA) | PCR | 1.5 h | Blood cultures | Staphylococcus species/S. aureus | mecA | n/a | n/a |
| Xpert MRSA/SA Nasal Complete Assay | Cepheid (Sunnyvale, CA, USA) | PCR | 1 h | Nasal swabs (infection control) | Staphylococcus species/S. aureus | mecA, SCCmec/attB | n/a | n/a |
| LightCycler MRSA Advanced Test | Roche, Pleasanton, CA, USA | PCR | 1.5 h | Nasal swabs (infection control) | Staphylococcus species/S. aureus | SCCmec/orfX + MREJ | n/a | n/a |
| Xpert MRSA | Cepheid (Sunnyvale, CA, USA) | PCR | 1 h | Nasal swabs (infection control) | Staphylococcus species/S. aureus | mecA, SCCmec/attB | n/a | n/a |
| BD GeneOhm MRSA Assay, formerly IDI-MRSA | Becton, Dickinson and Company (Sparks, MD, USA) | PCR | 2 h | Nasal swabs (infection control) | Staphylococcus species/S. aureus | SCCmec/orfX | n/a | n/a |
| COBAS MRSA/SA Test | Roche (Pleasanton, CA, USA) | PCR | 2 h | Nasal swabs (infection control) | Staphylococcus species/S. aureus | SCCmec/orfX + MREJ | n/a | n/a |
| Xpert MRSA NxG | Cepheid (Sunnyvale, CA, USA) | PCR | 45 min | Nasal swabs (infection control) | Staphylococcus species/S. aureus | mecA/C, SCCmec/orfX | n/a | n/a |
| BD Max MRSA Assay | Becton, Dickinson and Company (Sparks, MD, USA) | PCR | <2 h | Nasal swabs (infection control) | Staphylococcus species/S. aureus | SCCmec/orfX | n/a | n/a |
| MRSA/SA ELITe MBG | EliTechGroup Epoech Biosciences (Paris, Ile-de-France, France) | PCR | 2.5 h | Nasal swabs (infection control) | Staphylococcus species/S. aureus | mecA | n/a | n/a |
| NucliSENS EasyQ MRSA Assay | bioMerieux (Marcy-l’Étoile, France) | PCR | 3 h | Nasal swabs (infection control) | Staphylococcus species/S. aureus | Sccmec junction and mecA gene | n/a | n/a |
| BD GeneOhm MRSA ACP Assay | Becton, Dickinson and Company (Sparks, MD, USA) | PCR | <2 h | Nasal swabs (infection control) | Staphylococcus species/S. aureus | SCCmec/orfX | n/a | n/a |
| BD GeneOhm Van R Assay | Becton, Dickinson and Company (Sparks, MD, USA) | PCR | <2 h | Peri-anal and rectal swabs (infection control) | Enterococcus species | n/a | vanA/B | n/a |
| IMDx Van R for Abbott m2000 | Intelligent Medical Devices, Inc., (Waltham, MA, USA) | PCR | 3–4 h | Peri-rectal and rectal swabs, stool (infection control) | Enterococcus species | n/a | vanA/B | n/a |
| Xpert vanA Assay | Cepheid (Sunnyvale, CA, USA) | PCR | 45 min | Rectal swabs (infection control) | Enterococcus species | n/a | vanA | n/a |
| Xpert MRSA/SA SSTI Assay | Cepheid (Sunnyvale, CA, USA) | PCR | 1 h | Skin and soft tissue swabs | Staphylococcus species/S. aureus | mecA, SCCmec/attB | n/a | n/a |
| Xpert MTB/RIF (Ultra) Assay | Cepheid (Sunnyvale, CA, USA) | PCR | 2 h | Sputum | Mycobacterium tuberculosis | n/a | n/a | rpoB |
| Acuitas AMR Gene Panel | OpGen, Inc. (Rockville, MD, USA) | PCR | 2.5 h | Bacterial colonies | Enterococcus faecalis | n/a | vanA | n/a |
| Test | Manufacturer | Technology | Run Time | Specimen Type | Organism | Resistance Markers | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ΒLac | ESBL | CARBA | AMIN | FLQ | COL | SUL | ||||||
| Revogene Carba C (formerly GenePOC Carba assay) | Meridian Bioscience (Cincinnati, OH, USA) | PCR | 70 mins | Bacterial colonies (infection control) | Enterobacterales, P. aeruginosa, A. baumannii | n/a | n/a | blaIMP, blaNDM, blaVIM, blaOXA-48-like | n/a | n/a | n/a | n/a |
| Acuitas AMR Gene Panel | OpGen, Inc. (Rockville, MD, USA) | PCR | 2.5 h | Bacterial colonies | Enterobacterales, P. aeruginosa | blaCMY, DHA | blaOXA1, blaOXA-9, blaCTX-M-1, blaCTX-M-2, blaCTX-M-9, blaTEM, blaSHV, blaPER, VEB | blaKPC, blaIMP, blaNDM, blaVIM, blaOXA-48 | aac, aad, ant, aph, armA, RMT | gyrA | mcr-1 | Sulf1, Sulf2, DFR |
| Xpert Carba-R (GNR) | Cepheid (Sunnyvale, CA, USA) | PCR | 50 mins | Peri-rectal and rectal swabs, bacterial colonies (infection control) | Enterobacterales, P. aeruginosa, A. baumannii | n/a | n/a | blaKPC, blaIMP, blaNDM, blaVIM, blaOXA-48 | n/a | n/a | n/a | n/a |
| Test | Manufacturer | Technology | Run Time | Specimen Type | Organism | Resistance Markers | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| ESBL | CARBA | COL | MET | VAN | ||||||
| ePlex Blood Culture Identification Gram Negative Panel | GenMark Diagnostics (Carlsbad, CA, USA) | PCR | 1.5 h | Blood cultures | GN | blaCTX-M | blaKPC, blaIMP, blaNDM, blaOXA-23 blaOXA-48, bla VIM | n/a | n/a | n/a |
| ePlex Blood Culture Identification Gram Positive Panel | GenMark Diagnostics (Carlsbad, CA, USA) | PCR | 1.5 h | Blood cultures | GP | n/a | n/a | n/a | mecA/C | vanA/B |
| BioFire FilmArray Blood Culture Identification Panel | bioMerieux (Marcy-l’Étoile, France) | PCR | 1 h | Blood cultures | GP, GN, and yeast | n/a | blaKPC | n/a | mecA/C | vanA/B |
| BioFire FilmArray Blood Culture Identification 2 | bioMerieux (Marcy-l’Étoile, France) | PCR | 1 h | Blood cultures | GP, GN and, yeast | blaCTX-M | blaKPC, blaIMP, blaNDM, blaVIM, bla OXA-48-like | mcr-1 | mecA/C, mec A/C + MREJ | vanA/B |
| Verigene Gram-Positive Nuclei Acid Test | DiaSorin (Saluggia, Italy) | Microarray | 2.5 h | Blood cultures | GP | n/a | n/a | n/a | mecA | vanA/B |
| Verigene Gram-Negative Nuclei Acid Test | DiaSorin (Saluggia, Italy) | Microarray | 2 h | Blood cultures | GN | blaCTX-M | blaKPC, blaIMP, blaNDM, blaVIM, blaOXA-48-like | n/a | n/a | n/a |
| BioFire FilmArray Pneumonia Panel | bioMerieux (Marcy-l’Étoile, France) | PCR | 1 h | Sputum, endo-tracheal aspirate, BAL | GP, GN, atypical bacteria, and viruses | blaCTX-M | blaKPC, blaIMP, blaNDM, blaVIM, blaOXA-48-like, | n/a | mecA/C, mec A/C + MREJ | n/a |
| Unyvero LRT BAL Application | OpGen, Inc. (Rockville, MD, USA) | PCR | 5 h | BAL | GP and GN | blaCTX-M, blaTEM, | blaKPC, blaNDM, blaVIM, bla OXA-23, blaOXA-24, blaOXA-48, blaOXA-58 | n/a | mecA | n/a |
| BioFire FilmArray Joint Infection Panel | bioMerieux (Marcy-l’Étoile, France) | PCR | 1 h | Synovial fluid | GP and GN | blaCTX-M | blaKPC, blaIMP, blaNDM, blaVIM, blaOXA-48-like | n/a | mecA/C, mec A/C + MREJ | vanA/B |
| Characteristic | Rapid Phenotypic Methods | Rapid Genotypic Methods |
|---|---|---|
| Principle | Evaluating growth, bacterial cellular and/or metabolic changes in the presence of antibiotics | Detecting the gene or mutation associated with antimicrobial resistance |
| Sample | Positive blood cultures, isolated bacterial colonies | For diagnostic purposes: Positive blood cultures, skin and soft tissue swabs, sputum, endotracheal aspirate, bronchoalveolar lavage, synovial fluid, isolated bacterial colonies For infection prevention/control and surveillance purposes: Nasal/peri-anal/rectal swabs, stool, isolated bacterial colonies |
| Identification of organism | Not available, need prior knowledge | Syndromic panels can provide both identification and AST |
| AST result | Antibiotic with interpretations (susceptible, intermediate, susceptible dose-dependent, resistant) | Genetic element ‘detected’ or ‘not detected’ |
| MIC | Yes | No |
| Determining mechanism of resistance | No | Yes |
| Turnaround time | 2–7 h | 45 min–5 h |
| Adaptability | Easier to implement for new antibiotics on market | Harder to implement until a resistant mechanism is known for the antibiotic |
| Performance evaluation |
|
|
| Criteria | Considerations |
|---|---|
| Test | Performance characteristics |
| Inclusion of antibiotics on hospital formulary | |
| Need for organism identification | |
| Availability of on-panel organisms | |
| Availability of on-panel antimicrobial genes | |
| Singleplex versus multiplex targets | |
| Number of ‘drug-bug’ combinations | |
| Flexibility to add more antibiotics or extend the concentrations of antibiotics tested | |
| Breakpoint interpretations used | |
| Automation versus manual hands-on time needed | |
| Laboratory requirements | Availability of supplies and consumables |
| Space requirements | |
| Scale of throughput | |
| Integration into laboratory workflow | |
| Cost and reimbursement | |
| Difficulty and expertise needed | |
| Maintenance needs | |
| Clinical impact | Turnaround time |
| Reporting structure changes | |
| Impact on patient outcomes | |
| Clinical education needed | |
| Integration with active antimicrobial stewardship |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hattab, S.; Ma, A.H.; Tariq, Z.; Vega Prado, I.; Drobish, I.; Lee, R.; Yee, R. Rapid Phenotypic and Genotypic Antimicrobial Susceptibility Testing Approaches for Use in the Clinical Laboratory. Antibiotics 2024, 13, 786. https://doi.org/10.3390/antibiotics13080786
Hattab S, Ma AH, Tariq Z, Vega Prado I, Drobish I, Lee R, Yee R. Rapid Phenotypic and Genotypic Antimicrobial Susceptibility Testing Approaches for Use in the Clinical Laboratory. Antibiotics. 2024; 13(8):786. https://doi.org/10.3390/antibiotics13080786
Chicago/Turabian StyleHattab, Siham, Adrienne H. Ma, Zoon Tariq, Ilianne Vega Prado, Ian Drobish, Rachel Lee, and Rebecca Yee. 2024. "Rapid Phenotypic and Genotypic Antimicrobial Susceptibility Testing Approaches for Use in the Clinical Laboratory" Antibiotics 13, no. 8: 786. https://doi.org/10.3390/antibiotics13080786
APA StyleHattab, S., Ma, A. H., Tariq, Z., Vega Prado, I., Drobish, I., Lee, R., & Yee, R. (2024). Rapid Phenotypic and Genotypic Antimicrobial Susceptibility Testing Approaches for Use in the Clinical Laboratory. Antibiotics, 13(8), 786. https://doi.org/10.3390/antibiotics13080786






