Identifying Cell-Penetrating Peptides for Effectively Delivering Antimicrobial Molecules into Streptococcus suis
Abstract
1. Introduction
2. Results
2.1. CPPs Penetration into S. suis
2.2. CPPs Uptake Efficiency Analysis by Flow Cytometry
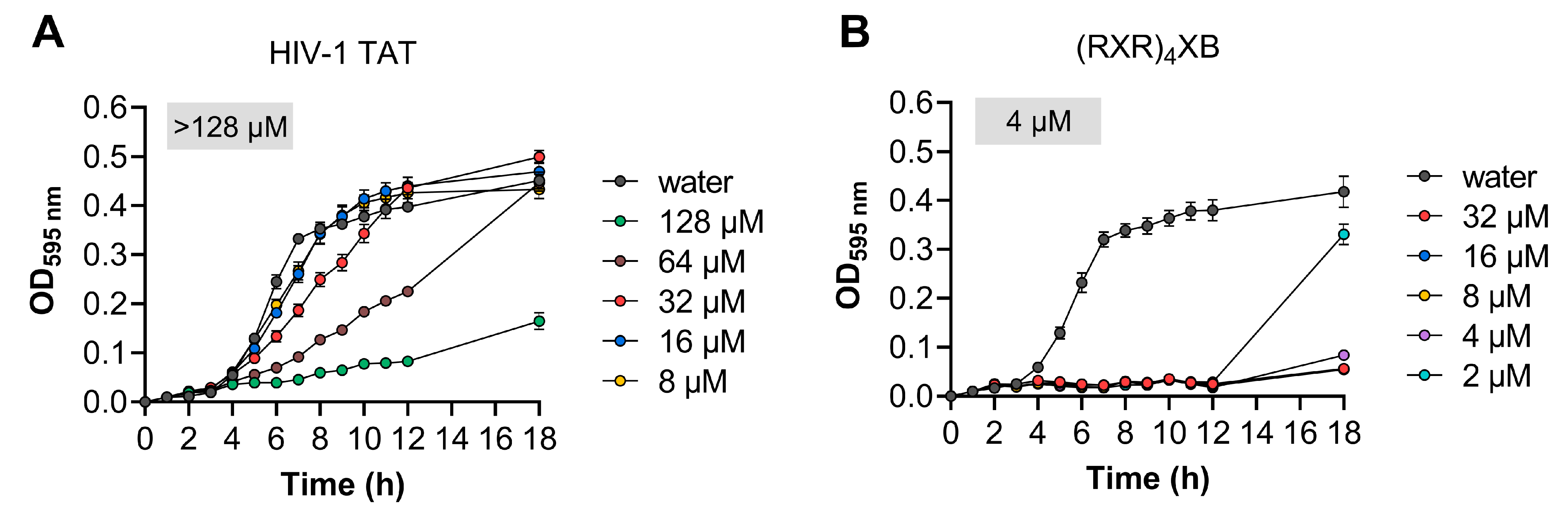
2.3. Toxicity Analysis of HIV-1 TAT and (RXR)4XB
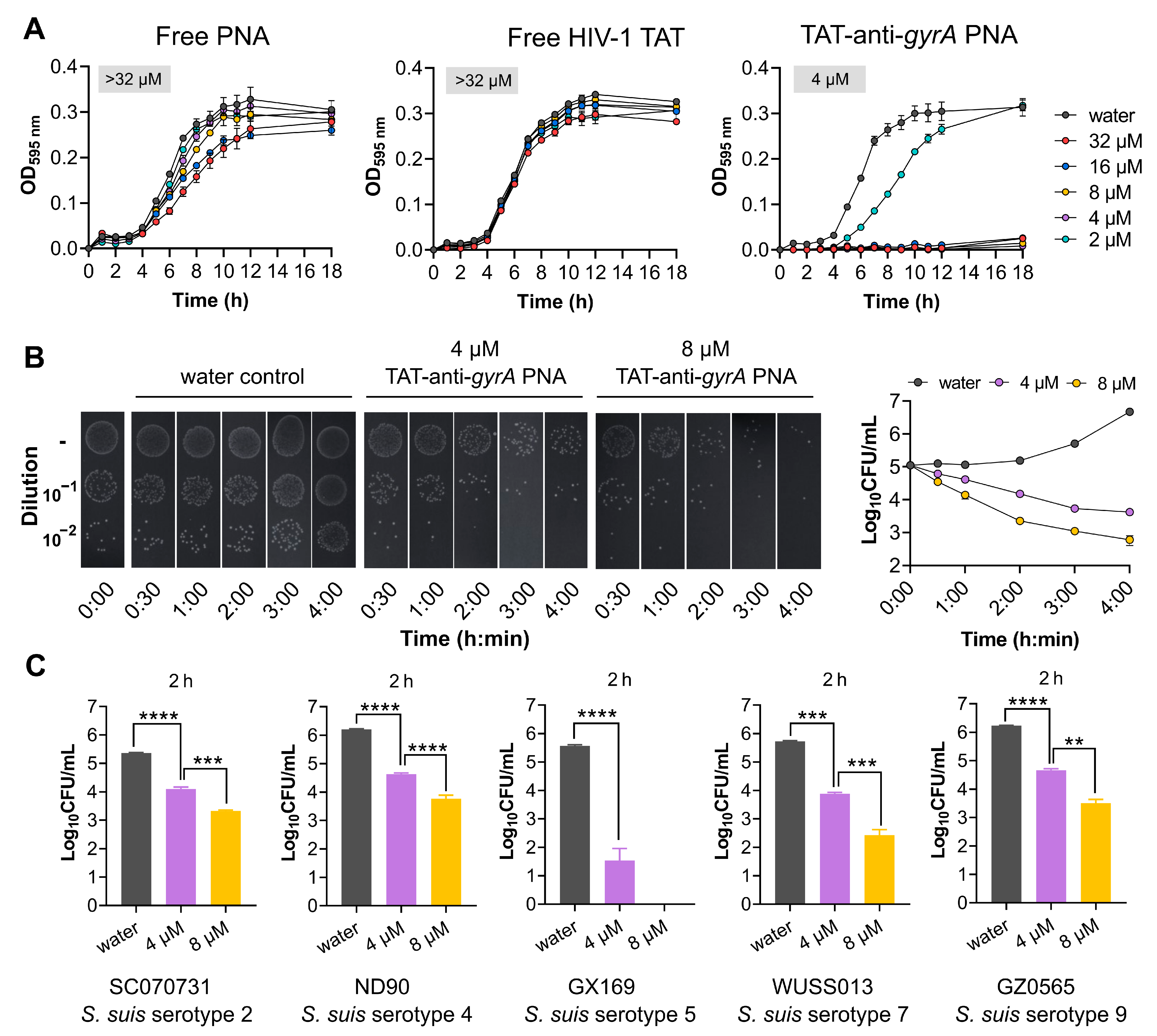
2.4. HIV-1 TAT-Coupled gyrA-Specific PNA Exhibits Bactericidal Activity
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains and Culture Conditions
4.2. CPPs Synthesis and Fluorescent Labeling
4.3. SR-SIM Analysis
4.4. Flow Cytometry Analysis
4.5. Synthesis of PNAs and CPP-PNA Conjugates
4.6. MIC Determination
4.7. Determination of Bactericidal Effects
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gori, A.; Lodigiani, G.; Colombarolli, S.G.; Bergamaschi, G.; Vitali, A. Cell Penetrating Peptides: Classification, Mechanisms, Methods of Study, and Applications. ChemMedChem 2023, 18, e202300236. [Google Scholar] [CrossRef] [PubMed]
- Zorko, M.; Langel, U. Cell-Penetrating Peptides. Methods Mol. Biol. 2022, 2383, 3–32. [Google Scholar] [CrossRef] [PubMed]
- Guidotti, G.; Brambilla, L.; Rossi, D. Cell-Penetrating Peptides: From Basic Research to Clinics. Trends Pharmacol. Sci. 2017, 38, 406–424. [Google Scholar] [CrossRef] [PubMed]
- Kotadiya, D.D.; Patel, P.; Patel, H.D. Cell-Penetrating Peptides: A Powerful Tool for Targeted Drug Delivery. Curr. Drug Deliv. 2024, 21, 368–388. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhang, Y.; Zhang, C.; Yu, H.; Ma, Y.; Li, Z.; Shi, N. Recent Advances of Cell-Penetrating Peptides and Their Application as Vectors for Delivery of Peptide and Protein-Based Cargo Molecules. Pharmaceutics 2023, 15, 2093. [Google Scholar] [CrossRef] [PubMed]
- Tietz, O.; Cortezon-Tamarit, F.; Chalk, R.; Able, S.; Vallis, K.A. Tricyclic cell-penetrating peptides for efficient delivery of functional antibodies into cancer cells. Nat. Chem. 2022, 14, 284–293. [Google Scholar] [CrossRef] [PubMed]
- Sauter, M.; Strieker, M.; Kleist, C.; Wischnjow, A.; Daniel, V.; Altmann, A.; Haberkorn, U.; Mier, W. Improving antibody-based therapies by chemical engineering of antibodies with multimeric cell-penetrating peptides for elevated intracellular delivery. J. Control. Release 2020, 322, 200–208. [Google Scholar] [CrossRef] [PubMed]
- Pifer, R.; Greenberg, D.E. Antisense antibacterial compounds. Transl. Res. 2020, 223, 89–106. [Google Scholar] [CrossRef]
- Barkowsky, G.; Abt, C.; Pohner, I.; Bieda, A.; Hammerschmidt, S.; Jacob, A.; Kreikemeyer, B.; Patenge, N. Antimicrobial Activity of Peptide-Coupled Antisense Peptide Nucleic Acids in Streptococcus pneumoniae. Microbiol. Spectr. 2022, 10, e00497-22. [Google Scholar] [CrossRef]
- Moustafa, D.A.; Wu, A.W.; Zamora, D.; Daly, S.M.; Sturge, C.R.; Pybus, C.; Geller, B.L.; Goldberg, J.B.; Greenberg, D.E. Peptide-Conjugated Phosphorodiamidate Morpholino Oligomers Retain Activity against Multidrug-Resistant Pseudomonas aeruginosa In Vitro and In Vivo. mBio 2021, 12, e02411-20. [Google Scholar] [CrossRef]
- Geller, B.L.; Marshall-Batty, K.; Schnell, F.J.; McKnight, M.M.; Iversen, P.L.; Greenberg, D.E. Gene-silencing antisense oligomers inhibit acinetobacter growth in vitro and in vivo. J. Infect. Dis. 2013, 208, 1553–1560. [Google Scholar] [CrossRef] [PubMed]
- Vogel, J. An RNA biology perspective on species-specific programmable RNA antibiotics. Mol. Microbiol. 2020, 113, 550–559. [Google Scholar] [CrossRef] [PubMed]
- Sully, E.K.; Geller, B.L. Antisense antimicrobial therapeutics. Curr. Opin. Microbiol. 2016, 33, 47–55. [Google Scholar] [CrossRef] [PubMed]
- El-Fateh, M.; Chatterjee, A.; Zhao, X. A systematic review of peptide nucleic acids (PNAs) with antibacterial activities: Efficacy, potential and challenges. Int. J. Antimicrob. Agents 2024, 63, 107083. [Google Scholar] [CrossRef] [PubMed]
- Eller, K.A.; Aunins, T.R.; Courtney, C.M.; Campos, J.K.; Otoupal, P.B.; Erickson, K.E.; Madinger, N.E.; Chatterjee, A. Facile accelerated specific therapeutic (FAST) platform develops antisense therapies to counter multidrug-resistant bacteria. Commun. Biol. 2021, 4, 331. [Google Scholar] [CrossRef]
- Good, L.; Nielsen, P.E. Antisense inhibition of gene expression in bacteria by PNA targeted to mRNA. Nat. Biotechnol. 1998, 16, 355–358. [Google Scholar] [CrossRef]
- Geller, B.L.; Li, L.; Martinez, F.; Sully, E.; Sturge, C.R.; Daly, S.M.; Pybus, C.; Greenberg, D.E. Morpholino oligomers tested in vitro, in biofilm and in vivo against multidrug-resistant Klebsiella pneumoniae. J. Antimicrob. Chemother. 2018, 73, 1611–1619. [Google Scholar] [CrossRef] [PubMed]
- Barkowsky, G.; Lemster, A.L.; Pappesch, R.; Jacob, A.; Kruger, S.; Schroder, A.; Kreikemeyer, B.; Patenge, N. Influence of Different Cell-Penetrating Peptides on the Antimicrobial Efficiency of PNAs in Streptococcus pyogenes. Mol. Ther. Nucleic Acids 2019, 18, 444–454. [Google Scholar] [CrossRef]
- Patenge, N.; Pappesch, R.; Krawack, F.; Walda, C.; Mraheil, M.A.; Jacob, A.; Hain, T.; Kreikemeyer, B. Inhibition of Growth and Gene Expression by PNA-peptide Conjugates in Streptococcus pyogenes. Mol. Ther. Nucleic Acids 2013, 2, e132. [Google Scholar] [CrossRef]
- Popella, L.; Jung, J.; Popova, K.; Ethurica-Mitic, S.; Barquist, L.; Vogel, J. Global RNA profiles show target selectivity and physiological effects of peptide-delivered antisense antibiotics. Nucleic Acids Res. 2021, 49, 4705–4724. [Google Scholar] [CrossRef]
- Tripathy, S.; Sahu, S.K. FtsZ inhibitors as a new genera of antibacterial agents. Bioorg. Chem. 2019, 91, 103169. [Google Scholar] [CrossRef]
- Abushahba, M.F.; Mohammad, H.; Thangamani, S.; Hussein, A.A.; Seleem, M.N. Impact of different cell penetrating peptides on the efficacy of antisense therapeutics for targeting intracellular pathogens. Sci. Rep. 2016, 6, 20832. [Google Scholar] [CrossRef]
- Popella, L.; Jung, J.; Do, P.T.; Hayward, R.J.; Barquist, L.; Vogel, J. Comprehensive analysis of PNA-based antisense antibiotics targeting various essential genes in uropathogenic Escherichia coli. Nucleic Acids Res. 2022, 50, 6435–6452. [Google Scholar] [CrossRef]
- Inoue, G.; Toyohara, D.; Mori, T.; Muraoka, T. Critical Side Chain Effects of Cell-Penetrating Peptides for Transporting Oligo Peptide Nucleic Acids in Bacteria. ACS Appl. Bio Mater. 2021, 4, 3462–3468. [Google Scholar] [CrossRef]
- Murray, G.G.R.; Hossain, A.; Miller, E.L.; Bruchmann, S.; Balmer, A.J.; Matuszewska, M.; Herbert, J.; Hadjirin, N.F.; Mugabi, R.; Li, G.; et al. The emergence and diversification of a zoonotic pathogen from within the microbiota of intensively farmed pigs. Proc. Natl. Acad. Sci. USA 2023, 120, e2307773120. [Google Scholar] [CrossRef] [PubMed]
- Goyette-Desjardins, G.; Auger, J.P.; Xu, J.; Segura, M.; Gottschalk, M. Streptococcus suis, an important pig pathogen and emerging zoonotic agent-an update on the worldwide distribution based on serotyping and sequence typing. Emerg. Microbes Infect. 2014, 3, e45. [Google Scholar] [CrossRef]
- Segura, M.; Aragon, V.; Brockmeier, S.L.; Gebhart, C.; Greeff, A.; Kerdsin, A.; O’Dea, M.A.; Okura, M.; Salery, M.; Schultsz, C.; et al. Update on Streptococcus suis Research and Prevention in the Era of Antimicrobial Restriction: 4th International Workshop on S. suis. Pathogens 2020, 9, 374. [Google Scholar] [CrossRef]
- Zhu, J.; Wang, J.; Kang, W.; Zhang, X.; Kerdsin, A.; Yao, H.; Zheng, H.; Wu, Z. Streptococcus suis serotype 4: A population with the potential pathogenicity in humans and pigs. Emerg. Microbes Infect. 2024, 13, 2352435. [Google Scholar] [CrossRef] [PubMed]
- Hatrongjit, R.; Fittipaldi, N.; Jenjaroenpun, P.; Wongsurawat, T.; Visetnan, S.; Zheng, H.; Gottschalk, M.; Kerdsin, A. Genomic comparison of two Streptococcus suis serotype 1 strains recovered from porcine and human disease cases. Sci. Rep. 2023, 13, 5380. [Google Scholar] [CrossRef]
- Liang, P.; Wang, M.; Gottschalk, M.; Vela, A.I.; Estrada, A.A.; Wang, J.; Du, P.; Luo, M.; Zheng, H.; Wu, Z. Genomic and pathogenic investigations of Streptococcus suis serotype 7 population derived from a human patient and pigs. Emerg. Microbes Infect. 2021, 10, 1960–1974. [Google Scholar] [CrossRef]
- Petrocchi-Rilo, M.; Martinez-Martinez, S.; Aguaron-Turrientes, A.; Roca-Martinez, E.; Garcia-Iglesias, M.J.; Perez-Fernandez, E.; Gonzalez-Fernandez, A.; Herencia-Lagunar, E.; Gutierrez-Martin, C.B. Anatomical Site, Typing, Virulence Gene Profiling, Antimicrobial Susceptibility and Resistance Genes of Streptococcus suis Isolates Recovered from Pigs in Spain. Antibiotics 2021, 10, 707. [Google Scholar] [CrossRef] [PubMed]
- Tan, M.F.; Tan, J.; Zeng, Y.B.; Li, H.Q.; Yang, Q.; Zhou, R. Antimicrobial resistance phenotypes and genotypes of Streptococcus suis isolated from clinically healthy pigs from 2017 to 2019 in Jiangxi Province, China. J. Appl. Microbiol. 2021, 130, 797–806. [Google Scholar] [CrossRef] [PubMed]
- Matajira, C.E.C.; Moreno, L.Z.; Poor, A.P.; Gomes, V.T.M.; Dalmutt, A.C.; Parra, B.M.; Oliveira, C.H.; Barbosa, M.R.F.; Sato, M.I.Z.; Calderaro, F.F.; et al. Streptococcus suis in Brazil: Genotypic, Virulence, and Resistance Profiling of Strains Isolated from Pigs between 2001 and 2016. Pathogens 2019, 9, 31. [Google Scholar] [CrossRef] [PubMed]
- Yongkiettrakul, S.; Maneerat, K.; Arechanajan, B.; Malila, Y.; Srimanote, P.; Gottschalk, M.; Visessanguan, W. Antimicrobial susceptibility of Streptococcus suis isolated from diseased pigs, asymptomatic pigs, and human patients in Thailand. BMC Vet. Res. 2019, 15, 5. [Google Scholar] [CrossRef] [PubMed]
- Vaara, M.; Porro, M. Group of peptides that act synergistically with hydrophobic antibiotics against gram-negative enteric bacteria. Antimicrob. Agents Chemother. 1996, 40, 1801–1805. [Google Scholar] [CrossRef] [PubMed]
- Vives, E.; Brodin, P.; Lebleu, B. A truncated HIV-1 Tat protein basic domain rapidly translocates through the plasma membrane and accumulates in the cell nucleus. J. Biol. Chem. 1997, 272, 16010–16017. [Google Scholar] [CrossRef] [PubMed]
- Abes, R.; Moulton, H.M.; Clair, P.; Yang, S.T.; Abes, S.; Melikov, K.; Prevot, P.; Youngblood, D.S.; Iversen, P.L.; Chernomordik, L.V.; et al. Delivery of steric block morpholino oligomers by (R-X-R)4 peptides: Structure-activity studies. Nucleic Acids Res. 2008, 36, 6343–6354. [Google Scholar] [CrossRef] [PubMed]
- MacNair, C.R.; Rutherford, S.T.; Tan, M.W. Alternative therapeutic strategies to treat antibiotic-resistant pathogens. Nat. Rev. Microbiol. 2024, 22, 262–275. [Google Scholar] [CrossRef]
- Demidov, V.V.; Potaman, V.N.; Frank-Kamenetskii, M.D.; Egholm, M.; Buchard, O.; Sonnichsen, S.H.; Nielsen, P.E. Stability of peptide nucleic acids in human serum and cellular extracts. Biochem. Pharmacol. 1994, 48, 1310–1313. [Google Scholar] [CrossRef]
- Good, L.; Awasthi, S.K.; Dryselius, R.; Larsson, O.; Nielsen, P.E. Bactericidal antisense effects of peptide-PNA conjugates. Nat. Biotechnol. 2001, 19, 360–364. [Google Scholar] [CrossRef]
- Nekhotiaeva, N.; Awasthi, S.K.; Nielsen, P.E.; Good, L. Inhibition of Staphylococcus aureus gene expression and growth using antisense peptide nucleic acids. Mol. Ther. 2004, 10, 652–659. [Google Scholar] [CrossRef] [PubMed]
- Tavakoli, M.; Hashemi, A.; Vaezjalali, M.; Mohammadzadeh, M.; Goudarzi, H. Inhibition of growth and gene expression in Staphylococcus aureus by anti-gyrA peptide nucleic acid. Future Microbiol. 2019, 14, 1123–1132. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; He, Y.; Xia, Y.; Wang, L.; Liang, S. Inhibition of gene expression and growth of multidrug-resistant Acinetobacter baumannii by antisense peptide nucleic acids. Mol. Biol. Rep. 2014, 41, 7535–7541. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, D.J.; Kim, D.T.; Steinman, L.; Fathman, C.G.; Rothbard, J.B. Polyarginine enters cells more efficiently than other polycationic homopolymers. J. Pept. Res. 2000, 56, 318–325. [Google Scholar] [CrossRef] [PubMed]
- Howard, J.J.; Sturge, C.R.; Moustafa, D.A.; Daly, S.M.; Marshall-Batty, K.R.; Felder, C.F.; Zamora, D.; Yabe-Gill, M.; Labandeira-Rey, M.; Bailey, S.M.; et al. Inhibition of Pseudomonas aeruginosa by Peptide-Conjugated Phosphorodiamidate Morpholino Oligomers. Antimicrob. Agents Chemother. 2017, 61, e01938-16. [Google Scholar] [CrossRef] [PubMed]
- Daly, S.M.; Sturge, C.R.; Felder-Scott, C.F.; Geller, B.L.; Greenberg, D.E. MCR-1 Inhibition with Peptide-Conjugated Phosphorodiamidate Morpholino Oligomers Restores Sensitivity to Polymyxin in Escherichia coli. mBio 2017, 8, e01315-17. [Google Scholar] [CrossRef]
- Aunins, T.R.; Erickson, K.E.; Chatterjee, A. Transcriptome-based design of antisense inhibitors potentiates carbapenem efficacy in CRE Escherichia coli. Proc. Natl. Acad. Sci. USA 2020, 117, 30699–30709. [Google Scholar] [CrossRef]
- Oh, E.; Zhang, Q.; Jeon, B. Target optimization for peptide nucleic acid (PNA)-mediated antisense inhibition of the CmeABC multidrug efflux pump in Campylobacter jejuni. J. Antimicrob. Chemother. 2014, 69, 375–380. [Google Scholar] [CrossRef] [PubMed]
- Goh, S.; Loeffler, A.; Lloyd, D.H.; Nair, S.P.; Good, L. Oxacillin sensitization of methicillin-resistant Staphylococcus aureus and methicillin-resistant Staphylococcus pseudintermedius by antisense peptide nucleic acids in vitro. BMC Microbiol. 2015, 15, 262. [Google Scholar] [CrossRef]
- Fleitas Martinez, O.; Cardoso, M.H.; Ribeiro, S.M.; Franco, O.L. Recent Advances in Anti-virulence Therapeutic Strategies with a Focus on Dismantling Bacterial Membrane Microdomains, Toxin Neutralization, Quorum-Sensing Interference and Biofilm Inhibition. Front. Cell Infect. Microbiol. 2019, 9, 74. [Google Scholar] [CrossRef]
- Rasko, D.A.; Sperandio, V. Anti-virulence strategies to combat bacteria-mediated disease. Nat. Rev. Drug Discov. 2010, 9, 117–128. [Google Scholar] [CrossRef] [PubMed]
- Muhlen, S.; Dersch, P. Anti-virulence Strategies to Target Bacterial Infections. Curr. Top. Microbiol. Immunol. 2016, 398, 147–183. [Google Scholar] [CrossRef]
- Daly, S.M.; Elmore, B.O.; Kavanaugh, J.S.; Triplett, K.D.; Figueroa, M.; Raja, H.A.; El-Elimat, T.; Crosby, H.A.; Femling, J.K.; Cech, N.B.; et al. omega-Hydroxyemodin limits Staphylococcus aureus quorum sensing-mediated pathogenesis and inflammation. Antimicrob. Agents Chemother. 2015, 59, 2223–2235. [Google Scholar] [CrossRef] [PubMed]
- Sully, E.K.; Malachowa, N.; Elmore, B.O.; Alexander, S.M.; Femling, J.K.; Gray, B.M.; DeLeo, F.R.; Otto, M.; Cheung, A.L.; Edwards, B.S.; et al. Selective chemical inhibition of agr quorum sensing in Staphylococcus aureus promotes host defense with minimal impact on resistance. PLoS Pathog. 2014, 10, e1004174. [Google Scholar] [CrossRef] [PubMed]
- Islam, M.M.; Odahara, M.; Yoshizumi, T.; Oikawa, K.; Kimura, M.; Su’etsugu, M.; Numata, K. Cell-Penetrating Peptide-Mediated Transformation of Large Plasmid DNA into Escherichia coli. ACS Synth. Biol. 2019, 8, 1215–1218. [Google Scholar] [CrossRef] [PubMed]
- Numata, K.; Kaplan, D.L. Silk-based gene carriers with cell membrane destabilizing peptides. Biomacromolecules 2010, 11, 3189–3195. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Dong, W.; Ma, J.; Zhang, Y.; Pan, Z.; Yao, H. Utilization of the ComRS system for the rapid markerless deletion of chromosomal genes in Streptococcus suis. Future Microbiol. 2019, 14, 207–222. [Google Scholar] [CrossRef] [PubMed]
- Zaccaria, E.; van Baarlen, P.; de Greeff, A.; Morrison, D.A.; Smith, H.; Wells, J.M. Control of competence for DNA transformation in streptococcus suis by genetically transferable pherotypes. PLoS ONE 2014, 9, e99394. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Wang, W.; Tang, M.; Shao, J.; Dai, C.; Zhang, W.; Fan, H.; Yao, H.; Zong, J.; Chen, D.; et al. Comparative genomic analysis shows that Streptococcus suis meningitis isolate SC070731 contains a unique 105K genomic island. Gene 2014, 535, 156–164. [Google Scholar] [CrossRef]
- Wu, Z.; Zhang, W.; Lu, C. Comparative proteome analysis of secreted proteins of Streptococcus suis serotype 9 isolates from diseased and healthy pigs. Microb. Pathog. 2008, 45, 159–166. [Google Scholar] [CrossRef]
- Aldred, K.J.; Kerns, R.J.; Osheroff, N. Mechanism of quinolone action and resistance. Biochemistry 2014, 53, 1565–1574. [Google Scholar] [CrossRef] [PubMed]
- Goltermann, L.; Nielsen, P.E. PNA Antisense Targeting in Bacteria: Determination of Antibacterial Activity (MIC) of PNA-Peptide Conjugates. Methods Mol. Biol. 2020, 2105, 231–239. [Google Scholar] [CrossRef] [PubMed]
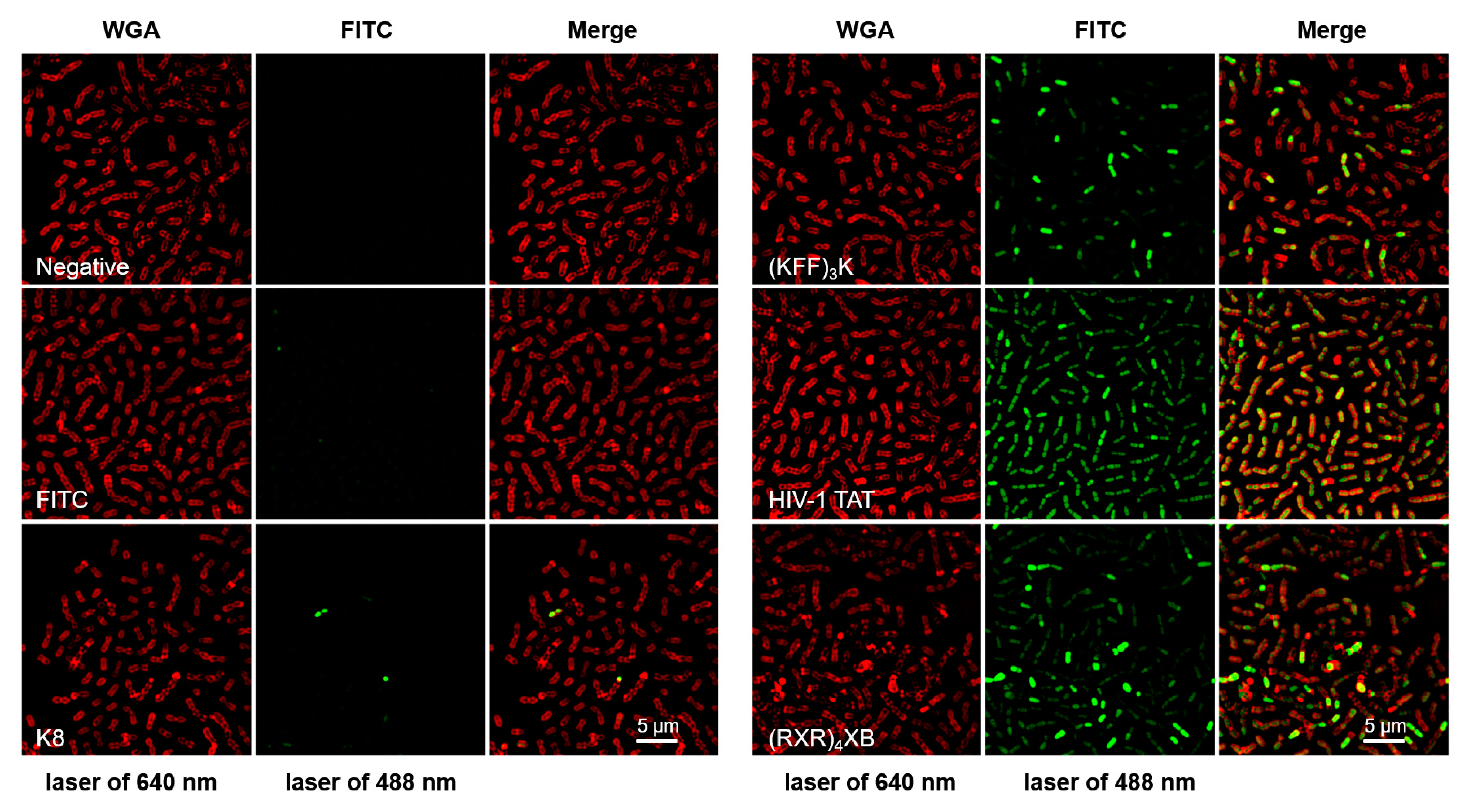
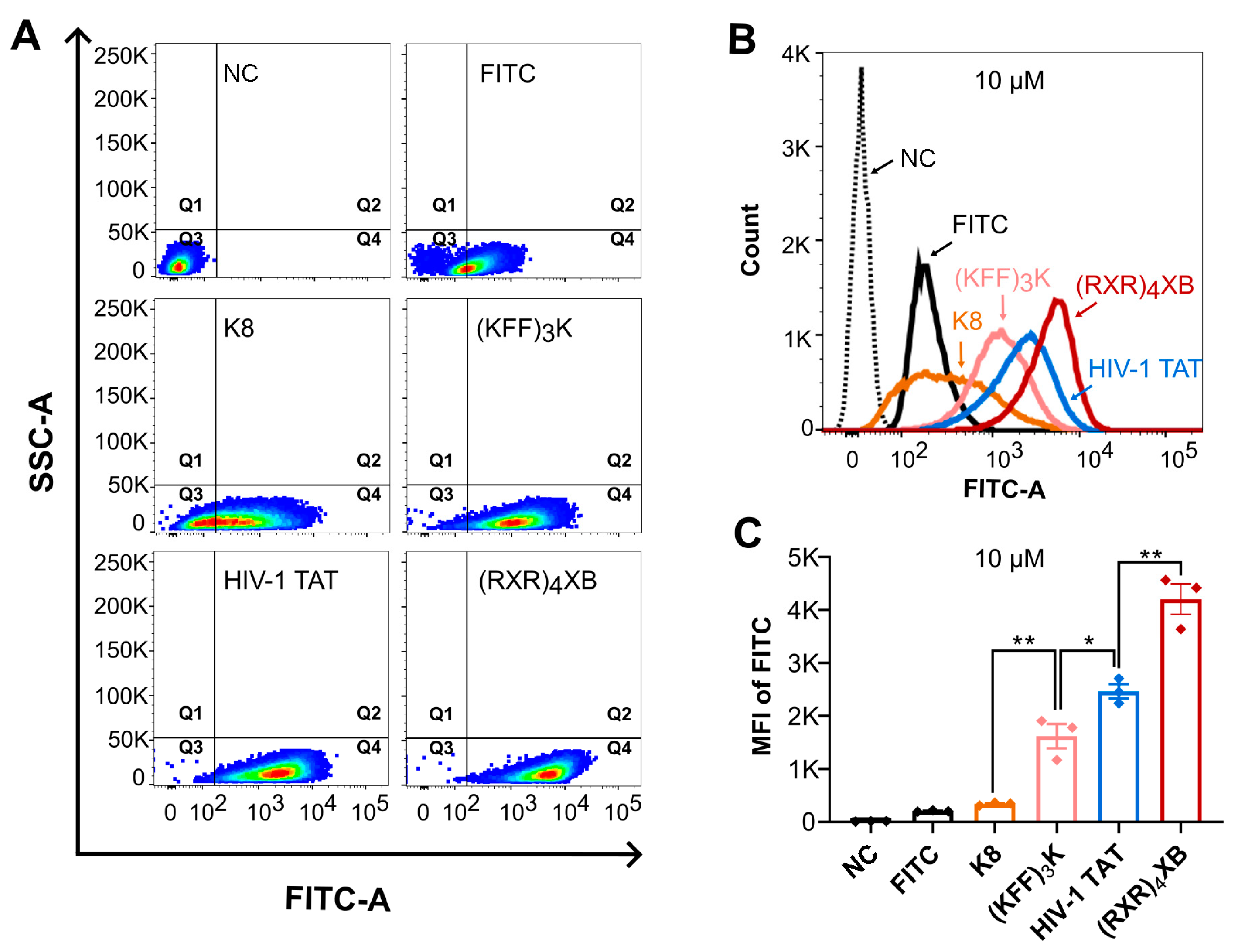

| Name | CPP Sequence | PNA Sequence | MIC (µM) | Reference |
|---|---|---|---|---|
| K8 | KKKKKKKK-NH2 | - | - | [18] |
| (KFF)3K | KFFKFFKFFK-NH2 | - | - | [35] |
| HIV-1 TAT | GRKKRRQRRRYK-NH2 | - | >128 | [36] |
| (RXR)4XB | RXRRXRRXRRXRXB-NH2 | - | 4 | [37] |
| Free PNA | - | ttgcattatatg | >32 | |
| TAT-anti-gyrA PNA | GRKKRRQRRRYK | ttgcattatatg | 4 |
| Treatment | 8 µM | 16 µM | 32 µM | 64 µM | ||||
|---|---|---|---|---|---|---|---|---|
| (Lg CFU) ± SD | Lg CFU Reduction | (Lg CFU) ± SD | Lg CFU Reduction | (Lg CFU) ± SD | Lg CFU Reduction | (Lg CFU) ± SD | Lg CFU Reduction | |
| HIV-1 TAT | 6.37 ± 0.06 | −0.09 ns | 6.32 ± 0.09 | −0.04 ns | 6.05 ± 0.27 | 0.23 ns | 6.00 ± 0.25 | 0.28 ns |
| Water control | 6.28 ± 0.08 | 0 | ||||||
| (RXR)4XB | 5.01 ± 0.14 | 1.34 * | 4.86 ± 0.02 | 1.49 * | 4.72 ± 0.03 | 1.63 * | 4.58 ± 0.01 | 1.77 * |
| Water control | 6.35 ± 0.13 | 0 | ||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, J.; Liang, Z.; Yao, H.; Wu, Z. Identifying Cell-Penetrating Peptides for Effectively Delivering Antimicrobial Molecules into Streptococcus suis. Antibiotics 2024, 13, 725. https://doi.org/10.3390/antibiotics13080725
Zhu J, Liang Z, Yao H, Wu Z. Identifying Cell-Penetrating Peptides for Effectively Delivering Antimicrobial Molecules into Streptococcus suis. Antibiotics. 2024; 13(8):725. https://doi.org/10.3390/antibiotics13080725
Chicago/Turabian StyleZhu, Jinlu, Zijing Liang, Huochun Yao, and Zongfu Wu. 2024. "Identifying Cell-Penetrating Peptides for Effectively Delivering Antimicrobial Molecules into Streptococcus suis" Antibiotics 13, no. 8: 725. https://doi.org/10.3390/antibiotics13080725
APA StyleZhu, J., Liang, Z., Yao, H., & Wu, Z. (2024). Identifying Cell-Penetrating Peptides for Effectively Delivering Antimicrobial Molecules into Streptococcus suis. Antibiotics, 13(8), 725. https://doi.org/10.3390/antibiotics13080725







