New Insights into Pseudomonas spp.-Produced Antibiotics: Genetic Regulation of Biosynthesis and Implementation in Biotechnology
Abstract
1. Introduction
2. Mupirocin
2.1. Mechanism of Mupirocin Biosynthesis
- The mupQ, mupS, mupT, and mupW genes were essential for the production of mupirocin, whereas mupO, mupU, mupV, and macpE were essential for the production of PA-A but not PA-B [45]. In this work, it was assumed for the first time that PA-B is a precursor of PA-A.
- PA-C, previously assumed to be a precursor of PA-A, was formed by a minor parallel pathway [41]. Attempting to disable this pathway at the initial stage (ΔmupW) resulted in the loss of the ability to synthesize the major product (PA-A). Moreover, all the mutagenesis operations of the mupirocin cluster elements resulted in the loss of the ability to synthesize PA-A, but ΔmupC and ΔmupF retained PA-B production.
2.2. Regulation of Mupirocin Production
2.3. Mupirocin in Biotechnology
3. Gluconic Acid
3.1. Gluconic Acid Biosynthesis Pathway
3.2. Regulation of Gluconic Acid Production
3.3. Gluconic Acid in Biotechnology
4. PRN
4.1. Mechanism of PRN Biosynthesis
4.2. Regulation of PRN Production
4.3. Modifications of PRN Producers
4.4. PRN in Biotechnology
5. DAPG
5.1. Mechanism of DAPG Biosynthesis
5.2. Regulation of DAPG Biosynthesis
5.3. Producer Modifications
5.4. DAPG in Biotechnology
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Newman, D.J.; Cragg, G.M. Natural Products as Sources of New Drugs over the Last 25 Years. J. Nat. Prod. 2007, 70, 461–477. [Google Scholar] [CrossRef] [PubMed]
- Saadoun, I.; Gharaibeh, R. The Streptomyces flora of Badia region of Jordan and its potential as a source of antibiotics active against antibiotic-resistant bacteria. J. Arid Environ. 2003, 53, 365–371. [Google Scholar] [CrossRef]
- Zetola, N.; Francis, J.S.; Nuermberger, E.L.; Bishai, W.R. Community-acquired meticillin-resistant Staphylococcus aureus: An emerging threat. Lancet Infect. Dis. 2005, 5, 275–286. [Google Scholar] [CrossRef] [PubMed]
- Haas, D.; Keel, C. Regulation of antibiotic production in root-colonizing Pseudomonas spp. and relevance for biological control of plant disease. Annu. Rev. Phytopathol. 2003, 41, 117–153. [Google Scholar] [CrossRef]
- Del Rio, L.A.; Olivares, J.; Blesa, M.C.; Mayor, F. Antibiotics from Pseudomonas reptilivora I. Taxonomic Classification and Optimal Conditions of Fermentation for Antibiotic Production. Antimicrob. Agents Chemother. 1972, 2, 186–188. [Google Scholar] [CrossRef] [PubMed]
- Castro Tapia, M.P.; Madariaga Burrows, R.P.; Ruiz Sepúlveda, B.; Vargas Concha, M.; Vera Palma, C.; Moya-Elizondo, E.A. Antagonistic Activity of Chilean Strains of Pseudomonas protegens Against Fungi Causing Crown and Root Rot of Wheat (Triticum aestivum L.). Front. Plant Sci. 2020, 11, 951. [Google Scholar] [CrossRef] [PubMed]
- Furuya, N.; Yamasaki, S.; Nishioka, M.; Shiraishi, I.; Iiyama, K.; Matsuyama, N. Antimicrobial Activities of Pseudomonads against Plant Pathogenic Organisms and Efficacy of Pseudomonas aeruginosa ATCC7700 against Bacterial Wilt of Tomato. Jpn. J. Phytopathol. 1997, 63, 417–424. [Google Scholar] [CrossRef]
- Das, K.; Abrol, S.; Verma, R.; Annapragada, H.; Katiyar, N.; Senthilkumar, M. Pseudomonas. In Beneficial Microbes in Agro-Ecology; Elsevier: Amsterdam, The Netherlands, 2020; pp. 133–148. [Google Scholar] [CrossRef]
- Arrebola, E.; Tienda, S.; Vida, C.; de Vicente, A.; Cazorla, F.M. Fitness Features Involved in the Biocontrol Interaction of Pseudomonas chlororaphis with Host Plants: The Case Study of PcPCL1606. Front. Microbiol. 2019, 10, 719. [Google Scholar] [CrossRef]
- O’Callaghan, M. Microbial inoculation of seed for improved crop performance: Issues and opportunities. Appl. Microbiol. Biotechnol. 2016, 100, 5729–5746. [Google Scholar] [CrossRef]
- Stockwell, V.O.; Johnson, K.B.; Sugar, D.; Loper, J.E. Control of Fire Blight by Pseudomonas fluorescens A506 and Pantoea vagans C9-1 Applied as Single Strains and Mixed Inocula. Phytopathology® 2010, 100, 1330–1339. [Google Scholar] [CrossRef]
- Stockwell, V.O.; Stack, J.P. Using Pseudomonas spp. for Integrated Biological Control. Phytopathology® 2007, 97, 244–249. [Google Scholar] [CrossRef] [PubMed]
- Sahayaraj, K. (Ed.) Basic and Applied Aspects of Biopesticides; Springer: New Delhi, India, 2014. [Google Scholar] [CrossRef]
- Kumar, A. (Ed.) Microbial Biocontrol: Sustainable Agriculture and Phytopathogen Management; Springer International Publishing: Berlin/Heidelberg, Germany, 2022. [Google Scholar] [CrossRef]
- Anderson, J.A.; Staley, J.; Challender, M.; Heuton, J. Safety of Pseudomonas chlororaphis as a gene source for genetically modified crops. Transgenic Res. 2018, 27, 103–113. [Google Scholar] [CrossRef] [PubMed]
- Osdaghi, E.; Martins, S.J.; Ramos-Sepulveda, L.; Vieira, F.R.; Pecchia, J.A.; Beyer, D.M.; Bell, T.H.; Yang, Y.; Hockett, K.L.; Bull, C.T. 100 Years Since Tolaas: Bacterial Blotch of Mushrooms in the 21st Century. Plant Dis. 2019, 103, 2714–2732. [Google Scholar] [CrossRef]
- Sajben, E.; Manczinger, L.; Nagy, A.; Kredics, L.; Vágvölgyi, C. Characterization of pseudomonads isolated from decaying sporocarps of oyster mushroom. Microbiol. Res. 2011, 166, 255–267. [Google Scholar] [CrossRef] [PubMed]
- Manczinger, L.; Vágvölgyi, C.; Sajben, E.; Nagy, A.; Szöke-Kis, Z.; Nagy, A.; Turóczi, G.; Kovács, A. Wirkstoffe Gegen Pseudomona-Spezies als Verursacher von Fäulnisbefall bei der Pilzproduktion, Ihre Verwendung und Diese Enthaltende Zusammensetzungen. European Patent No. EP2,753,182B1, 30 August 2012. [Google Scholar]
- Fermor, T.R.; Henry, M.B.; Fenlon, J.S.; Glenister, M.J.; Lincoln, S.P.; Lynch, J.M. Development and application of a biocontrol system for bacterial blotch of the cultivated mushroom. Crop Prot. 1991, 10, 271–278. [Google Scholar] [CrossRef]
- Singh, M.; Srivastava, M.; Kumar, A.; Singh, A.K.; Pandey, K.D. Endophytic Bacteria in Plant Disease Management. In Microbial Endophytes; Elsevier: Amsterdam, The Netherlands, 2020; pp. 61–89. [Google Scholar] [CrossRef]
- Lindberg, G.D. Antibiotic Produced from the Microorganism (Pseudomonas lindbergii), Its Preparation and Method of Use. U.S. Patent No. US4,062,943, 1 February 1977. [Google Scholar]
- Farmer, S. Topical Compositions Containing Extracellular Products of Pseudomonas lindbergii and Emu oil. U.S. Patent No. US 6,645,506 B1, 26 August 1999. [Google Scholar]
- Farmer, S. Verbesserte Topische Zusammensetzungen mit Extrazellulären Produkten von Probiotischen Bakterien und Verwendungen Davon. European Patent No. EP 1 212 069 B1, 25 August 2000. [Google Scholar]
- Che, Y.; Qi, X.; Qu, W.; Shi, B.; Lin, Q.; Yao, H.; Zhang, Y.; Wei, T. Synthetic strategies of phenazine derivatives: A review. J. Heterocycl. Chem. 2022, 59, 969–996. [Google Scholar] [CrossRef]
- Yan, J.; Liu, W.; Cai, J.; Wang, Y.; Li, D.; Hua, H.; Cao, H. Advances in Phenazines over the Past Decade: Review of Their Pharmacological Activities, Mechanisms of Action, Biosynthetic Pathways and Synthetic Strategies. Mar. Drugs 2021, 19, 610. [Google Scholar] [CrossRef] [PubMed]
- Kaur, R.; Macleod, J.; Foley, W.; Nayudu, M. Gluconic acid: An antifungal agent produced by Pseudomonas species in biological control of take-all. Phytochemistry 2006, 67, 595–604. [Google Scholar] [CrossRef] [PubMed]
- Carruthers, F.L.; Shum-Thomas, T.; Conner, A.J.; Mahanty, H.K. The significance of antibiotic production by Pseudomonas aureofaciens PA 147-2 for biological control of Phytophthora megasperma root rot of asparagus. Plant Soil 1995, 170, 339–344. [Google Scholar] [CrossRef]
- Matthijs, S.; Vander Wauven, C.; Cornu, B.; Ye, L.; Cornelis, P.; Thomas, C.M.; Ongena, M. Antimicrobial properties of Pseudomonas strains producing the antibiotic mupirocin. Res. Microbiol. 2014, 165, 695–704. [Google Scholar] [CrossRef]
- Zhang, Y.; Tu, Q.; Yu, F.; Jing, X.; Bian, X.; Chen, H. Method for Screening Pseudomonas Protegens Mutant Strain, and Application Thereof in Biological Control. International Patent No. WO2018192507A1, 18 April 2018. [Google Scholar]
- Khoshnood, S.; Heidary, M.; Asadi, A.; Soleimani, S.; Motahar, M.; Savari, M.; Saki, M.; Abdi, M. A review on mechanism of action, resistance, synergism, and clinical implications of mupirocin against Staphylococcus aureus. Biomed. Pharmacother. 2019, 109, 1809–1818. [Google Scholar] [CrossRef] [PubMed]
- Troeman, D.P.R.; Van Hout, D.; Kluytmans, J.A.J.W. Antimicrobial approaches in the prevention of Staphylococcus aureus infections: A review. J. Antimicrob. Chemother. 2019, 74, 281–294. [Google Scholar] [CrossRef] [PubMed]
- Fuller, A.T.; Mellows, G.; Woolford, M.; Banks, G.T.; Barrow, K.D.; chain, e.b. Pseudomonic Acid: An Antibiotic produced by Pseudomonas fluorescens. Nature 1971, 234, 416–417. [Google Scholar] [CrossRef] [PubMed]
- Barrow, K.D.; Mellows, G. Antibiotics. U.S. Patent No. US3,977,943, 7 July 1975. [Google Scholar]
- Beecham Group PLC. The Antibiotic Pseudomonic Acid. United Kingdom Patent No. UK1,395,907, 9 June 1972.
- Class, Y.J.; DeShong, P. The Pseudomonic Acids. Chem. Rev. 1995, 95, 1843–1857. [Google Scholar] [CrossRef]
- Szell, V.; Lang, I.; Barta, I.; Tedges, A.; Albrecht, K.; Mozes, J.; Suto, N.; Szabo, I.; Petroczki, M.; Erdei, J.; et al. Process for the Preparation of Pseudomonic Acid A Antibiotic by Microbiological Method. U.S. Patent No. US20030100083A1, 12 November 2002. [Google Scholar]
- O’Donnell, J.A.; Gelone, S.P.; Safdar, A. Topical Antibacterials. In Mandell, Douglas, and Bennett’s Principles and Practice of Infectious Diseases; Elsevier: Amsterdam, The Netherlands, 2015; pp. 452–462.e2. [Google Scholar] [CrossRef]
- Hothersall, J.; Murphy, A.C.; Iqbal, Z.; Campbell, G.; Stephens, E.R.; Wu, J.; Cooper, H.; Atkinson, S.; Williams, P.; Crosby, J.; et al. Manipulation of quorum sensing regulation in Pseudomonas fluorescens NCIMB 10586 to increase mupirocin production. Appl. Microbiol. Biotechnol. 2011, 90, 1017–1026. [Google Scholar] [CrossRef] [PubMed]
- Whatling, C.A.; Hodgson, J.E.; Burnham, M.K.R.; Clarke, N.J.; Franklin, F.C.H.; Thomas, C.M. Identification of a 60 kb region of the chromosome of Pseudomonas fluorescens NCIB 10586 required for the biosynthesis of pseudomonic acid (mupirocin). Microbiology 1995, 141, 973–982. [Google Scholar] [CrossRef][Green Version]
- Tucaliuc, A.; Blaga, A.C.; Galaction, A.I.; Cascaval, D. Mupirocin: Applications and production. Biotechnol. Lett. 2019, 41, 495–502. [Google Scholar] [CrossRef] [PubMed]
- Gao, S.-S.; Hothersall, J.; Wu, J.; Murphy, A.C.; Song, Z.; Stephens, E.R.; Thomas, C.M.; Crump, M.P.; Cox, R.J.; Simpson, T.J.; et al. Biosynthesis of Mupirocin by Pseudomonas fluorescens NCIMB 10586 Involves Parallel Pathways. J. Am. Chem. Soc. 2014, 136, 5501–5507. [Google Scholar] [CrossRef]
- Fritz, E.; Fekete, A.; Lintelmann, J.; Schmitt-Kopplin, P.; Meckenstock, R.U. Isolation of two Pseudomonas strains producing pseudomonic acid A. Syst. Appl. Microbiol. 2009, 32, 56–64. [Google Scholar] [CrossRef]
- Haines, A.S.; Kendrew, S.G.; Crowhurst, N.; Stephens, E.R.; Connolly, J.; Hothersall, J.; Miller, C.E.; Collis, A.J.; Huckle, B.D.; Thomas, C.M. High quality genome annotation and expression visualisation of a mupirocin-producing bacterium. PLoS ONE 2022, 17, e0268072. [Google Scholar] [CrossRef]
- Hothersall, J.; Wu, J.; Rahman, A.S.; Shields, J.A.; Haddock, J.; Johnson, N.; Cooper, S.M.; Stephens, E.R.; Cox, R.J.; Crosby, J.; et al. Mutational Analysis Reveals That All Tailoring Region Genes Are Required for Production of Polyketide Antibiotic Mupirocin by Pseudomonas fluorescens. J. Biol. Chem. 2007, 282, 15451–15461. [Google Scholar] [CrossRef]
- Cooper, S.M.; Laosripaiboon, W.; Rahman, A.S.; Hothersall, J.; El-Sayed, A.K.; Winfield, C.; Crosby, J.; Cox, R.J.; Simpson, T.J.; Thomas, C.M. Shift to Pseudomonic Acid B Production in P. fluorescens NCIMB10586 by Mutation of Mupirocin Tailoring Genes mupO, mupU, mupV, and macpE. Chem. Biol. 2005, 12, 825–833. [Google Scholar] [CrossRef]
- Caldwell, C.J.; Hynes, R.K.; Boyetchko, S.M.; Korber, D.R. Colonization and bioherbicidal activity on green foxtail by Pseudomonas fluorescens BRG100 in a pesta formulation. Can. J. Microbiol. 2012, 58, 1–9. [Google Scholar] [CrossRef]
- Dumonceaux, T.J.; Town, J.; Links, M.G.; Boyetchko, S. High-Quality Draft Genome Sequence of Pseudomonas sp. BRG100, a Strain with Bioherbicidal Properties against Setaria viridis (Green Foxtail) and Other Pests of Agricultural Signifi-cance. Genome Announc. 2014, 2, e00995-14. [Google Scholar] [CrossRef]
- Barrish, J.C.; Lee, H.L.; Mitt, T.; Pizzolato, G.; Baggiolini, E.G.; Uskokovic, M.R. Total synthesis of pseudomonic acid C. J. Org. Chem. 1988, 53, 4282–4295. [Google Scholar] [CrossRef]
- Lazar, R.G.; Blaga, A.C.; Dragoi, E.N.; Galaction, A.I.; Cascaval, D. Application of reactive extraction for the separation of pseudomonic acids: Influencing factors, interfacial mechanism, and process modelling. Can. J. Chem. Eng. 2022, 100, S246–S257. [Google Scholar] [CrossRef]
- Snider, B.B.; Phillips, G.B. Total synthesis of (+-)-pseudomonic acids A and C. J. Am. Chem. Soc. 1982, 104, 1113–1114. [Google Scholar] [CrossRef]
- Sridhar, Y.; Srihari, P. A unified strategy for the synthesis of the C1–C14 fragment of marinolic acids, mupirocins, pseudomonic acids and thiomarinols: Total synthesis of pseudomonic acid methyl monate C. Org. Biomol. Chem. 2014, 12, 2950. [Google Scholar] [CrossRef]
- El-Sayed, A.K.; Hothersall, J.; Thomas, C.M. Quorum-sensing-dependent regulation of biosynthesis of the polyketide antibiotic mupirocin in Pseudomonas fluorescens NCIMB 10586 The GenBank accession numbers for the sequences determined in this work are AF318063 (mupA), AF318064 (mupR) and AF318065 (mupI). Microbiology 2001, 147, 2127–2139. [Google Scholar] [CrossRef] [PubMed]
- Macioszek, M. Biosynthesis of Mupirocin by Pseudomonas fluorescens NCIMB 10586. Ph.D. Thesis, The University of Birmingham, Birmingham, UK, 2009. [Google Scholar]
- Dwivedi, D.; Johri, B.N. Antifungals from fluorescent pseudomonads: Biosynthesis and regulation. Curr. Sci. 2003, 85, 1693–1703. [Google Scholar]
- Gurney, R. Biosynthesis of the Antibiotic Mupirocin by Pseudomonas fluorescens NCIMB 10586. Ph.D. Thesis, The University of Birmingham, Birmingham, UK, 2012. [Google Scholar]
- Gulyas, E.; Balogh, G.; Erdei, J.; Seress, P. PH Controlled Fermentation Process for Pseudomonic Acid Production. U.S. Patent No. 7439,045 B2, 21 June 2002. [Google Scholar]
- Conly, J.M.; Johnston, B.L. Mupirocin—Are We in Danger of Losing It? Can. J. Infect. Dis. 2002, 13, 157–159. [Google Scholar] [CrossRef]
- Cern, A.; Connolly, K.L.; Jerse, A.E.; Barenholz, Y. In Vitro Susceptibility of Neisseria gonorrhoeae Strains to Mupirocin, an Antibiotic Reformulated for Parenteral Administration in Nanoliposomes. Antimicrob. Agents Chemother. 2018, 62, e02377-17. [Google Scholar] [CrossRef] [PubMed]
- Hughes, J.; Mellows, G. Inhibition of isoleucyl-transfer ribonucleic acid synthetase in Echerichia coli by pseudomonic acid. Biochem. J. 1978, 176, 305–318. [Google Scholar] [CrossRef]
- Lamb, Y.J. Overview of the role of mupirocin. J. Hosp. Infect. 1991, 19, 27–30. [Google Scholar] [CrossRef]
- Parenti, M.A.; Hatfield, S.M.; Leyden, J.J. Mupirocin: A topical antibiotic with a unique structure and mechanism of action. Clin. Pharm. 1987, 6, 761–770. [Google Scholar]
- Savage, P.; Milner, S.M. Expanding the spectrum of activity of mupirocin to include gram-negative bacteria using cationic steroid antibiotics. J. Am. Acad. Dermatol. 2005, 52, P7. [Google Scholar] [CrossRef]
- Oduro-Yeboah, J. Pharmaceutical Formulations Containing Pseudomonic Acid. U.S. Patent No. 4,524,075, 26 May 1983. [Google Scholar]
- Hunter, P.; Berry, V.; Oduro-Yeboah, J.; Orr, N. Treatment of Fungal Infections. U.S. Patent No. 4,790,989, 10 April 1987. [Google Scholar]
- Zimmerman, H.L. Pharmaceutical and Veterinary Compositions of Mupirocin and Methods for Their Preparation. U.S. Patent No. 6,025,389, 25 July 1997. [Google Scholar]
- Lavon, I.; Zeevi, A.; Cherkez, S.; Arkin, M.; Abu-Gnim, C.; Raechav, Y.; Kaspi, J. Pharmaceutical Compositions Con-Taining Amorphous Mupirocin. European Patent Application No. 1 174 133 A1, 23 January 2002. [Google Scholar]
- Curzons, A.D. Lithium Pseudomonate and Its Preparation. Canada Patent No. CA1,115,699A, 4 May 1979. [Google Scholar]
- Curzons, A.D. Crystalline Lithium Pseudomonete. U.S. Patent No. US4,786,742, 8 August 1986. [Google Scholar]
- Baker, G.H.; Beal, M. Crystalline Calcium Pseudomonate. U.S. Patent No. US4,916,155, 13 April 1989. [Google Scholar]
- Baker, G.H.; Beal, M. Process for Preparing Crystalline Calcium Pseudomonate. U.S. Patent No. US5,191,093, 12 March 1990. [Google Scholar]
- Baker, G.H.; Beal, M. Compounds. U.S. Patent No. US5,436,266, 28 February 1994. [Google Scholar]
- Orr, N.A.; Greenway, M.J. Pharmaceutical Composition. U.S. Patent No. US4,879,287, 21 December 1988. [Google Scholar]
- Weisman, A.; Kaspi, J.; Cherkez, S. Stable Amorphous Calcium Pseudomonate and Processes for the Preparation Thereof. European Patent No. EP 1 384 721 A1, 23 July 2003. [Google Scholar]
- Pappa, K.A. The clinical development of mupirocin. J. Am. Acad. Dermatol. 1990, 22, 873–879. [Google Scholar] [CrossRef]
- Bull, D.N.; Kempe, L.L. Kinetics of the conversion of glucose to gluconic acid by Pseudomonas ovalis. Biotechnol. Bioeng. 1970, 12, 273–290. [Google Scholar] [CrossRef][Green Version]
- de Werra, P.; Péchy-Tarr, M.; Keel, C.; Maurhofer, M. Role of Gluconic Acid Production in the Regulation of Biocontrol Traits of Pseudomonas fluorescens CHA0. Appl. Environ. Microbiol. 2009, 75, 4162–4174. [Google Scholar] [CrossRef] [PubMed]
- Galet, J.; Deveau, A.; Hôtel, L.; Leblond, P.; Frey-Klett, P.; Aigle, B. Gluconic acid-producing Pseudomonas sp. prevent γ-actinorhodin biosynthesis by Streptomyces coelicolor A3(2). Arch. Microbiol. 2014, 196, 619–627. [Google Scholar] [CrossRef][Green Version]
- Kaur, R.; Nayudu, M. A Method of Controlling Fungal Pathogens, and Agents Useful for Same. U.S. Patent No. US 2007/0274973 A1, 7 August 2006. [Google Scholar]
- Yu, K.; Liu, Y.; Tichelaar, R.; Savant, N.; Lagendijk, E.; van Kuijk, S.J.L.; Stringlis, I.A.; van Dijken, A.J.H.; Pieterse, C.M.J.; Bakker, P.A.H.M.; et al. Rhizosphere-Associated Pseudomonas Suppress Local Root Immune Responses by Gluconic Acid-Mediated Lowering of Environmental pH. Curr. Biol. 2019, 29, 3913–3920.e4. [Google Scholar] [CrossRef] [PubMed]
- Whiting, P.H.; Midgley, M.; Dawes, E.A. The regulation of transport of glucose, gluconate and 2-oxogluconate and of glucose catabolism in Pseudomonas aeruginosa. Biochem. J. 1976, 154, 659–668. [Google Scholar] [CrossRef] [PubMed]
- Goldstein, A.H. Recent Progress in Understanding the Molecular Genetics and Biochemistry of Calcium Phosphate Solubilization by Gram Negative Bacteria. Biol. Agric. Hortic. 1995, 12, 185–193. [Google Scholar] [CrossRef]
- Ghose, T.K.; Ghosh, P. Kinetic analysis of gluconic acid production by Pseudomonas ovalis. J. Appl. Chem. Biotechnol. 2007, 26, 768–777. [Google Scholar] [CrossRef]
- Constantinides, A.; Rai, V.R. Mathematical Modeling and Optimization of Gluconic Acid Fermentation. AIChE Symp. Ser. 1973, 69, 114. [Google Scholar]
- Nyiri, L.K.; Toth, G.M. Space Biosynthesis Systems; Report No. 102110176FD. 1976. Available online: https://ntrs.nasa.gov/api/citations/19770009736/downloads/19770009736.pdf (accessed on 20 April 2024).
- Illmer, P.; Schinner, F. Solubilization of inorganic phosphates by microorganisms isolated from forest soils. Soil Biol. Biochem. 1992, 24, 389–395. [Google Scholar] [CrossRef]
- Di Simine, C.D.; Sayer, J.A.; Gadd, G.M. Solubilization of zinc phosphate by a strain of Pseudomonas fluorescens isolated from a forest soil. Biol. Fertil. Soils 1998, 28, 87–94. [Google Scholar] [CrossRef]
- Ramachandran, S.; Fontanille, P.; Pandey, A.; Larroche, C. Gluconic Acid: Properties, Applications and Microbial Production. Food Technol. Biotechnol. 2006, 44, 185–195. [Google Scholar]
- Trivedi, P.; Sa, T. Pseudomonas corrugata (NRRL B-30409) Mutants Increased Phosphate Solubilization, Organic Acid Production, and Plant Growth at Lower Temperatures. Curr. Microbiol. 2008, 56, 140–144. [Google Scholar] [CrossRef] [PubMed]
- Zia, M.A.; Khalil-ur-Rahman; Saeed, M.K.; Andaleeb, F.; Rajoka, M.I.; Sheikh, M.A.; Khan, I.A.; Khan, A.I. Thermal Characterization of Purified Glucose Oxidase from A Newly Isolated Aspergillus Niger UAF-1. J. Clin. Biochem. Nutr. 2007, 41, 132–138. [Google Scholar] [CrossRef]
- Frederick, K.R.; Tung, J.; Emerick, R.S.; Masiarz, F.R.; Chamberlain, S.H.; Vasavada, A.; Rosenberg, S.; Chakraborty, S.; Schopfer, L.M.; Schopter, L.M. Glucose oxidase from Aspergillus niger. Cloning, gene sequence, secretion from Saccharomyces cerevisiae and kinetic analysis of a yeast-derived enzyme. J. Biol. Chem. 1990, 265, 3793–3802. [Google Scholar] [CrossRef]
- Dokter, P.; Frank, J.; Duine, J.A. Purification and characterization of quinoprotein glucose dehydrogenase from Acinetobacter calcoaceticus L.M.D. 79.41. Biochem. J. 1986, 239, 163–167. [Google Scholar] [CrossRef] [PubMed]
- Pronk, J.T.; van Schie, B.J.; van Dijken, J.P.; Kuenen, J.G. Energization of solute transport by PQQ-dependent glucose dehydrogenase in membrane vesicles of Acinetobacter species. Antonie Van Leeuwenhoek 1985, 51, 560. [Google Scholar] [CrossRef]
- Schie, B.J.; Dijken, J.P.; Kuenen, J.G. Non-coordinated synthesis of glucose dehydrogenase and its prosthetic group PQQ in Acinetobacter and Pseudomonas species. FEMS Microbiol. Lett. 1984, 24, 133–138. [Google Scholar] [CrossRef]
- Velizarov, S.; Beschkov, V. Production of free gluconic acid by cells of Gluconobacter oxydans. Biotechnol. Lett. 1994, 16, 715–720. [Google Scholar] [CrossRef]
- Khan, S.; Somerville, D.; Frese, M.; Nayudu, M. Environmental gut bacteria in European honey bees (Apis mellifera) from Australia and their relationship to the chalkbrood disease. PLoS ONE 2020, 15, e0238252. [Google Scholar] [CrossRef]
- Kremmydas, G.F.; Tampakaki, A.P.; Georgakopoulos, D.G. Characterization of the Biocontrol Activity of Pseudomonas fluorescens Strain X Reveals Novel Genes Regulated by Glucose. PLoS ONE 2013, 8, e61808. [Google Scholar] [CrossRef] [PubMed]
- An, R.; Moe, L.A. Regulation of Pyrroloquinoline Quinone-Dependent Glucose Dehydrogenase Activity in the Model Rhizosphere-Dwelling Bacterium Pseudomonas putida KT2440. Appl. Environ. Microbiol. 2016, 82, 4955–4964. [Google Scholar] [CrossRef] [PubMed]
- Wei, Q.; Ran, T.; Ma, C.; He, J.; Xu, D.; Wang, W. Crystal Structure and Function of PqqF Protein in the Pyrroloquinoline Quinone Biosynthetic Pathway. J. Biol. Chem. 2016, 291, 15575–15587. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.-Q.; Bonnot, F.; Imsand, E.M.; RoseFigura, J.M.; Sjölander, K.; Klinman, J.P. Distribution and Properties of the Genes Encoding the Biosynthesis of the Bacterial Cofactor, Pyrroloquinoline Quinone. Biochemistry 2012, 51, 2265–2275. [Google Scholar] [CrossRef]
- Morales, G.; Ugidos, A.; Rojo, F. Inactivation of the Pseudomonas putida cytochrome o ubiquinol oxidase leads to a significant change in the transcriptome and to increased expression of the CIO and cbb3-1 terminal oxidases. Environ. Microbiol. 2006, 8, 1764–1774. [Google Scholar] [CrossRef] [PubMed]
- Miller, S.H.; Browne, P.; Prigent-Combaret, C.; Combes-Meynet, E.; Morrissey, J.P.; O’Gara, F. Biochemical and genomic comparison of inorganic phosphate solubilization in Pseudomonas species. Environ. Microbiol. Rep. 2010, 2, 403–411. [Google Scholar] [CrossRef] [PubMed]
- Foster, P.L. In Vivo Mutagenesis; Elsevier: Amsterdam, The Netherlands, 1991; pp. 114–125. [Google Scholar] [CrossRef]
- Brian, P.; Riggle, P.J.; Santos, R.A.; Champness, W.C. Global negative regulation of Streptomyces coelicolor antibiotic synthesis mediated by an absA-encoded putative signal transduction system. J. Bacteriol. 1996, 178, 3221–3231. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Magnolo, S.K.; Leenutaphong, D.L.; DeModena, J.A.; Curtis, J.E.; Bailey, J.E.; Galazzo, J.L.; Hughes, D.E. Actinorhodin Production by Streptomyces coelicolor and Growth of Streptomyces lividans Are Improved by the Expression of a Bacterial Hemoglobin. Nat. Biotechnol. 1991, 9, 473–476. [Google Scholar] [CrossRef] [PubMed]
- Thompson, I.P.; Lilley, A.K.; Ellis, R.J.; Bramwell, P.A.; Bailey, M.J. Survival, Colonization and Dispersal of Genetically Modified Pseudomonas fluorescens SBW25 in the Phytosphere of Field Grown Sugar Beet. Nat. Biotechnol. 1995, 13, 1493–1497. [Google Scholar] [CrossRef]
- Compeau, G.; Al-Achi, B.J.; Platsouka, E.; Levy, S.B. Survival of rifampin-resistant mutants of Pseudomonas fluorescens and Pseudomonas putida in soil systems. Appl. Environ. Microbiol. 1988, 54, 2432–2438. [Google Scholar] [CrossRef] [PubMed]
- Ramette, A.; Frapolli, M.; Saux, M.F.-L.; Gruffaz, C.; Meyer, J.-M.; Défago, G.; Sutra, L.; Moënne-Loccoz, Y. Pseudomonas protegens sp. nov.; widespread plant-protecting bacteria producing the biocontrol compounds 2,4-diacetylphloroglucinol and pyoluteorin. Syst. Appl. Microbiol. 2011, 34, 180–188. [Google Scholar] [CrossRef] [PubMed]
- Holloway, B.W.; Romling, U.; Tummler, B. Genomic mapping of Pseudomonas aeruginosa PAO. Microbiology 1994, 140, 2907–2929. [Google Scholar] [CrossRef] [PubMed]
- Ponraj, P.; Shankar, M.; Ilakkiam, D.; Rajendhran, J.; Gunasekaran, P. Influence of periplasmic oxidation of glucose on pyoverdine synthesis in Pseudomonas putida S11. Appl. Microbiol. Biotechnol. 2013, 97, 5027–5041. [Google Scholar] [CrossRef]
- Vyas, P.; Gulati, A. Organic acid production in vitro and plant growth promotion in maize under controlled environment by phosphate-solubilizing fluorescent Pseudomonas. BMC Microbiol. 2009, 9, 174. [Google Scholar] [CrossRef]
- Chernin, L.; Brandis, A.; Ismailov, Z.; Chet, I. Pyrrolnitrin Production by an Enterobacter agglomerans Strain with a Broad Spectrum of Antagonistic Activity Towards Fungal and Bacterial Phytopathogens. Curr. Microbiol. 1996, 32, 208–212. [Google Scholar] [CrossRef]
- Arima, K.; Imanaka, H.; Kousaka, M.; Fukuta, A.; Tamura, G. Pyrrolnitrin, a New Antibiotic Substance, Produced by Pseudomonas. Agric. Biol. Chem. 1964, 28, 575–576. [Google Scholar] [CrossRef]
- Hammer, P.E.; Hill, D.S.; Lam, S.T.; Van Pée, K.H.; Ligon, J.M. Four genes from Pseudomonas fluorescens that encode the biosynthesis of pyrrolnitrin. Appl. Environ. Microbiol. 1997, 63, 2147–2154. [Google Scholar] [CrossRef] [PubMed]
- Kirner, S.; Hammer, P.E.; Hill, D.S.; Altmann, A.; Fischer, I.; Weislo, L.J.; Lanahan, M.; van Pée, K.-H.; Ligon, J.M. Functions Encoded by Pyrrolnitrin Biosynthetic Genes from Pseudomonas fluorescens. J. Bacteriol. 1998, 180, 1939–1943. [Google Scholar] [CrossRef] [PubMed]
- Kirner, S.; Krauss, S.; Sury, G.; Lam, S.T.; Ligon, J.M.; van Pee, K.-H. The non-haem chloroperoxidase from Pseudomonas fluorescens and its relationship to pyrrolnitrin biosynthesis. Microbiology 1996, 142, 2129–2135. [Google Scholar] [CrossRef]
- Hammer, P.E.; Burd, W.; Hill, D.S.; Ligon, J.M.; Pée, K.-H. Conservation of the pyrrolnitrin biosynthetic gene cluster among six pyrrolnitrin-producing strains. FEMS Microbiol. Lett. 1999, 180, 39–44. [Google Scholar] [CrossRef] [PubMed]
- Huang, R.; Feng, Z.; Chi, X.; Sun, X.; Lu, Y.; Zhang, B.; Lu, R.; Luo, W.; Wang, Y.; Miao, J.; et al. Pyrrolnitrin is more essential than phenazines for Pseudomonas chlororaphis G05 in its suppression of Fusarium graminearum. Microbiol. Res. 2018, 215, 55–64. [Google Scholar] [CrossRef]
- Nandi, M.; Selin, C.; Brassinga, A.K.C.; Belmonte, M.F.; Fernando, W.G.D.; Loewen, P.C.; de Kievit, T.R. Pyrrolnitrin and Hydrogen Cyanide Production by Pseudomonas chlororaphis Strain PA23 Exhibits Nematicidal and Repellent Activity against Caenorhabditis elegans. PLoS ONE 2015, 10, e0123184. [Google Scholar] [CrossRef] [PubMed]
- Park, J.Y.; Oh, S.A.; Anderson, A.J.; Neiswender, J.; Kim, J.-C.; Kim, Y.C. Production of the antifungal compounds phenazine and pyrrolnitrin from Pseudomonas chlororaphis O6 is differentially regulated by glucose. Lett. Appl. Microbiol. 2011, 52, 532–537. [Google Scholar] [CrossRef]
- Cartwright, D.K.; Chilton, W.S.; Benson, D.M. Pyrrolnitrin and phenazine production by Pseudomonas cepacia, strain 5.5B, a biocontrol agent of Rhizoctonia solani. Appl. Microbiol. Biotechnol. 1995, 43, 211–216. [Google Scholar] [CrossRef]
- Lively, D.H.; Gorman, M.; Haney, M.E.; Mabe, J.A. Metabolism of tryptophans by Pseudomonas aureofaciens. I. Biosynthesis of pyrrolnitrin. Antimicrob. Agents Chemother. 1966, 6, 462–469. [Google Scholar]
- Hamill, R.; Elander, R.; Mabe, J.; Gorman, M. Metabolism of tryptophans by Pseudomonas aureofaciens. V. Conversion of tryptophan to pyrrolnitrin. Antimicrob. Agents Chemother. 1967, 7, 388–396. [Google Scholar]
- Pawar, S.; Chaudhari, A.; Prabha, R.; Shukla, R.; Singh, D.P. Microbial Pyrrolnitrin: Natural Metabolite with Immense Practical Utility. Biomolecules 2019, 9, 443. [Google Scholar] [CrossRef]
- De Laurentis, W.; Khim, L.; Anderson, J.L.R.; Adam, A.; Phillips, R.S.; Chapman, S.K.; van Pee, K.-H.; Naismith, J.H. The Second Enzyme in Pyrrolnitrin Biosynthetic Pathway Is Related to the Heme-Dependent Dioxygenase Superfamily. Biochemistry 2007, 46, 12393–12404. [Google Scholar] [CrossRef]
- van Pée, K.-H.; Salcher, O.; Lingens, F. Formation of Pyrrolnitrin and 3-(2-Amino-3-chlorophenyl)pyrrole from 7-Chlorotryptophan. Angew. Chem. Int. Ed. Engl. 1980, 19, 828–829. [Google Scholar] [CrossRef]
- Chang, C.J.; Floss, H.G.; Hook, D.J.; Mabe, J.A.; Manni, P.E.; Martin, L.L.; Schröder, K.; Shieh, T.L. The biosynthesis of the antibiotic pyrrolnitrin by Pseudomonas aureofaciens. J. Antibiot. 1981, 34, 555–566. [Google Scholar] [CrossRef]
- van Pée, K.-H. Biosynthesis of halogenated metabolites by bacteria. Annu. Rev. Microbiol. 1996, 50, 375–399. [Google Scholar] [CrossRef]
- Wiesner, W.; van Pée, K.H.; Lingens, F. Purification and characterization of a novel bacterial non-heme chloroperoxidase from Pseudomonas pyrrocinia. J. Biol. Chem. 1988, 263, 13725–13732. [Google Scholar] [CrossRef]
- Keller, S.; Wage, T.; Hohaus, K.; Hölzer, M.; Eichhorn, E.; van Pée, K.-H. Purification and Partial Characterization of Tryptophan 7-Halogenase (PrnA) from Pseudomonas fluorescens. Angew. Chem. Int. Ed. 2000, 39, 2300–2302. [Google Scholar] [CrossRef]
- Singh, R.N.; Singh, R.P.; Sharma, A.; Saxena, A.K. Modeling of PrnD protein from Pseudomonas fluorescens RajNB11 and its comparative structural analysis with PrnD proteins expressed in Burkholderia and Serratia. Turk. J. Biol. 2016, 40, 623–633. [Google Scholar] [CrossRef]
- de Souza, J.T.; Arnould, C.; Deulvot, C.; Lemanceau, P.; Gianinazzi-Pearson, V.; Raaijmakers, J.M. Effect of 2,4-Diacetylphloroglucinol on Pythium: Cellular Responses and Variation in Sensitivity Among Propagules and Species. Phytopathology 2003, 93, 966–975. [Google Scholar] [CrossRef]
- Laville, J.; Voisard, C.; Keel, C.; Maurhofer, M.; Défago, G.; Haas, D. Global control in Pseudomonas fluorescens mediating antibiotic synthesis and suppression of black root rot of tobacco. Proc. Natl. Acad. Sci. USA 1992, 89, 1562–1566. [Google Scholar] [CrossRef]
- Sarniguet, A.; Kraus, J.; Henkels, M.D.; Muehlchen, A.M.; Loper, J.E. The sigma factor sigma s affects antibiotic production and biological control activity of Pseudomonas fluorescens Pf-5. Proc. Natl. Acad. Sci. USA 1995, 92, 12255–12259. [Google Scholar] [CrossRef]
- Pfender, W.F. A Genomic Region from Pseudomonas fluorescens Pf-5 Required for Pyrrolnitrin Production and Inhibition of Pyrenophora tritici-repentis in Wheat Straw. Phytopathology 1993, 83, 1223. [Google Scholar] [CrossRef]
- Manuel, J.; Selin, C.; Fernando, W.G.D.; de Kievit, T. Stringent response mutants of Pseudomonas chlororaphis PA23 exhibit enhanced antifungal activity against Sclerotinia sclerotiorum in vitro. Microbiology 2012, 158, 207–216. [Google Scholar] [CrossRef]
- Wu, X.; Chi, X.; Wang, Y.; Zhang, K.; Kai, L.; He, Q.; Tang, J.; Wang, K.; Sun, L.; Hao, X.; et al. vfr, A Global Regulatory Gene, is Required for Pyrrolnitrin but not for Phenazine-1-carboxylic Acid Biosynthesis in Pseudomonas chlororaphis G05. Plant Pathol. J. 2019, 35, 351–361. [Google Scholar] [CrossRef]
- West, S.E.; Sample, A.K.; Runyen-Janecky, L.J. The vfr gene product, required for Pseudomonas aeruginosa exotoxin A and protease production, belongs to the cyclic AMP receptor protein family. J. Bacteriol. 1994, 176, 7532–7542. [Google Scholar] [CrossRef][Green Version]
- Ogura, K.; Matsui, H.; Yamamoto, M.; Noutoshi, Y.; Toyoda, K.; Taguchi, F.; Ichinose, Y. Vfr targets promoter of genes encoding methyl-accepting chemotaxis protein in Pseudomonas syringae pv. tabaci 6605. Biochem. Biophys. Rep. 2021, 26, 100944. [Google Scholar] [CrossRef]
- Fuchs, E.L.; Brutinel, E.D.; Jones, A.K.; Fulcher, N.B.; Urbanowski, M.L.; Yahr, T.L.; Wolfgang, M.C. The Pseudomonas aeruginosa Vfr regulator controls global virulence factor expression through cyclic AMP-dependent and -independent mechanisms. J. Bacteriol. 2010, 192, 3553–3564. [Google Scholar] [CrossRef]
- Keum, Y.S.; Lee, H.R.; Kim, J.-H. Effects of Pesticides on the Bacterial Production of Pyrrolnitrin. J. Agric. Food Chem. 2010, 58, 5531–5537. [Google Scholar] [CrossRef]
- Salcher, O.; Lingens, F. Isolation and Characterization of a Mutant of Pseudomonas aureofaciens ATCC 15926 with an Increased Capacity for Synthesis of Pyrrolnitrin. Microbiology 1980, 118, 509–513. [Google Scholar] [CrossRef]
- Auden, J.; Gruner, J.; Nüesch, J.; Knüsel, F. Some Statistical Methods in Nutrient Medium Optimalisation. Pathobiology 1967, 30, 858–866. [Google Scholar] [CrossRef]
- Olson, E.S.; Richards, J.H. Structure of the orange pigment from Pseudomonas aureofaciens. J. Org. Chem. 1967, 32, 2887–2890. [Google Scholar] [CrossRef]
- Zhang, J.; Mavrodi, D.V.; Yang, M.; Thomashow, L.S.; Mavrodi, O.V.; Kelton, J.; Weller, D.M. Pseudomonas synxantha 2-79 Transformed with Pyrrolnitrin Biosynthesis Genes Has Improved Biocontrol Activity Against Soilborne Pathogens of Wheat and Canola. Phytopathology 2020, 110, 1010–1017. [Google Scholar] [CrossRef]
- Loper, J.E.; Hassan, K.A.; Mavrodi, D.V.; Davis, E.W.; Lim, C.K.; Shaffer, B.T.; Elbourne, L.D.H.; Stockwell, V.O.; Hartney, S.L.; Breakwell, K.; et al. Comparative Genomics of Plant-Associated Pseudomonas spp.: Insights into Diversity and Inheritance of Traits Involved in Multitrophic Interactions. PLoS Genet. 2012, 8, e1002784. [Google Scholar] [CrossRef]
- Ligon, J.; Hill, D.; Lam, S.; Gaffney, T.; Torkewitz, N. Genetically Modified Pseudomonas Strans with Enhanced Biocontrol Activity. U.S. Patent No. US 5,756,087, 6 December 1996. [Google Scholar]
- Janisiewicz, W.; Roitman, J. Biological Control of Postharvest Rots in Fruits Using Pseudomonas cepacia and Pyrrolnitrin Produced Therefrom. U.S. Patent No. US 4,975,277, 2 August 1988. [Google Scholar]
- Gnanamanickam, S. Pseudomonas Bacterium. U.S. Patent Application No. US 2010/0093538A1, 29 September 2009. [Google Scholar]
- Roberts, D.P.; McKenna, L.F.; Lakshman, D.K.; Meyer, S.L.F.; Kong, H.; de Souza, J.T.; Lydon, J.; Baker, C.J.; Buyer, J.S.; Chung, S. Suppression of damping-off of cucumber caused by Pythium ultimum with live cells and extracts of Serratia marcescens N4-5. Soil Biol. Biochem. 2007, 39, 2275–2288. [Google Scholar] [CrossRef]
- Envall, M. On the difference between mono-, holo-, and paraphyletic groups: A consistent distinction of process and pattern. Biol. J. Linn. Soc. 2008, 94, 217–220. [Google Scholar] [CrossRef]
- Almario, J.; Bruto, M.; Vacheron, J.; Prigent-Combaret, C.; Moënne-Loccoz, Y.; Muller, D. Distribution of 2,4-Diacetylphloroglucinol Biosynthetic Genes among the Pseudomonas spp. Reveals Unexpected Polyphyletism. Front. Microbiol. 2017, 8, 1218. [Google Scholar] [CrossRef]
- Shanahan, P.; Glennon, J.D.; Crowley, J.J.; Donnelly, D.F.; O’Gara, F. Liquid chromatographic assay of microbially derived phloroglucinol antibiotics for establishing the biosynthetic route to production, and the factors affecting their regulation. Anal. Chim. Acta 1993, 272, 271–277. [Google Scholar] [CrossRef]
- Bangera, M.G.; Thomashow, L.S. Identification and Characterization of a Gene Cluster for Synthesis of the Polyketide Antibiotic 2,4-Diacetylphloroglucinol from Pseudomonas fluorescens Q2-87. J. Bacteriol. 1999, 181, 3155–3163. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Zhao, H.; Wu, X.; Zhang, L.-Q. The Oxidoreductase DsbA1 negatively influences 2,4-diacetylphloroglucinol biosynthesis by interfering the function of Gcd in Pseudomonas fluorescens 2P24. BMC Microbiol. 2020, 20, 39. [Google Scholar] [CrossRef] [PubMed]
- Nowak-Thompson, B.; Gould, S.J.; Kraus, J.; Loper, J.E. Production of 2,4-diacetylphloroglucinol by the biocontrol agent Pseudomonas fluorescens Pf-5. Can. J. Microbiol. 1994, 40, 1064–1066. [Google Scholar] [CrossRef]
- Jing, X.; Cui, Q.; Li, X.; Yin, J.; Ravichandran, V.; Pan, D.; Fu, J.; Tu, Q.; Wang, H.; Bian, X.; et al. Engineering Pseudomonas protegens Pf-5 to improve its antifungal activity and nitrogen fixation. Microb. Biotechnol. 2020, 13, 118–133. [Google Scholar] [CrossRef] [PubMed]
- Shanahan, P.; O’Sullivan, D.J.; Simpson, P.; Glennon, J.D.; O’Gara, F. Isolation of 2,4-Diacetylphloroglucinol from a Fluorescent Pseudomonad and Investigation of Physiological Parameters Influencing Its Production. Appl. Environ. Microbiol. 1992, 58, 353–358. [Google Scholar] [CrossRef] [PubMed]
- Achkar, J.; Xian, M.; Zhao, H.; Frost, J.W. Biosynthesis of Phloroglucinol. J. Am. Chem. Soc. 2005, 127, 5332–5333. [Google Scholar] [CrossRef] [PubMed]
- Hutchinson, C.R.; Fujii, I. Polyketide synthase gene manipulation: A Structure-Function Approach in Engineering Novel Antibiotics. Annu. Rev. Microbiol. 1995, 49, 201–238. [Google Scholar] [CrossRef]
- McDaniel, R.; Ebert-Khosla, S.; Hopwood, D.A.; Khosla, C. Engineered Biosynthesis of Novel Polyketides. Science 1993, 262, 1546–1550. [Google Scholar] [CrossRef] [PubMed]
- Gupta, P.; Dash, P.K.; Sanjay, T.D.; Pradhan, S.K.; Sreevathsa, R.; Rai, R. Cloning and Molecular Characterization of the phlD Gene Involved in the Biosynthesis of “Phloroglucinol”, a Compound with Antibiotic Properties from Plant Growth Promoting Bacteria Pseudomonas spp. Antibiotics 2023, 12, 260. [Google Scholar] [CrossRef]
- Balthazar, C.; St-Onge, R.; Léger, G.; Lamarre, S.G.; Joly, D.L.; Filion, M. Pyoluteorin and 2,4-diacetylphloroglucinol are major contributors to Pseudomonas protegens Pf-5 biocontrol against Botrytis cinerea in cannabis. Front. Microbiol. 2022, 13, 945498. [Google Scholar] [CrossRef] [PubMed]
- Kidarsa, T.A.; Goebel, N.C.; Zabriskie, T.M.; Loper, J.E. Phloroglucinol mediates cross-talk between the pyoluteorin and 2,4-diacetylphloroglucinol biosynthetic pathways in Pseudomonas fluorescens Pf-5. Mol. Microbiol. 2011, 81, 395–414. [Google Scholar] [CrossRef]
- Zhang, Q.X.; Kong, X.W.; Li, S.Y.; Chen, X.J.; Chen, X.J. Antibiotics of Pseudomonas protegens FD6 are essential for biocontrol activity. Australas. Plant Pathol. 2020, 49, 307–317. [Google Scholar] [CrossRef]
- Dash, P.K.; Gupta, P.; Panwar, B.S.; Rai, R. Isolation, cloning and characterization of phlB gene from an Indian strain of Gram negative soil bacteria Pseudomonas fluorescens. Indian J. Exp. Biol. 2020, 58, 412–419. [Google Scholar] [CrossRef]
- Liang, F.; Zhang, B.; Yang, Q.; Zhang, Y.; Zheng, D.; Zhang, L.; Yan, Q.; Wu, X. Cyclic-di-GMP Regulates the Quorum-Sensing System and Biocontrol Activity of Pseudomonas fluorescens 2P24 through the RsmA and RsmE Proteins. Appl. Environ. Microbiol. 2020, 86, e02016-20. [Google Scholar] [CrossRef] [PubMed]
- Landa, B.B.; Mavrodi, O.V.; Raaijmakers, J.M.; McSpadden Gardener, B.B.; Thomashow, L.S.; Weller, D.M. Differential Ability of Genotypes of 2,4-Diacetylphloroglucinol-Producing Pseudomonas fluorescens Strains to Colonize the Roots of Pea Plants. Appl. Environ. Microbiol. 2002, 68, 3226–3237. [Google Scholar] [CrossRef] [PubMed]
- Mavrodi, O.V.; McSpadden Gardener, B.B.; Mavrodi, D.V.; Bonsall, R.F.; Weller, D.M.; Thomashow, L.S. Genetic Diversity of phlD from 2,4-Diacetylphloroglucinol-Producing Fluorescent Pseudomonas spp. Phytopathology 2001, 91, 35–43. [Google Scholar] [CrossRef] [PubMed]
- Weller, D.M.; Landa, B.B.; Mavrodi, O.V.; Schroeder, K.L.; De La Fuente, L.; Blouin Bankhead, S.; Allende Molar, R.; Bonsall, R.F.; Mavrodi, D.V.; Thomashow, L.S. Role of 2,4-Diacetylphloroglucinol-Producing Fluorescent Pseudomonas spp. in the Defense of Plant Roots. Plant Biol. 2007, 9, 4–20. [Google Scholar] [CrossRef] [PubMed]
- Bangera, G.M. Characterization of a Genomic Locus Required for Synthesis of the Antibiotic 2,4-diacetylphloroglucinol by the Biological Control Agent Pseudomonas fluorescens Q2-87. Mol. Plant-Microbe Interact. 1996, 9, 083. [Google Scholar] [CrossRef] [PubMed]
- Abbas, A.; McGuire, J.E.; Crowley, D.; Baysse, C.; Dow, M.; O’Gara, F. The putative permease PhlE of Pseudomonas fluorescens F113 has a role in 2,4-diacetylphloroglucinol resistance and in general stress tolerance. Microbiology 2004, 150, 2443–2450. [Google Scholar] [CrossRef]
- Bottiglieri, M.; Keel, C. Characterization of PhlG, a Hydrolase That Specifically Degrades the Antifungal Compound 2,4-Diacetylphloroglucinol in the Biocontrol Agent Pseudomonas fluorescens CHA0. Appl. Environ. Microbiol. 2006, 72, 418–427. [Google Scholar] [CrossRef] [PubMed]
- Yan, X.; Yang, R.; Zhao, R.-X.; Han, J.-T.; Jia, W.-J.; Li, D.-Y.; Wang, Y.; Zhang, N.; Wu, Y.; Zhang, L.-Q.; et al. Transcriptional Regulator PhlH Modulates 2,4-Diacetylphloroglucinol Biosynthesis in Response to the Biosynthetic Intermediate and End Product. Appl. Environ. Microbiol. 2017, 83, e01419-17. [Google Scholar] [CrossRef]
- Zhao, M.-M.; Lyu, N.; Wang, D.; Wu, X.-G.; Zhao, Y.-Z.; Zhang, L.-Q.; Zhou, H.-Y. PhlG mediates the conversion of DAPG to MAPG in Pseudomonas fluorescens 2P24. Sci. Rep. 2020, 10, 4296. [Google Scholar] [CrossRef]
- He, Y.-X.; Huang, L.; Xue, Y.; Fei, X.; Teng, Y.-B.; Rubin-Pitel, S.B.; Zhao, H.; Zhou, C.-Z. Crystal Structure and Computational Analyses Provide Insights into the Catalytic Mechanism of 2,4-Diacetylphloroglucinol Hydrolase PhlG from Pseudomonas fluorescens. J. Biol. Chem. 2010, 285, 4603–4611. [Google Scholar] [CrossRef] [PubMed]
- Saitou, H.; Watanabe, M.; Maruyama, K. Molecular and Catalytic Properties of 2,4-Diacetylphloroglucinol Hydrolase (PhlG) from Pseudomonas sp. YGJ3. Biosci. Biotechnol. Biochem. 2012, 76, 1239–1241. [Google Scholar] [CrossRef]
- Abbas, A.; Morrissey, J.P.; Marquez, P.C.; Sheehan, M.M.; Delany, I.R.; O’Gara, F. Characterization of Interactions between the Transcriptional Repressor PhlF and Its Binding Site at the phlA Promoter in Pseudomonas fluorescens F113. J. Bacteriol. 2002, 184, 3008–3016. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Liu, J.; Zhang, W.; Zhang, L. Multiple-Level Regulation of 2,4-Diacetylphloroglucinol Production by the Sigma Regulator PsrA in Pseudomonas fluorescens 2P24. PLoS ONE 2012, 7, e50149. [Google Scholar] [CrossRef] [PubMed]
- Hansen, M.L.; He, Z.; Wibowo, M.; Jelsbak, L. A Whole-Cell Biosensor for Detection of 2,4-Diacetylphloroglucinol (DAPG)-Producing Bacteria from Grassland Soil. Appl. Environ. Microbiol. 2021, 87, e01400-20. [Google Scholar] [CrossRef] [PubMed]
- Schnider-Keel, U.; Seematter, A.; Maurhofer, M.; Blumer, C.; Duffy, B.; Gigot-Bonnefoy, C.; Reimmann, C.; Notz, R.; Défago, G.; Haas, D.; et al. Autoinduction of 2,4-Diacetylphloroglucinol Biosynthesis in the Biocontrol Agent Pseudomonas fluorescens CHA0 and Repression by the Bacterial Metabolites Salicylate and Pyoluteorin. J. Bacteriol. 2000, 182, 1215–1225. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, A.; Saitou, H.; Mori, T.; Matano, I.; Sugisaki, H.; Maruyama, K. Molecular and Catalytic Properties of Monoacetylphloroglucinol Acetyltransferase from Pseudomonas sp. YGJ3. Biosci. Biotechnol. Biochem. 2012, 76, 559–566. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Yu, X.; Yan, X.; Zhang, M.; Zhang, L.; He, Y. Flavonoids repress the production of antifungal 2,4-DAPG but potentially facilitate root colonization of the rhizobacterium Pseudomonas fluorescens. Environ. Microbiol. 2020, 22, 5073–5089. [Google Scholar] [CrossRef]
- Delany, I.; Sheehan, M.M.; Fenton, A.; Bardin, S.; Aarons, S.; O’Gara, F. Regulation of production of the antifungal metabolite 2,4-diacetylphloroglucinol in Pseudomonas fluorescens F113: Genetic analysis of phlF as a transcriptional repressor The GenBank accession number for the sequence reported in this paper is AF129856. Microbiology 2000, 146, 537–546. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, B.; Wu, H.; Wu, X.; Yan, Q.; Zhang, L.-Q. Pleiotropic effects of RsmA and RsmE proteins in Pseudomonas fluorescens 2P24. BMC Microbiol. 2020, 20, 191. [Google Scholar] [CrossRef] [PubMed]
- Biessy, A.; Filion, M. Phloroglucinol Derivatives in Plant-Beneficial Pseudomonas spp.: Biosynthesis, Regulation, and Functions. Metabolites 2021, 11, 182. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Gu, G.-Q.; Chen, W.; Gao, L.-J.; Wu, X.-H.; Zhang, L.-Q. The outer membrane protein OprF and the sigma factor SigX regulate antibiotic production in Pseudomonas fluorescens 2P24. Microbiol. Res. 2018, 206, 159–167. [Google Scholar] [CrossRef] [PubMed]
- Xiao-Gang, W.; Hui-Mei, D.; Tao, T.; Nan, Y.; Hong-You, Z.; Li-Qun, Z. Effect of the hfq gene on 2,4-diacetylphloroglucinol production and the PcoI/PcoR quorum-sensing system in Pseudomonas fluorescens 2P24. FEMS Microbiol. Lett. 2010, 309, 16–24. [Google Scholar] [CrossRef] [PubMed]
- Dong, Q.; Yan, Q.; Zhang, B.; Zhang, L.; Wu, X. Effect of the Monothiol Glutaredoxin GrxD on 2,4-Diacetylphloroglucinol Biosynthesis and Biocontrol Activity of Pseudomonas fluorescens 2P24. Front. Microbiol. 2022, 13, 920793. [Google Scholar] [CrossRef] [PubMed]
- Humair, B.; González, N.; Mossialos, D.; Reimmann, C.; Haas, D. Temperature-responsive sensing regulates biocontrol factor expression in Pseudomonas fluorescens CHA0. ISME J. 2009, 3, 955–965. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Zhang, W.; Wu, X.; Zhang, L. [Effect of retS gene on biosynthesis of 2, 4-diacetyl-phloroglucinol in Pseudomonas fluorescens 2P24]. Wei Sheng Wu Xue Bao = Acta Microbiol. Sin. 2013, 53, 118–126. [Google Scholar]
- He, Y.; Suzuki, S.; Aono, T.; Oyaizu, H. Importance of 2,4-DAPG in the biological control of brown patch by Pseudomonas fluorescens HP72 and newly identified genes involved in 2,4-DAPG biosynthesis. Soil Sci. Plant Nutr. 2004, 50, 1287–1293. [Google Scholar] [CrossRef]
- Zhou, T.-T.; Li, C.-Y.; Chen, D.; Wu, K.; Shen, Q.-R.; Shen, B. phlF− mutant of Pseudomonas fluorescens J2 improved 2,4-DAPG biosynthesis and biocontrol efficacy against tomato bacterial wilt. Biol. Control 2014, 78, 1–8. [Google Scholar] [CrossRef]
- Aarons, S.; Abbas, A.; Adams, C.; Fenton, A.; O’Gara, F. A Regulatory RNA (PrrB RNA) Modulates Expression of Secondary Metabolite Genes in Pseudomonas fluorescens F113. J. Bacteriol. 2000, 182, 3913–3919. [Google Scholar] [CrossRef]
- Heeb, S.; Itoh, Y.; Nishijyo, T.; Schnider, U.; Keel, C.; Wade, J.; Walsh, U.; O’Gara, F.; Haas, D. Small, Stable Shuttle Vectors Based on the Minimal pVS1 Replicon for Use in Gram-Negative, Plant-Associated Bacteria. Mol. Plant-Microbe Interact. 2000, 13, 232–237. [Google Scholar] [CrossRef] [PubMed]
- Kwak, Y.-S.; Han, S.; Thomashow, L.S.; Rice, J.T.; Paulitz, T.C.; Kim, D.; Weller, D.M. Saccharomyces cerevisiae Genome-Wide Mutant Screen for Sensitivity to 2,4-Diacetylphloroglucinol, an Antibiotic Produced by Pseudomonas fluorescens. Appl. Environ. Microbiol. 2011, 77, 1770–1776. [Google Scholar] [CrossRef] [PubMed]
- Troppens, D.M.; Dmitriev, R.I.; Papkovsky, D.B.; O’Gara, F.; Morrissey, J.P. Genome-wide investigation of cellular targets and mode of action of the antifungal bacterial metabolite 2,4-diacetylphloroglucinol in Saccharomyces cerevisiae. FEMS Yeast Res. 2013, 13, 322–334. [Google Scholar] [CrossRef] [PubMed]
- Stepanov, A.A.; Vasilchenko, A.S. 2,4-Diacetylphloroglucinol against Candida albicans: Biofilm Formation, Aspartyl Protease Production and Ultrastructure Changes. bioRxiv 2022. [Google Scholar] [CrossRef]
- Schouten, A.; van den Berg, G.; Edel-Hermann, V.; Steinberg, C.; Gautheron, N.; Alabouvette, C.; de Vos, C.H. (Ric), Lemanceau, P.; Raaijmakers, J.M. Defense Responses of Fusarium oxysporum to 2,4-Diacetylphloroglucinol, a Broad-Spectrum Antibiotic Produced by Pseudomonas fluorescens. Mol. Plant-Microbe Interact. 2004, 17, 1201–1211. [Google Scholar] [CrossRef] [PubMed]
- Suresh, P.; Varathraju, G.; Shanmugaiah, V.; Almaary, K.S.; Elbadawi, Y.B.; Mubarak, A. Partial purification and characterization of 2, 4-diacetylphloroglucinol producing Pseudomonas fluorescens VSMKU3054 against bacterial wilt disease of tomato. Saudi J. Biol. Sci. 2021, 28, 2155–2167. [Google Scholar] [CrossRef] [PubMed]
- Mazzola, M.; Fujimoto, D.K.; Thomashow, L.S.; Cook, R.J. Variation in Sensitivity of Gaeumannomyces graminis to Antibiotics Produced by Fluorescent Pseudomonas spp. and Effect on Biological Control of Take-All of Wheat. Appl. Environ. Microbiol. 1995, 61, 2554–2559. [Google Scholar] [CrossRef] [PubMed]
- Keel, C. Suppression of Root Diseases by Pseudomonas fluorescens CHA0: Importance of the Bacterial Secondary Metabolite 2,4-Diacetylphloroglucinol. Mol. Plant-Microbe Interact. 1992, 5, 4. [Google Scholar] [CrossRef]
- Isnansetyo, A.; Horikawa, M.; Kamei, Y. In vitro anti-methicillin-resistant Staphylococcus aureus activity of 2,4-diacetylphloroglucinol produced by Pseudomonas sp. AMSN isolated from a marine alga. J. Antimicrob. Chemother. 2001, 47, 724–725. [Google Scholar] [CrossRef]
- Julian, W.T.; Vasilchenko, A.V.; Shpindyuk, D.D.; Poshvina, D.V.; Vasilchenko, A.S. Bacterial-Derived Plant Protection Metabolite 2,4-Diacetylphloroglucinol: Effects on Bacterial Cells at Inhibitory and Subinhibitory Concentrations. Biomolecules 2020, 11, 13. [Google Scholar] [CrossRef]
- Velusamy, P.; Immanuel, J.E.; Gnanamanickam, S.S.; Thomashow, L. Biological control of rice bacterial blight by plant-associated bacteria producing 2,4-diacetylphloroglucinol. Can. J. Microbiol. 2006, 52, 56–65. [Google Scholar] [CrossRef] [PubMed]
- Meyer, S.L.F.; Everts, K.L.; Gardener, B.M.; Masler, E.P.; Abdelnabby, H.M.E.; Skantar, A.M. Assessment of DAPG-producing Pseudomonas fluorescens for Management of Meloidogyne incognita and Fusarium oxysporum on Watermelon. J. Nematol. 2016, 48, 43–53. [Google Scholar] [CrossRef] [PubMed]
- Siddiqui, I.A.; Shaukat, S.S. Plant Species, Host Age and Host Genotype Effects on Meloidogyne incognita Biocontrol by Pseudomonas fluorescens Strain CHA0 and its Genetically-Modified Derivatives. J. Phytopathol. 2003, 151, 231–238. [Google Scholar] [CrossRef]
- Siddiqui, I.A.; Shaukat, S.S. Suppression of root-knot disease by Pseudomonas fluorescens CHA0 in tomato: Importance of bacterial secondary metabolite, 2,4-diacetylpholoroglucinol. Soil Biol. Biochem. 2003, 35, 1615–1623. [Google Scholar] [CrossRef]
- Cronin, D.; Moenne-Loccoz, Y.; Fenton, A.; Dunne, C.; Dowling, D.N.; O’gara, F. Role of 2,4-Diacetylphloroglucinol in the Interactions of the Biocontrol Pseudomonad Strain F113 with the Potato Cyst Nematode Globodera rostochiensis. Appl. Environ. Microbiol. 1997, 63, 1357–1361. [Google Scholar] [CrossRef] [PubMed]
- Asolkar, R.; Cordova-Kreylos, A.L.; Todd, C. Control of Phytopathogenic Microorganisms with Pseudomonas sp. and Substances and Compositions Derived Therefrom. Canadian Patent No. CA2865237C, Patent of PRC No. CN104254611A, 21 August 2014. [Google Scholar]
- Huang, H.; Asolkar, R.; Marrone, P.; Cordova-Kreylos, A.L. Use of Proteins Isolated from Pseudomonas to Control Molluscs. U.S. Patent No. US 8,728,754 B1, 15 March 2013. [Google Scholar]
- Frost, J.W. Biosynthesis of Phloroglucinol and Preparation of 1,3-Dihydroxybenzene Therefrom. Patent of PRC No. CN101084311A, 10 November 2005. [Google Scholar]
- Raaijmakers, J.M.; Weller, D.M.; Thomashow, L.S.; Cook, R.J. Biocontrol Agents for Take-All. U.S. Patent No. US 6,447,770 B1, 14 September 1999. [Google Scholar]
- Thomashow, L.S.; Bangera, M.; Weller, D.M.; Cook, J.R. Sequences for Production of 2,4-Diacetylphloroglucinol and Methods. International Patent No. WO1997001572A1, 26 June 1996. [Google Scholar]
- Barea, J.M.; Andrade, G.; Bianciotto, V.; Dowling, D.; Lohrke, S.; Bonfante, P.; O’Gara, F.; Azcon-Aguilar, C. Impact on Arbuscular Mycorrhiza Formation of Pseudomonas Strains Used as Inoculants for Biocontrol of Soil-Borne Fungal Plant Pathogens. Appl. Environ. Microbiol. 1998, 64, 2304–2307. [Google Scholar] [CrossRef] [PubMed]
- Resca, R.; Basaglia, M.; Poggiolini, S.; Vian, P.; Bardin, S.; Walsh, U.F.; Enriquez Barreiros, C.M.; O’Gara, F.; Nuti, M.P.; Casella, S.; et al. An integrated approach for the evaluation of biological control of the complex Polymyxa betae/Beet Necrotic Yellow Vein Virus, by means of seed inoculants. Plant Soil 2001, 232, 215–226. [Google Scholar] [CrossRef]
- Girlanda, M.; Perotto, S.; Moenne-Loccoz, Y.; Bergero, R.; Lazzari, A.; Defago, G.; Bonfante, P.; Luppi, A.M. Impact of Biocontrol Pseudomonas fluorescens CHA0 and a Genetically Modified Derivative on the Diversity of Culturable Fungi in the Cucumber Rhizosphere. Appl. Environ. Microbiol. 2001, 67, 1851–1864. [Google Scholar] [CrossRef]
- Natsch, A.; Keel, C.; Hebecker, N.; Laasik, E.; Défago, G. Impact of Pseudomonas fluorescens strain CHA0 and a derivative with improved biocontrol activity on the culturable resident bacterial community on cucumber roots. FEMS Microbiol. Ecol. 1998, 27, 365–380. [Google Scholar] [CrossRef]
- Niemann, S.; Keel, C.; Pühler, A.; Selbitschka, W. Biocontrol strain Pseudomonas fluorescens CHA0 and its genetically modified derivative with enhanced biocontrol capability exert comparable effects on the structure of a Sinorhizobium meliloti population in gnotobiotic systems. Biol. Fertil. Soils 1997, 25, 240–244. [Google Scholar] [CrossRef]
- Huang, Z.; Thomashow, L.S.; Mavrodi, D.V.; Raaijmakers, J.M.; Weller, D.M.; Cook, J.R. Transgenic Strains of Pseudomonas for Biocontrol of Plant Root Diseases. U.S. Patent No. US 6,277,625 B1, 18 December 1997. [Google Scholar]
- Gangwar, A.; Kumar, P.; Singh, R.; Kush, P. Recent Advances in Mupirocin Delivery Strategies for the Treatment of Bacterial Skin and Soft Tissue Infection. Future Pharmacol. 2021, 1, 80–103. [Google Scholar] [CrossRef]
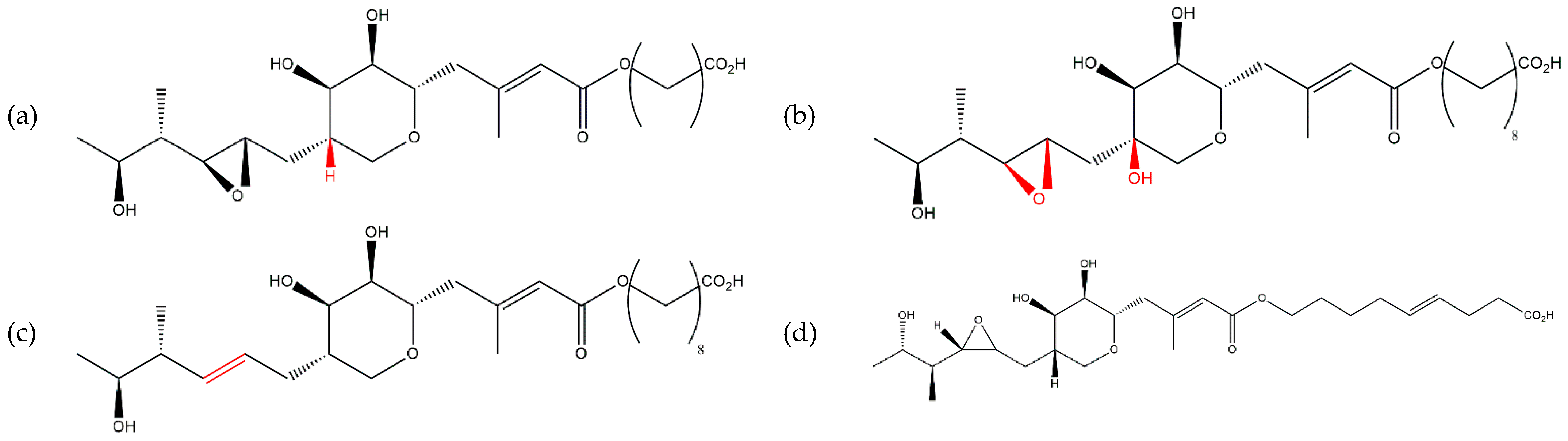



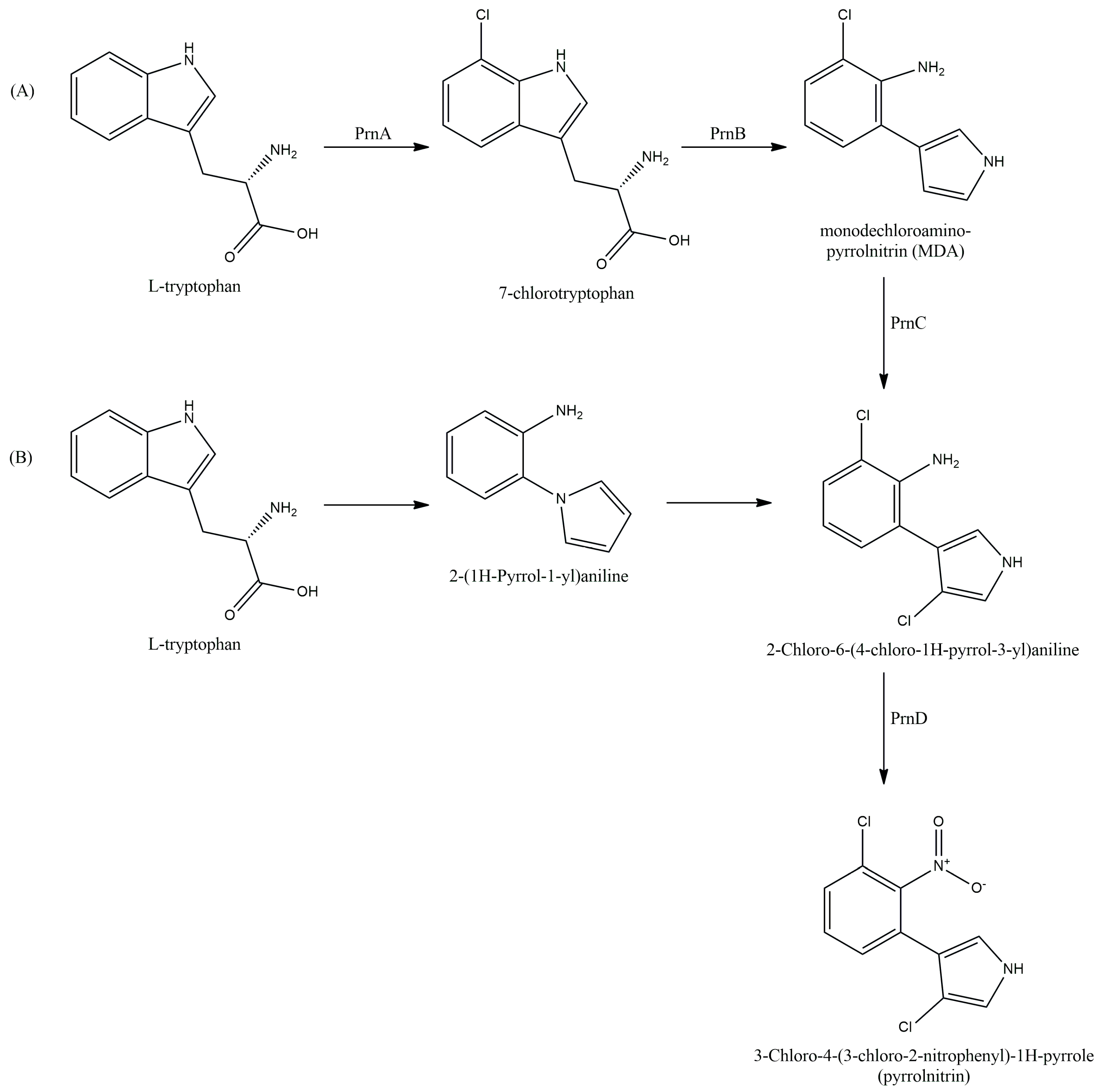
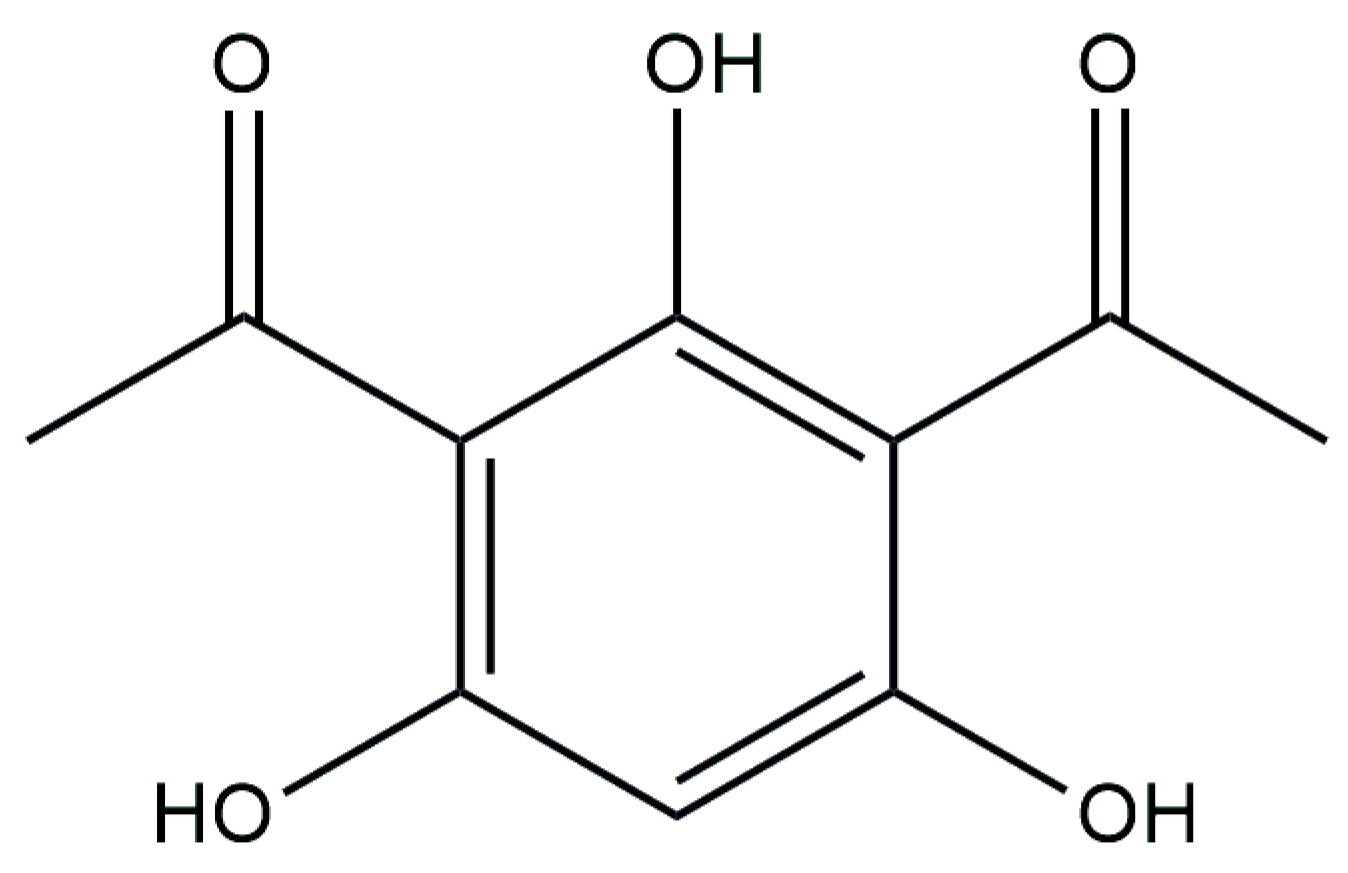
| Producer Strain | Mixture Composition | Component Ratio | Reference |
|---|---|---|---|
| Pseudomonas fluorescens NCIB 10586 | Pseudomonic acid A, its isomer with a cis double bond in the cis position between the carbon atoms C2 and C3, and pseudomonic acid B | 4.5:4.5:1 | [33,34] |
| Pseudomonic acids A, B, C and D | 90:8:1:1 | [35] | |
| Pseudomonas fluorescens Y-11633 | Pseudomonic acids A and B, and two components with unknown structures | 9:0.5:0.5 | [36] |
| Pseudomonas sp. No 19/26 | The main component is pseudomonic acid A, and minor amounts of pseudomonic acids B and C are also present | ND | [36] |
| Module | Composition of Module | Corresponding Genes | Reference |
|---|---|---|---|
| I | Large polyketide synthases (PKS) | Type I multifunctional gene mmpA | [44,45] |
| Type I multifunctional gene mmpD | |||
| Small PKS | Trans-acyltransferase mmpC | ||
| ORF | mupA | ||
| ORF | mupB | ||
| II | Small PKSs | mmpE | [40,41] |
| mmpF | |||
| 27 single ORFs | mupC-X and macpA-E |
| Gene | Role in Mupirocin Production | Reference |
|---|---|---|
| mupI | Essential for generating of N-(3-oxodecanoyl) homoserine lactone (3-O-C10-HSL) | [53] |
| mupR | 3-O-C10-HSL binds to MupR, thereby activating the promoter |
| Brand Name | The Main Active Ingredient | Additional Ingredients | Target Organisms |
|---|---|---|---|
| SAPHIRE | Fludioxonil, ND | - | Microdochium spp., Fusarium, Septoria |
| BERET | Fludioxonil, 25 g/L | - | Microdochium spp., Fusarium, Septoria |
| CELEST | Fludioxonil, 25 g/L | Thiamethoxam, 262.5 g/L, difenoconazole, 25 g/L | Microdochium nivale, Fusarium, Septoria |
| MAXIM | Fludioxonil, 25 g/L | Mefenoxam, 10 g/L | Rhizoctonia solani, Fusarium, Helminthosporium |
| GALBAS | Fenpiclonil, ND | - | Rhizoctonia solani, Helminthosporium solani, Fusarium solani f. sp. coeruleum, Fusarium sulphureum |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Baukova, A.; Bogun, A.; Sushkova, S.; Minkina, T.; Mandzhieva, S.; Alliluev, I.; Jatav, H.S.; Kalinitchenko, V.; Rajput, V.D.; Delegan, Y. New Insights into Pseudomonas spp.-Produced Antibiotics: Genetic Regulation of Biosynthesis and Implementation in Biotechnology. Antibiotics 2024, 13, 597. https://doi.org/10.3390/antibiotics13070597
Baukova A, Bogun A, Sushkova S, Minkina T, Mandzhieva S, Alliluev I, Jatav HS, Kalinitchenko V, Rajput VD, Delegan Y. New Insights into Pseudomonas spp.-Produced Antibiotics: Genetic Regulation of Biosynthesis and Implementation in Biotechnology. Antibiotics. 2024; 13(7):597. https://doi.org/10.3390/antibiotics13070597
Chicago/Turabian StyleBaukova, Alexandra, Alexander Bogun, Svetlana Sushkova, Tatiana Minkina, Saglara Mandzhieva, Ilya Alliluev, Hanuman Singh Jatav, Valery Kalinitchenko, Vishnu D. Rajput, and Yanina Delegan. 2024. "New Insights into Pseudomonas spp.-Produced Antibiotics: Genetic Regulation of Biosynthesis and Implementation in Biotechnology" Antibiotics 13, no. 7: 597. https://doi.org/10.3390/antibiotics13070597
APA StyleBaukova, A., Bogun, A., Sushkova, S., Minkina, T., Mandzhieva, S., Alliluev, I., Jatav, H. S., Kalinitchenko, V., Rajput, V. D., & Delegan, Y. (2024). New Insights into Pseudomonas spp.-Produced Antibiotics: Genetic Regulation of Biosynthesis and Implementation in Biotechnology. Antibiotics, 13(7), 597. https://doi.org/10.3390/antibiotics13070597












