Prevalence of Infections and Antimicrobial Resistance of ESKAPE Group Bacteria Isolated from Patients Admitted to the Intensive Care Unit of a County Emergency Hospital in Romania
Abstract
1. Introduction
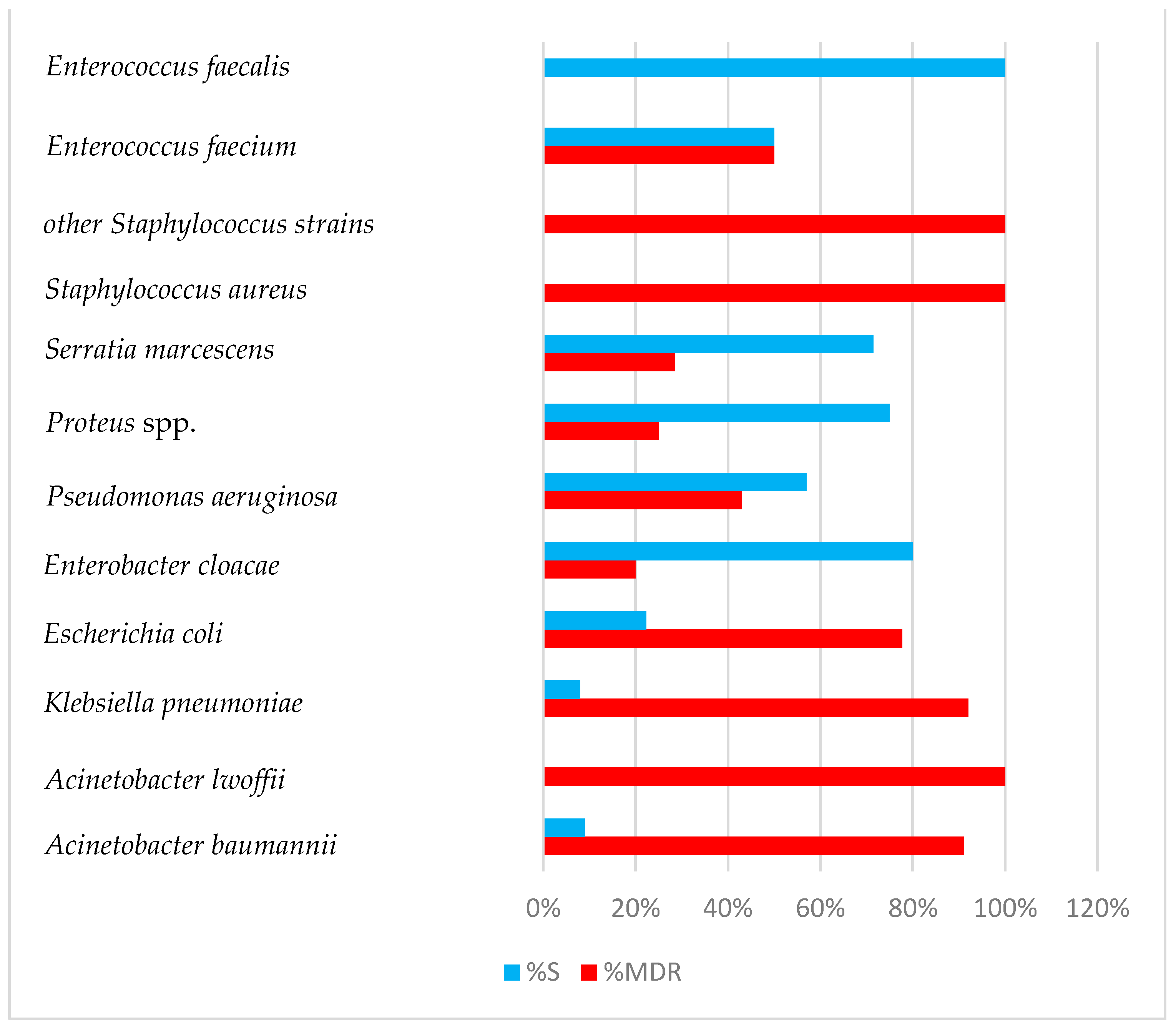
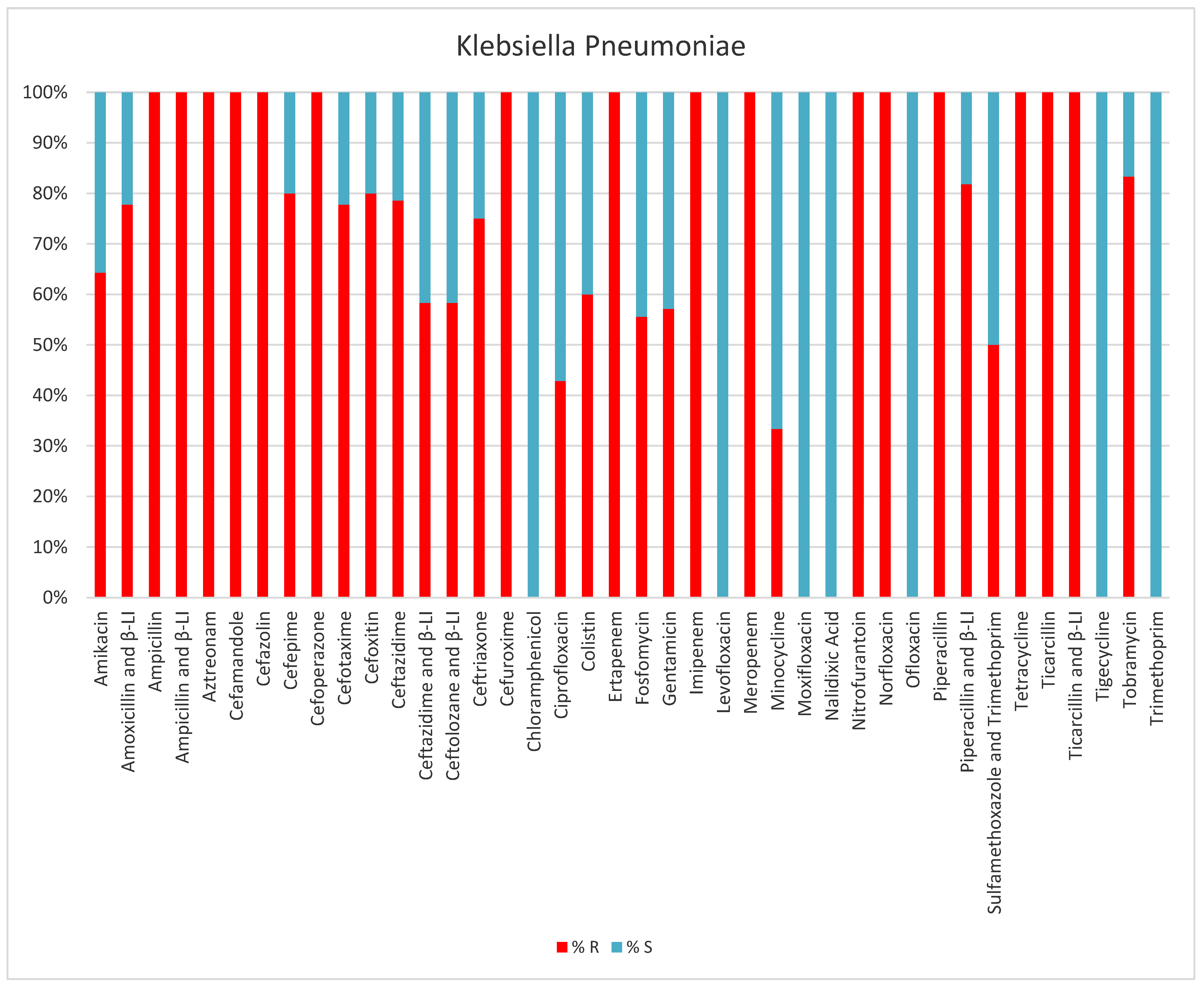
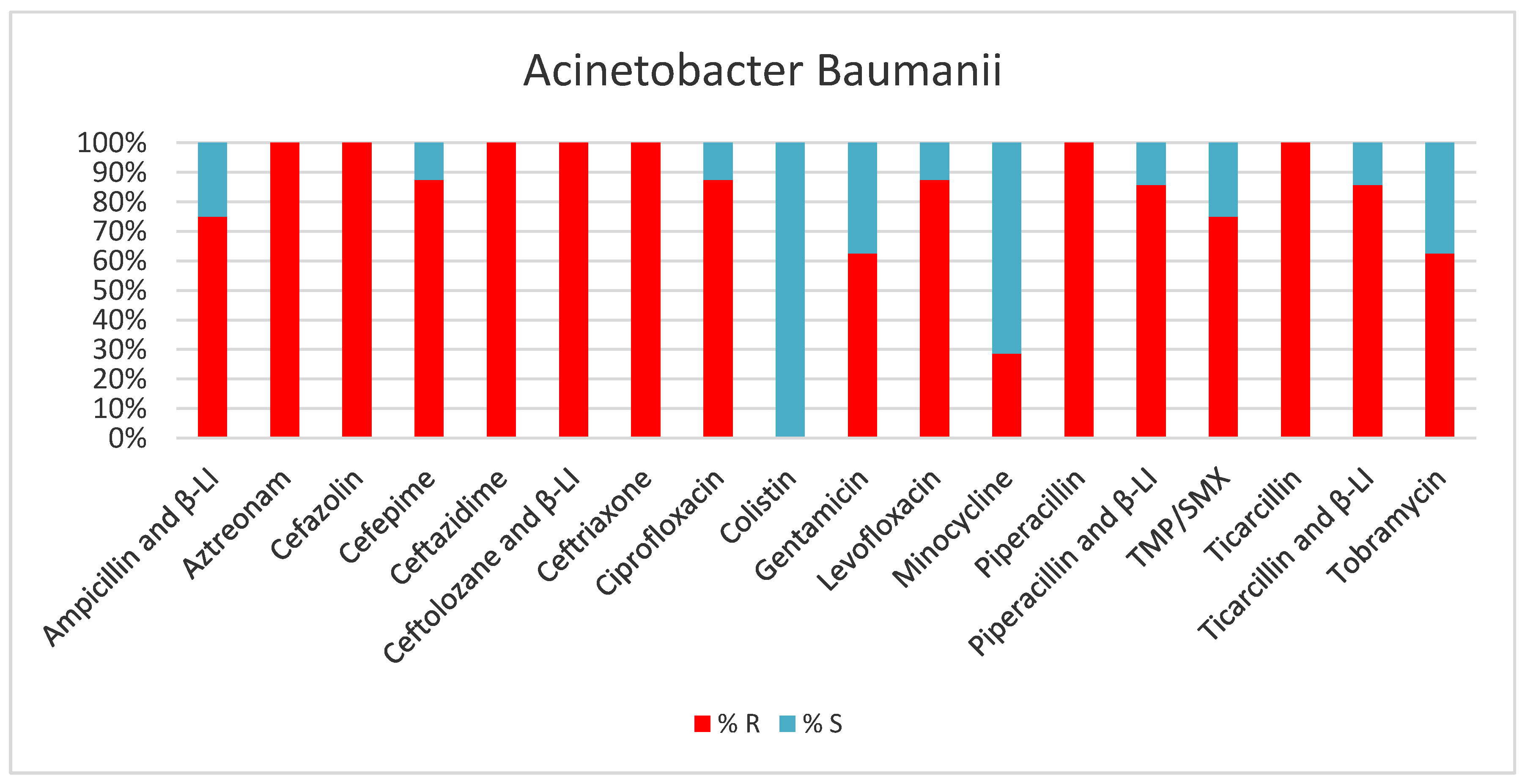
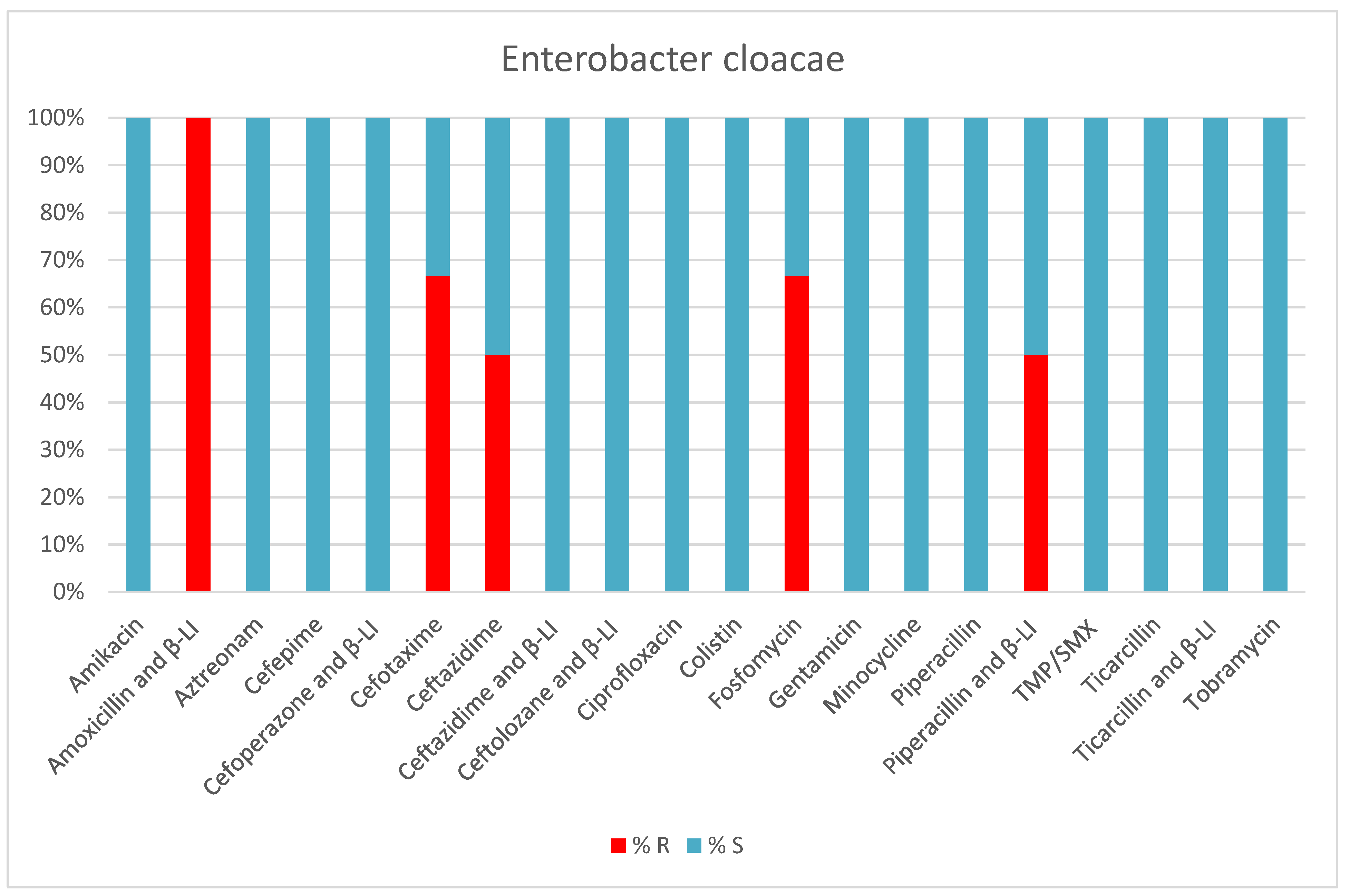
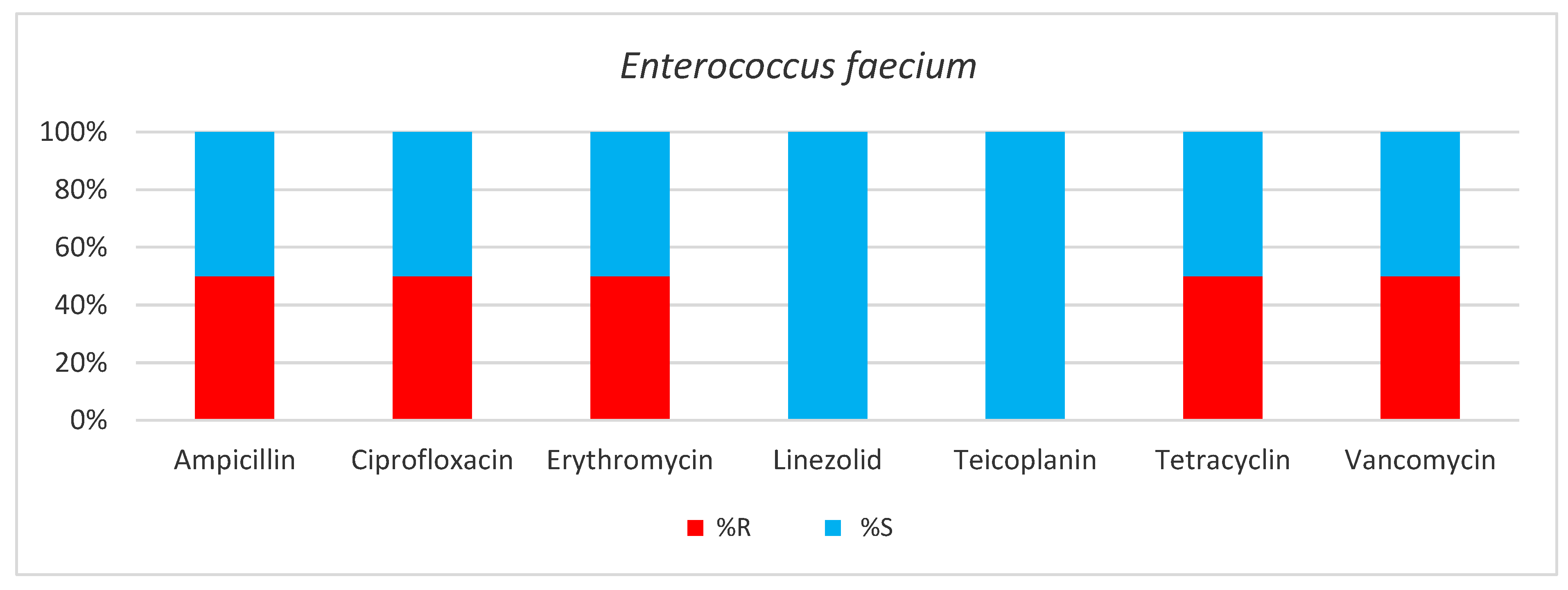
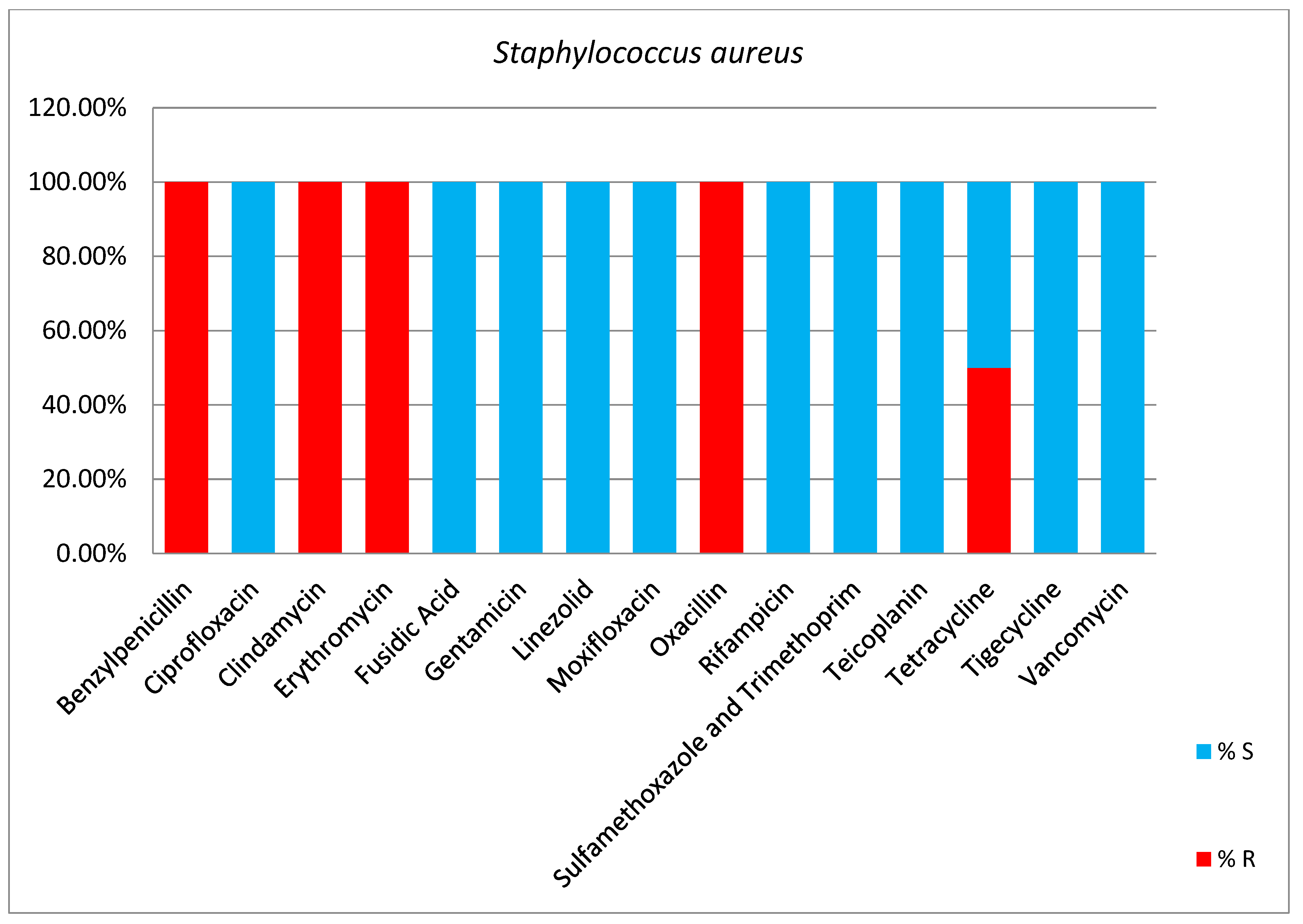
2. Results
3. Discussion
4. Materials and Methods
4.1. Sampling and Bacterial Isolation
4.2. Antimicrobial Susceptibility Testing
- -
- The Kirby-Bauer disk diffusion susceptibility test;
- -
- The dilution method—allows the precise establishment of the minimum inhibitory concentration (MIC).
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AES | Advanced Expert System |
| β-LI | beta-lactamase inhibitor |
| AMR | Antimicrobial resistance |
| CLSI | Institute for Clinical and Laboratory Standards |
| CVC | Central venous catheter |
| CRKP | Carbapenem-resistant Klebsiella pneumoniae |
| DSB | Dry surface biofilms |
| EPS | Extracellular polymeric substances |
| ESLB | Extended spectrum beta-lactamase |
| GLASS | Global Antimicrobial Resistance and Use Surveillance System |
| ICU | Intensive care unit |
| KPC-KP | Carbapenemase-producing Klebsiella pneumoniae |
| MDR | Multidrug-resistant |
| MIC | Minimum inhibitory concentration |
| MIU | Motility indole urea |
| MRSA | Methicillin-resistant Staphylococcus aureus |
| PDR | Pandrug-resistant |
| PVC | Polyvinil chloride |
| R | Resistance |
| S | Sensitivity |
| TMP/SMX | Trimethoprim and Sulfamethoxazole |
| TSI | Triple sugar iron |
| VAP | Ventilator-associated pneumonia |
| VISA | Vancomycin-intermediate Staphylococcus aureus |
| VRE | Vancomycin-resistant Enterococcus faecium |
| VRSA | Vancomycin-resistant Staphylococcus aureus |
| WHO | World Health Organization |
| XDR | Extensively drug-resistant |
References
- Ma, J.; Song, X.; Li, M.; Yu, Z.; Cheng, W.; Yu, Z.; Zhang, W.; Zhang, Y.; Shen, A.; Sun, H.; et al. Global spread of carbapenem-resistant Enterobacteriaceae: Epidemiological features, resistance mechanisms, detection and therapy. Microbiol. Res. 2023, 266, 127249. [Google Scholar] [CrossRef] [PubMed]
- Ranjbar, R.; Alam, M.; Antimicrobial Resistance Collaborators (2022). Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Evid. Based Nurs. 2023, 27, 16. [Google Scholar] [CrossRef] [PubMed]
- Santajit, S.; Indrawattana, N. Mechanisms of Antimicrobial Resistance in ESKAPE Pathogens. BioMed. Res. Int. 2016, 2475067. [Google Scholar] [CrossRef] [PubMed]
- Mahmood, H.Y.; Jamshidi, S.; Sutton, J.M.; Rahman, K.M. Current Advances in Developing Inhibitors of Bacterial Multidrug Efflux Pumps. Curr. Med. Chem. 2016, 23, 1062–1081. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Jean, S.S.; Lee, W.S.; Lam, C.; Hsu, C.W.; Chen, R.J.; Hsueh, P.R. Carbapenemase-producing Gram-negative bacteria: Current epidemics, antimicrobial susceptibility and treatment options. Future Microbiol. 2015, 10, 407–425, Erratum in Future Microbiol. 2017, 12, 456. [Google Scholar] [CrossRef] [PubMed]
- Halat, D.H.; Moubareck, C.A. The Current Burden of Carbapenemases: Review of Significant Properties and Dissemination among Gram-Negative Bacteria. Antibiotics 2020, 9, 186. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Aurilio, C.; Sansone, P.; Barbarisi, M.; Pota, V.; Giaccari, L.G.; Coppolino, F.; Barbarisi, A.; Passavanti, M.B.; Pace, M.C. Mechanisms of Action of Carbapenem Resistance. Antibiotics 2022, 11, 421. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Acman, M.; Wang, R.; van Dorp, L.; Shaw, L.P.; Wang, Q.; Luhmann, N.; Yin, Y.; Sun, S.; Chen, H.; Wang, H.; et al. Role of mobile genetic elements in the global dissemination of the carbapenem resistance gene blaNDM. Nat. Commun. 2022, 13, 1131. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Mukherjee, S.; Bhadury, P.; Mitra, S.; Naha, S.; Saha, B.; Dutta, S.; Basu, S. Hypervirulent Klebsiella pneumoniae Causing Neonatal Bloodstream Infections: Emergence of NDM-1-Producing Hypervirulent ST11-K2 and ST15-K54 Strains Possessing pLVPK-Associated Markers. Microbiol. Spectr. 2023, 11, e0412122. [Google Scholar] [CrossRef] [PubMed]
- Suhay-Garcia, B.; Perez-Garcia, M.T. Present and future of carbapenem-resistant Enterobacteriaceae (CRE) infections. Antibiotics 2019, 8, 122. [Google Scholar] [CrossRef] [PubMed]
- Bassetti, M.; Peghin, M. How to manage KPC infections. Ther. Adv. Infect. Dis. 2020, 7, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Surveillance of Antimicrobial Resistance in Europe 2017. Available online: https://www.ecdc.europa.eu/en/publications-data/surveillance-antimicrobial-resistance-europe-2017 (accessed on 2 March 2024).
- Carbapenem- and/or Colistin-Resistant Klebsiella pneumoniae in Greece: Molecular Follow-up Survey 2022. Available online: https://www.ecdc.europa.eu/en/publications-data/carbapenem-andor-colistin-resistant-klebsiella-pneumoniae-greece-molecular-follow (accessed on 4 March 2024).
- Kyriakidis, I.; Vasileiou, E.; Pana, Z.D.; Tragiannidis, A. Acinetobacter baumannii Antibiotic Resistance Mechanisms. Pathogens 2021, 10, 373. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Verdial, C.; Serrano, I.; Tavares, L.; Gil, S.; Oliveira, M. Mechanisms of Antibiotic and Biocide Resistance That Contribute to Pseudomonas aeruginosa Persistence in the Hospital Environment. Biomedicines 2023, 11, 1221. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Karaman, R.; Jubeh, B.; Breijyeh, Z. Resistance of Gram-Positive Bacteria to Current Antibacterial Agents and Overcoming Approaches. Molecules 2020, 25, 2888. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Kakoullis, L.; Papachristodoulou, E.; Chra, P.; Panos, G. Mechanisms of Antibiotic Resistance in Important Gram-Positive and Gram-Negative Pathogens and Novel Antibiotic Solutions. Antibiotics 2021, 10, 415. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Tornimbene, B.; Eremin, S.; Abednego, R.; Abualas, E.O.; Boutiba, I.; Egwuenu, A.; Fuller, W.; Gahimbare, L.; Githii, S.; Kasambara, W.; et al. Global Antimicrobial Resistance and Use Surveillance System on the African continent: Early implementation 2017–2019. Afr. J. Lab. Med. 2022, 11, 1594. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Tsioutis, C.; Eichel, V.M.; Mutters, N.T. Transmission of Klebsiella pneumoniae carbapenemase (KPC)-producing Klebsiella pneumoniae: The role of infection control. J. Antimicrob. Chemother. 2021, 76, i4–i11. [Google Scholar] [CrossRef] [PubMed]
- Magiorakos, A.P.; Burns, K.; Baño, J.R.; Borg, M.; Daikos, G.; Dumpis, U.; Lucet, J.C.; Moro, M.L.; Tacconelli, E.; Simonsen, G.S.; et al. Infection prevention and control measures and tools for the prevention of entry of carbapenem-resistant Enterobacteriaceae into healthcare settings: Guidance from the European Centre for Disease Prevention and Control. Antimicrob. Resist. Infect. Control 2017, 6, 113. [Google Scholar] [CrossRef] [PubMed]
- Centers for Disease Control and Prevention. Facility Guidance for Control of Carbapenem-resistant Enterobacteriaceae (CRE)—November 2015 Update-CRE Toolkit. Available online: https://www.cdc.gov/hai/pdfs/cre/CRE-guidance-508.pdf (accessed on 3 February 2024).
- World Health Organization. Guidelines for the Prevention and Control of Carbapenem-Resistant Enterobacteriaceae, Acinetobacter baumannii and Pseudomonas aeruginosa in Health Care Facilities. 2017. Available online: https://www.who.int/infection-prevention/publications/guidelines-cre/en/ (accessed on 6 February 2024).
- Bassetti, M.; Giacobbe, D.; Giamarellou, H.; Viscoli, C.; Daikos, G.; Dimopoulos, G.; De Rosa, F.; Giamarellos-Bourboulis, E.; Rossolini, G.; Righi, E.; et al. Management of KPC-producing Klebsiella pneumoniae infections. Clin. Microbiol. Infect. 2018, 24, 133–144. [Google Scholar] [CrossRef]
- Vintila, B.I.; Arseniu, A.M.; Morgovan, C.; Butuca, A.; Bîrluțiu, V.; Dobrea, C.M.; Rus, L.L.; Ghibu, S.; Bereanu, A.S.; Arseniu, R.; et al. A Real-World Study on the Clinical Characteristics, Outcomes, and Relationship between Antibiotic Exposure and Clostridioides difficile Infection. Antibiotics 2024, 13, 144. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Reyes, J.; Aguilar, A.C.; Caicedo, A. Carbapenem-Resistant Klebsiella pneumoniae: Microbiology Key Points for Clinical Practice. Int. J. Gen. Med. 2019, 12, 437–446. [Google Scholar] [CrossRef] [PubMed]
- Brunke, M.S.; Konrat, K.; Schaudinn, C.; Piening, B.; Pfeifer, Y.; Becker, L.; Schwebke, I.; Arvand, M. Tolerance of biofilm of a carbapenem-resistant Klebsiella pneumoniae involved in a duodenoscopy-associated outbreak to the disinfectant used in reprocessing. Antimicrob. Resist. Infect. Control 2022, 11, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Dan, B.; Dai, H.; Zhou, D.; Tong, H.; Zhu, M. Relationship between Drug Resistance Characteristics and Biofilm Formation in Klebsiella pneumoniae Strains. Infect. Drug Resist. 2023, ume 16, 985–998. [Google Scholar] [CrossRef]
- Folliero, V.; Franci, G.; Dell’annunziata, F.; Giugliano, R.; Foglia, F.; Sperlongano, R.; De Filippis, A.; Finamore, E.; Galdiero, M. Evaluation of Antibiotic Resistance and Biofilm Production among Clinical Strain Isolated from Medical Devices. Int. J. Microbiol. 2021, 2021, 1–11. [Google Scholar] [CrossRef]
- Flemming, H.-C.; Wingender, J.; Szewzyk, U.; Steinberg, P.; Rice, S.A.; Kjelleberg, S. Biofilms: An emergent form of bacterial life. Nat. Rev. Microbiol. 2016, 14, 563–575. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, D.; Shivapriya, P.M.; Gautam, P.K.; Misra, K.; Sahoo, A.K.; Samanta, S.K. A Review on Basic Biology of Bacterial Biofilm Infections and Their Treatments by Nanotechnology-Based Approaches. Proc. Natl. Acad. Sci. India Sect. B: Biol. Sci. 2019, 90, 243–259. [Google Scholar] [CrossRef]
- Ramasamy, M.; Lee, J. Recent Nanotechnology Approaches for Prevention and Treatment of Biofilm-Associated Infections on Medical Devices. BioMed. Res. Int. 2016, 1851242. [Google Scholar] [CrossRef] [PubMed]
- Maiti, P.; Chatterjee, S.; Kundu, A.; Dey, R. Biofilms on indwelling urologic devices: Microbes and antimicrobial management prospect. Ann. Med. Health Sci. Res. 2014, 4, 100–104. [Google Scholar] [CrossRef] [PubMed]
- Codru, I.R.; Sava, M.; Vintilă, B.I.; Bereanu, A.S.; Bîrluțiu, V. A Study on the Contributions of Sonication to the Identification of Bacteria Associated with Intubation Cannula Biofilm and the Risk of Ventilator-Associated Pneumonia. Medicina 2023, 59, 1058. [Google Scholar] [CrossRef]
- Yin, W.; Wang, Y.; Liu, L.; He, J. Biofilms: The Microbial “Protective Clothing” in Extreme Environments. Int. J. Mol. Sci. 2019, 20, 3423. [Google Scholar] [CrossRef]
- Chung, P.Y. The emerging problems of Klebsiella pneumoniae infections: Carbapenem resistance and biofilm formation. FEMS Microbiol. Lett. 2016, 363, fnw219. [Google Scholar] [CrossRef] [PubMed]
- Rahdar, H.A.; Malekabad, E.S.; Dadashi, A.-R.; Takei, E.; Keikha, M.; Kazemian, H.; Karami-Zarandi, M. Correlation between biofilm formation and carbapenem resistance among clinical isolates of Klebsiella pneumoniae. Ethiop. J. Health Sci. 1970, 29, 745–750. [Google Scholar] [CrossRef] [PubMed]
- Thoendel, M.; Kavanaugh, J.S.; Flack, C.E.; Horswill, A.R. Peptide signaling in the staphylococci. Chem. Rev. 2011, 111, 117–151. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Rasamiravaka, T.; Labtani, Q.; Duez, P.; El Jaziri, M. The formation of biofilms by Pseudomonas aeruginosa: A review of the natural and synthetic compounds interfering with control mechanisms. Biomed. Res. Int. 2015, 2015, 759348. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Jamal, M.; Ahmad, W.; Andleeb, S.; Jalil, F.; Imran, M.; Nawaz, M.A.; Hussain, T.; Ali, M.; Rafiq, M.; Kamil, M.A. Bacterial biofilm and associated infections. J. Chin. Med. Assoc. 2018, 81, 7–11. [Google Scholar] [CrossRef] [PubMed]
- Schulze, A.; Mitterer, F.; Pombo, J.P.; Schild, S. Biofilms by bacterial human pathogens: Clinical relevance—Development, composition and regulation—Therapeutical strategies. Microb. Cell. 2021, 8, 28–56. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Tambyah, P.A. Catheter-associated urinary tract infections: Diagnosis and prophylaxis. Int. J. Antimicrob. Agents 2004, 24 (Suppl. S1), S44–S48. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.K.; Schaefer, A.L.; Parsek, M.R.; Moninger, T.O.; Welsh, M.J.; Greenberg, E.P. Quorum-sensing signals indicate that cystic fibrosis lungs are infected with bacterial biofilms. Nature 2000, 407, 762–764. [Google Scholar] [CrossRef] [PubMed]
- Jones, R.N. Microbial etiologies of hospital-acquired bacterial pneumonia and ventilator-associated bacterial pneumonia. Clin. Infect. Dis. 2010, 51 (Suppl. S1), S81–S87. [Google Scholar] [CrossRef] [PubMed]
- American Thoracic Society Documents. Guidelines for the management of adults with hospital-acquired, ventilator-associated, and healthcare-associated pneumonia. Am. J. Respir. Crit. Care Med. 2005, 171, 388–416. [Google Scholar] [CrossRef] [PubMed]
- Melsen, W.G.; Rovers, M.; Koeman, M.; Bonten, M.J.M. Estimating the attributable mortality of ventilator-associated pneumonia from randomized prevention studies. Crit. Care Med. 2011, 39, 2736–2742. [Google Scholar] [CrossRef] [PubMed]
- Mohammadi, M.; Saffari, M.; Siadat, S.D.; Hejazi, S.H.; Shayestehpour, M.; Motallebi, M.; Eidi, M. Isolation, characterization, therapeutic potency, and genomic analysis of a novel bacteriophage vB_KshKPC-M against carbapenemase-producing Klebsiella pneumoniae strains (CRKP) isolated from Ventilator-associated pneumoniae (VAP) infection of COVID-19 patients. Ann. Clin. Microbiol. Antimicrob. 2023, 22, 1–25. [Google Scholar] [CrossRef]
- Bassetti, M.; Carnelutti, A.; Peghin, M. Patient specific risk stratification for antimicrobial resistance and possible treatment strategies in gram-negative bacterial infections. Expert Rev. Anti-Infect. Ther. 2017, 15, 55–65. [Google Scholar] [CrossRef]
- Vasoo, S.; Barreto, J.N.; Tosh, P.K. Emerging issues in gram-negative bacterial resistance: An update for the practicing clinician. Mayo Clin. Proc. 2015, 90, 395–403. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.-R.; Lee, J.H.; Park, K.S.; Kim, Y.B.; Jeong, B.C.; Lee, S.H. Global Dissemination of Carbapenemase-Producing Klebsiella pneumoniae: Epidemiology, Genetic Context, Treatment Options, and Detection Methods. Front. Microbiol. 2016, 7, 895. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Chen, M.; Ou, Q.; Zheng, L.; Chen, X.; Mao, G.; Fang, J.; Jin, D.; Tang, X. Carbapenem-resistant hypermucoviscous Klebsiella pneumoniae clinical isolates from a tertiary hospital in China: Antimicrobial susceptibility, resistance phenotype, epidemiological characteristics, microbial virulence, and risk factors. Front. Cell. Infect. Microbiol. 2022, 12, 1083009. [Google Scholar] [CrossRef] [PubMed]
- Bereanu, A.-S.; Vintilă, B.I.; Bereanu, R.; Codru, I.R.; Hașegan, A.; Olteanu, C.; Săceleanu, V.; Sava, M. TiO2 Nanocomposite Coatings and Inactivation of Carbapenemase-Producing Klebsiella Pneumoniae Biofilm—Opportunities and Challenges. Microorganisms 2024, 12, 684. [Google Scholar] [CrossRef]
- Hasegan, A.; Totan, M.; Antonescu, E.; Bumbu, A.G.; Pantis, C.; Furau, C.; (Urducea), C.B.; Grigore, N. Prevalence of Urinary Tract Infections in Children and Changes in Sensitivity to Antibiotics of E. coli Strains. Rev. de Chim. 2019, 70, 3788–3792. [Google Scholar] [CrossRef]
- Pourmehdiabadi, A.; Nobakht, M.S.; Balajorshari, B.H.; Yazdi, M.R.; Amini, K. Investigating the effects of zinc oxide and titanium dioxide nanoparticles on the formation of biofilm and persister cells in Klebsiella pneumoniae. J. Basic Microbiol. 2023. [Google Scholar] [CrossRef]
- Araújo, B.F.; Ferreira, M.L.; de Campos, P.A.; Royer, S.; Gonçalves, I.R.; Batistão, D.W.d.F.; Fernandes, M.R.; Cerdeira, L.T.; de Brito, C.S.; Lincopan, N.; et al. Hypervirulence and biofilm production in KPC-2-producing Klebsiella pneumoniae CG258 isolated in Brazil. J. Med. Microbiol. 2018, 67, 523–528. [Google Scholar] [CrossRef]
- Stallbaum, L.R.; Pruski, B.B.; Amaral, S.C.; de Freitas, S.B.; Wozeak, D.R.; Hartwig, D.D. Phenotypic and molecular evaluation of biofilm formation in Klebsiella pneumoniae carbapenemase (KPC) isolates obtained from a hospital of Pelotas, RS, Brazil. J. Med. Microbiol. 2021, 70, 001451. [Google Scholar] [CrossRef]
- Santiago, A.J.; Burgos-Garay, M.L.; Kartforosh, L.; Mazher, M.; Donlan, R.M. Bacteriophage treatment of carbapenemase-producing Klebsiella pneumoniae in a multispecies biofilm: A potential biocontrol strategy for healthcare facilities. AIMS Microbiol. 2020, 6, 43–63. [Google Scholar] [CrossRef] [PubMed]
- Yazgan, B.; Türkel, I.; Güçkan, R.; Kılınç, K.; Yıldırım, T. Comparison of biofilm formation and efflux pumps in ESBL and carbapenemase producing Klebsiella pneumoniae. J. Infect. Dev. Ctries 2018, 12, 156–163. [Google Scholar] [CrossRef] [PubMed]
- Booq, R.Y.; Abutarboush, M.H.; Alolayan, M.A.; Huraysi, A.A.; Alotaibi, A.N.; Alturki, M.I.; Alshammari, M.K.; Bakr, A.A.; Alquait, A.A.; Tawfik, E.A.; et al. Identification and Characterization of Plasmids and Genes from Carbapenemase-Producing Klebsiella pneumoniae in Makkah Province, Saudi Arabia. Antibiotics 2022, 11, 1627. [Google Scholar] [CrossRef]
- D’apolito, D.; Arena, F.; Conte, V.; De Angelis, L.H.; Di Mento, G.; Carreca, A.P.; Cuscino, N.; Russelli, G.; Iannolo, G.; Barbera, F.; et al. Phenotypical and molecular assessment of the virulence potential of KPC-3-producing Klebsiella pneumoniae ST392 clinical isolates. Microbiol. Res. 2020, 240, 126551. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Liu, P.-P.; Wang, L.-H.; Wei, D.-D.; Wan, L.-G.; Zhang, W. Capsular Polysaccharide Types and Virulence-Related Traits of Epidemic KPC-Producing Klebsiella pneumoniae Isolates in a Chinese University Hospital. Microb. Drug Resist. 2017, 23, 901–907. [Google Scholar] [CrossRef]
- Mahmud, Z.H.; Uddin, S.Z.; Moniruzzaman, M.; Ali, S.; Hossain, M.; Islam, T.; Costa, D.T.D.; Islam, M.R.; Islam, S.; Hassan, Z.; et al. Healthcare Facilities as Potential Reservoirs of Antimicrobial Resistant Klebsiella pneumoniae: An Emerging Concern to Public Health in Bangladesh. Pharmaceuticals 2022, 15, 1116. [Google Scholar] [CrossRef] [PubMed]
- Silva, N.B.S.; Alves, P.G.V.; Marques, L.d.A.; Silva, S.F.; Faria, G.d.O.; de Araújo, L.B.; Pedroso, R.d.S.; Penatti, M.P.A.; Menezes, R.d.P.; Röder, D.v.D.d.B. Quantification of biofilm produced by clinical, environment and hands’ isolates Klebsiella species using colorimetric and classical methods. J. Microbiol. Methods 2021, 185, 106231. [Google Scholar] [CrossRef]
- Papalini, C.; Sabbatini, S.; Monari, C.; Mencacci, A.; Francisci, D.; Perito, S.; Pasticci, M.B. In vitro antibacterial activity of ceftazidime/avibactam in combination against planktonic and biofilm carbapenemase-producing Klebsiella pneumoniae isolated from blood. J. Glob. Antimicrob. Resist. 2020, 23, 4–8. [Google Scholar] [CrossRef] [PubMed]
- Kerbauy, G.; Vivan, A.C.; Simões, G.C.; Simionato, A.S.; Pelisson, M.; Vespero, E.C.; Costa, S.F.; Andrade, C.G.d.J.; Barbieri, D.M.; Mello, J.C.; et al. Effect of a Metalloantibiotic Produced by Pseudomonas aeruginosa on Klebsiella pneumoniae Carbapenemase (KPC)-producing K. pneumoniae. Curr. Pharm. Biotechnol. 2016, 17, 389–397. [Google Scholar] [CrossRef]
- Perez-Palacios, P.; Gual-De-Torrella, A.; Delgado-Valverde, M.; Oteo-Iglesias, J.; Hidalgo-Díaz, C.; Pascual, Á.; Fernández-Cuenca, F. Transfer of plasmids harbouring blaOXA-48-like carbapenemase genes in biofilm-growing Klebsiella pneumoniae: Effect of biocide exposure. Microbiol. Res. 2021, 254, 126894. [Google Scholar] [CrossRef] [PubMed]
- Dey, D.; Ghosh, S.; Ray, R.; Hazra, B. Polyphenolic Secondary Metabolites Synergize the Activity of Commercial Antibiotics against Clinical Isolates of β-Lactamase-producing Klebsiella pneumoniae. Phytotherapy Res. 2015, 30, 272–282. [Google Scholar] [CrossRef] [PubMed]
- Bai, J.; Liu, Y.; Kang, J.; Song, Y.; Yin, D.; Wang, S.; Guo, Q.; Wang, J.; Duan, J. Antibiotic resistance and virulence characteristics of four carbapenem-resistant Klebsiella pneumoniae strains coharbouring blaKPC and blaNDM based on whole genome sequences from a tertiary general teaching hospital in central China between 2019 and 2021. Microb. Pathog. 2023, 175, 105969. [Google Scholar] [CrossRef] [PubMed]
- Larcher, R.; Laffont-Lozes, P.; Naciri, T.; Bourgeois, P.-M.; Gandon, C.; Magnan, C.; Pantel, A.; Sotto, A. Continuous infusion of meropenem–vaborbactam for a KPC-3-producing Klebsiella pneumoniae bloodstream infection in a critically ill patient with augmented renal clearance. Infection 2023, 51, 1835–1840. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Zhang, H.; Xu, M.; Liu, Y.; Yuan, B.; Lin, Y.; Shen, F. Within-Host Resistance and Virulence Evolution of a Hypervirulent Carbapenem-Resistant Klebsiella pneumoniae ST11 Under Antibiotic Pressure. Infect. Drug Resist. 2023, ume 16, 7255–7270. [Google Scholar] [CrossRef]
- Thorarinsdottir, H.R.; Kander, T.; Holmberg, A.; Petronis, S.; Klarin, B. Biofilm formation on three different endotracheal tubes: A prospective clinical trial. Crit. Care 2020, 24, 1–12. [Google Scholar] [CrossRef]
- Di Martino, P.; Cafferini, N.; Joly, B.; Darfeuille-Michaud, A. Klebsiella pneumoniae type 3 pili facilitate adherence and biofilm formation on abiotic surfaces. Res. Microbiol. 2003, 154, 9–16. [Google Scholar] [CrossRef] [PubMed]
- MladenoviĆ, K.G.; GrujoviĆ, M.Ž.; NikodijeviĆ, D.D.; ČomiĆ, L.R. The hydrophobicity of enterobacteria and their co-aggregation with Enterococcus faecalis isolated from Serbian cheese. Biosci. Microbiota Food Health 2020, 39, 227–233. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Krasowska, A.; Sigler, K. How microorganisms use hydrophobicity and what does this mean for human needs? Front. Cell. Infect. Microbiol. 2014, 4, 112. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Pintavirooj, C.; Vongmanee, N.; Sukjee, W.; Sangma, C.; Visitsattapongse, S. Biosensors for Klebsiella pneumoniae with Molecularly Imprinted Polymer (MIP) Technique. Sensors 2022, 22, 4638. [Google Scholar] [CrossRef]
- Sabença, C.; Costa, E.; Sousa, S.; Barros, L.; Oliveira, A.; Ramos, S.; Igrejas, G.; Torres, C.; Poeta, P. Evaluation of the Ability to Form Biofilms in KPC-Producing and ESBL-Producing Klebsiella pneumoniae Isolated from Clinical Samples. Antibiotics 2023, 12, 1143. [Google Scholar] [CrossRef] [PubMed]
- Ochońska, D.; Ścibik, L.; Brzychczy-Włoch, M. Biofilm Formation of Clinical Klebsiella pneumoniae Strains Isolated from Tracheostomy Tubes and Their Association with Antimicrobial Resistance, Virulence and Genetic Diversity. Pathogens 2021, 10, 1345. [Google Scholar] [CrossRef]
- Vintila, B.I.; Arseniu, A.M.; Morgovan, C.; Butuca, A.; Sava, M.; Bîrluțiu, V.; Rus, L.L.; Ghibu, S.; Bereanu, A.S.; Codru, I.R.; et al. A Pharmacovigilance Study Regarding the Risk of Antibiotic-Associated Clostridioides difficile Infection Based on Reports from the EudraVigilance Database: Analysis of Some of the Most Used Antibiotics in Intensive Care Units. Pharmaceuticals 2023, 16, 1585. [Google Scholar] [CrossRef] [PubMed]
- Dutt, Y.; Dhiman, R.; Singh, T.; Vibhuti, A.; Gupta, A.; Pandey, R.P.; Raj, V.S.; Chang, C.M.; Priyadarshini, A. The Association between Biofilm Formation and Antimicrobial Resistance with Possible Ingenious Bio-Remedial Approaches. Antibiotics 2022, 11, 930. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Urban-Chmiel, R.; Marek, A.; Stępień-Pyśniak, D.; Wieczorek, K.; Dec, M.; Nowaczek, A.; Osek, J. Antibiotic Resistance in Bacteria-A Review. Antibiotics 2022, 11, 1079. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Pontes, J.T.C.; Toledo Borges, A.B.; Roque-Borda, C.A.; Pavan, F.R. Antimicrobial Peptides as an Alternative for the Eradication of Bacterial Biofilms of Multi-Drug Resistant Bacteria. Pharmaceutics 2022, 14, 642. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Vickery, K.; Deva, A.; Jacombs, A.; Allan, J.; Valente, P.; Gosbell, I. Presence of biofilm containing viable multiresistant organisms despite terminal cleaning on clinical surfaces in an intensive care unit. J. Hosp. Infect. 2012, 80, 52–55. [Google Scholar] [CrossRef] [PubMed]
- Ledwoch, K.; Dancer, S.; Otter, J.; Kerr, K.; Roposte, D.; Rushton, L.; Weiser, R.; Mahenthiralingam, E.; Muir, D.; Maillard, J.-Y. Beware biofilm! Dry biofilms containing bacterial pathogens on multiple healthcare surfaces; a multi-centre study. J. Hosp. Infect. 2018, 100, e47–e56. [Google Scholar] [CrossRef] [PubMed]
- Costa, D.; Johani, K.; Melo, D.; Lopes, L.; Lima, L.L.; Tipple, A.; Hu, H.; Vickery, K. Biofilm contamination of high-touched surfaces in intensive care units: Epidemiology and potential impacts. Lett. Appl. Microbiol. 2019, 68, 269–276. [Google Scholar] [CrossRef]
- Hu, H.; Johani, K.; Gosbell, I.; Jacombs, A.; Almatroudi, A.; Whiteley, G.; Deva, A.; Jensen, S.; Vickery, K. Intensive care unit environmental surfaces are contaminated by multidrug-resistant bacteria in biofilms: Combined results of conventional culture, pyrosequencing, scanning electron microscopy, and confocal laser microscopy. J. Hosp. Infect. 2015, 91, 35–44. [Google Scholar] [CrossRef]
- Ababneh, Q.; Abulaila, S.; Jaradat, Z. Isolation of extensively drug resistant Acinetobacter baumannii from environmental surfaces inside intensive care units. Am. J. Infect. Control 2021, 50, 159–165. [Google Scholar] [CrossRef] [PubMed]
- Maillard, J.-Y.; Centeleghe, I. How biofilm changes our understanding of cleaning and disinfection. Antimicrob. Resist. Infect. Control 2023, 12, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Centeleghe, I.; Norville, P.; Hughes, L.; Maillard, J.-Y. Klebsiella pneumoniae survives on surfaces as a dry biofilm. Am. J. Infect. Control 2023, 51, 1157–1162. [Google Scholar] [CrossRef] [PubMed]



| Klebsiella pneumoniae | 62 |
|---|---|
| tracheal aspirate | 26 |
| without resistance pattern | 5 |
| Positive-MDR | 4 |
| Positive-ESBL | 3 |
| Positive-PDR | 2 |
| Positive-XDR | 12 |
| Catheter-tip | 5 |
| Positive-PDR | 2 |
| Positive-XDR | 2 |
| Positive -MDR | 1 |
| Pharyngeal exudate | 2 |
| Positive-XDR | 2 |
| Surgical wound, abscess, ulcer, etc. | 8 |
| Positive-PDR | 1 |
| Positive-XDR | 6 |
| Positive MDR | 1 |
| Blood | 11 |
| Positive-ESLB | 2 |
| Positive-MDR | 1 |
| Positive-XDR | 8 |
| Urine | 10 |
| Positive-MDR | 4 |
| Positive-XDR | 6 |
| Acinetobacter baumannii | 33 |
|---|---|
| tracheal aspirate | 18 |
| without resistance pattern | 3 |
| Positive-MDR | 4 |
| Positive-XDR | 11 |
| Pharyngeal exudate | 5 |
| Positive-MDR | 1 |
| Positive-XDR | 4 |
| Peritoneal fluid | 2 |
| Positive-MDR | 1 |
| Positive-XDR | 1 |
| Surgical wound, abscess, ulcer, etc. | 3 |
| Positive-MDR | 1 |
| Positive-XDR | 2 |
| Blood | 2 |
| Positive-XDR | 2 |
| Urine | 3 |
| Positive-XDR | 3 |
| Pseudomonas aeruginosa | 14 |
|---|---|
| Tracheal aspirate | 9 |
| without resistance pattern | 4 |
| Positive-MDR | 5 |
| Pharyngeal exudate | 1 |
| without resistance pattern | 1 |
| Surgical wounds, abscesses, ulcers, etc. | 3 |
| without resistance pattern | 2 |
| Positive-MDR | 1 |
| Urine | 1 |
| without resistance pattern | 1 |
| Escherichia coli | 9 |
|---|---|
| Tracheal aspirate | 3 |
| without resistance pattern | 1 |
| Positive-ESBL | 1 |
| Positive-XDR | 1 |
| Surgical wounds, abscesses, ulcers, etc. | 1 |
| without resistance pattern | 1 |
| Urine | 5 |
| Positive-ESBL | 5 |
| Enterobacter cloacae | 5 |
|---|---|
| Tracheal aspirate | 3 |
| without resistance pattern | 2 |
| Positive-ESBL | 1 |
| Surgical wounds, abscesses, ulcers, etc. | 1 |
| without resistance pattern | 1 |
| Blood | 1 |
| without resistance pattern | 1 |
| Enterococcus faecium | 4 |
|---|---|
| Surgical wounds, abscesses, ulcers, etc. | 3 |
| negative VRE | 1 |
| positive-VRE | 2 |
| Urine | 1 |
| negative VRE | 1 |
| Staphylococcus aureus | 3 |
|---|---|
| Tracheal aspirate Positive-MRSA | 1 |
| Blood Positive-MRSA | 1 |
| Surgical wounds, abscesses, ulcers, etc. Positive-MRSA | 1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bereanu, A.-S.; Bereanu, R.; Mohor, C.; Vintilă, B.I.; Codru, I.R.; Olteanu, C.; Sava, M. Prevalence of Infections and Antimicrobial Resistance of ESKAPE Group Bacteria Isolated from Patients Admitted to the Intensive Care Unit of a County Emergency Hospital in Romania. Antibiotics 2024, 13, 400. https://doi.org/10.3390/antibiotics13050400
Bereanu A-S, Bereanu R, Mohor C, Vintilă BI, Codru IR, Olteanu C, Sava M. Prevalence of Infections and Antimicrobial Resistance of ESKAPE Group Bacteria Isolated from Patients Admitted to the Intensive Care Unit of a County Emergency Hospital in Romania. Antibiotics. 2024; 13(5):400. https://doi.org/10.3390/antibiotics13050400
Chicago/Turabian StyleBereanu, Alina-Simona, Rareș Bereanu, Cosmin Mohor, Bogdan Ioan Vintilă, Ioana Roxana Codru, Ciprian Olteanu, and Mihai Sava. 2024. "Prevalence of Infections and Antimicrobial Resistance of ESKAPE Group Bacteria Isolated from Patients Admitted to the Intensive Care Unit of a County Emergency Hospital in Romania" Antibiotics 13, no. 5: 400. https://doi.org/10.3390/antibiotics13050400
APA StyleBereanu, A.-S., Bereanu, R., Mohor, C., Vintilă, B. I., Codru, I. R., Olteanu, C., & Sava, M. (2024). Prevalence of Infections and Antimicrobial Resistance of ESKAPE Group Bacteria Isolated from Patients Admitted to the Intensive Care Unit of a County Emergency Hospital in Romania. Antibiotics, 13(5), 400. https://doi.org/10.3390/antibiotics13050400









