Automating Predictive Phage Therapy Pharmacology
Abstract
1. Introduction
2. Predictive Phage Therapy Pharmacology
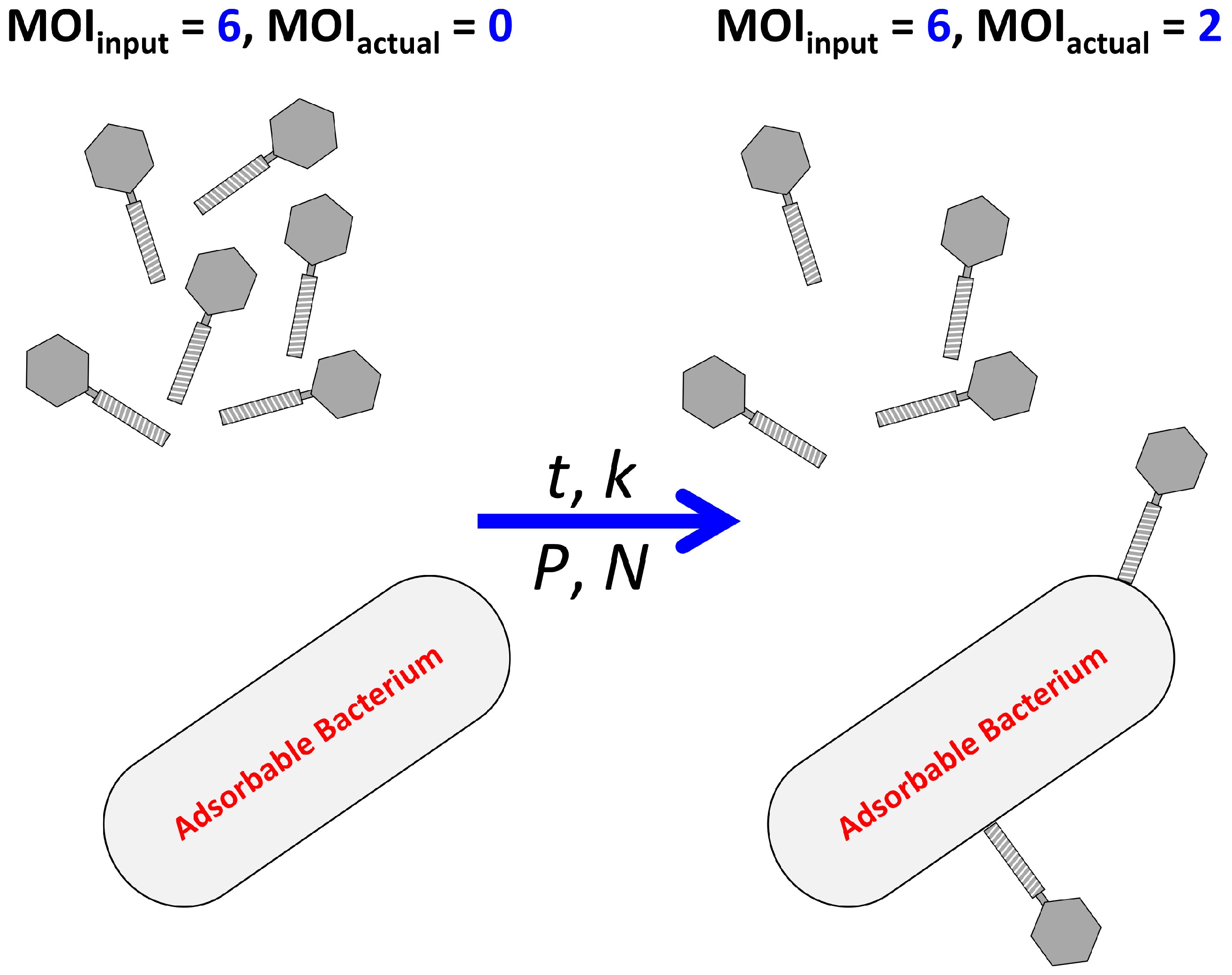
2.1. Multiplicity of Infection
2.1.1. MOIinput vs. MOIactual
2.1.2. Predicting MOIactual
2.1.3. Running the Calculator
2.2. Bacterial Likelihood of Being Phage Adsorbed
2.2.1. Predicting Bacterial Adsorption Likelihood: p(Ac)
2.2.2. Running the Calculator
2.3. Bacterial Reduction Times
2.3.1. Bacterial Half-Lives: t0.5, and Also tMFT
2.3.2. Decimal Reduction Times: t0.1, plus t0.01
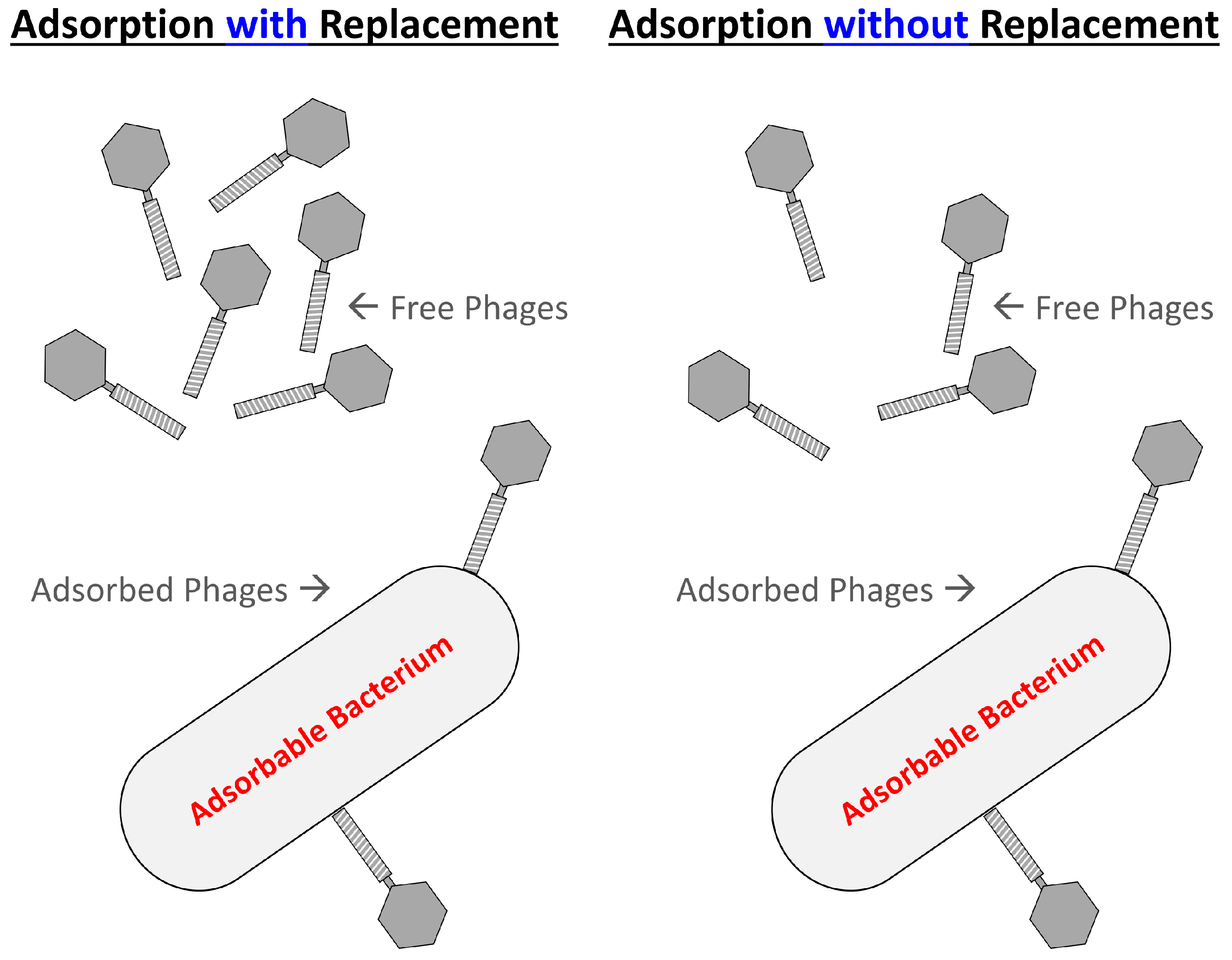
2.3.3. Phage Adsorption without Replacement
2.3.4. Running the Calculators
2.4. Inundative Phage Quantities
2.4.1. Inundative Phage Densities: IPDmin
2.4.2. Inundative Phage Number: IPNmin
2.4.3. Running the Calculator
2.5. Poisson Distributions
2.5.1. Predicting Bacterial Survival
2.5.2. Killing Titers: PK
2.5.3. Running the Calculators
2.6. Active Treatments
- Low bacterial concentrations without clumping and lower starting phage titers. In the case of low bacterial concentrations and no bacterial clumping, phage population growth likely is mostly irrelevant, since in situ phage replication will not be expected to have a substantial impact on more “global” phage titers. That is, bacteria are present in insufficient quantities to produce relatively large concentrations of new phages across environments. Still, these circumstances, given sufficient environmental mixing, are easily modelled mathematically.
- Low bacterial concentrations with clumping and lower starting phage titers. With spatial structure in combination with bacteria being found in clonal clusters—but bacteria nonetheless overall found at low concentrations—phage in situ replication could in fact be relevant, though not globally, and the mathematics portraying such situations is not straightforward. I describe this latter scenario as a locally active treatment [32].
- 3.
- Higher bacterial concentrations without clumping and higher starting phage titers. First is the noted passive treatment in which phage in situ replication is not required to achieve desired levels of bacterial eradication, e.g., as due to the employment of inundative phage concentrations (Section 2.4). This is because sufficient quantities of phages have been supplied via phage dosing alone.
- 4.
- Higher bacterial concentrations without clumping and lower starting phage titers. Second is what I have described as globally active treatment [32]. Here, the assumption is that phage virions are free to diffuse relatively rapidly about environments or otherwise be readily moved about, such as within blood. Therefore, phages produced in one location can give rise to sufficient increases in phage titers, i.e., to inundative densities (Section 2.4.1) throughout a phage-treated environment.
2.6.1. Considering Phage Population Growth
2.6.2. Considering Bacterial Population Growth
2.6.3. Running the Calculator
2.6.4. Additional Approaches to Predicting In Situ Efficacy, from In Vitro Characteristics
3. Discussion
4. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A. Improving the Realism of Phage–Bacteria Chemostat Modeling
References
- Górski, A.; Międzybrodzki, R.; Borysowski, J. Phage Therapy: A Practical Approach; Springer Nature Switzerland AG: Cham, Switzerland, 2019. [Google Scholar]
- Pirnay, J.-P.; Ferry, T.; Resch, G. Recent progress toward the implementation of phage therapy in Western medicine. FEMS Microbiol. Rev. 2022, 46, fuab040. [Google Scholar] [CrossRef] [PubMed]
- Baral, B. Phages against killer superbugs: An enticing strategy against antibiotics-resistant pathogens. Front. Pharmacol. 2023, 14, 1036051. [Google Scholar] [CrossRef]
- Cahan, E. As superbugs flourish, bacteriophage therapy recaptures researchers’ interest. J. Am. Med. Assoc. 2023, 329, 781–784. [Google Scholar] [CrossRef] [PubMed]
- Diallo, K.; Dublanchet, A. A century of clinical use of phages: A literature review. Antibiotics 2023, 12, 751. [Google Scholar] [CrossRef] [PubMed]
- Hitchcock, N.M.; Devequi Gomes, N.D.; Shiach, J.; Valeria Saraiva, H.K.; Dantas Viana, B.J.; Alencar Pereira, R.L.; Coler, B.S.; Botelho Pereira, S.M.; Badaro, R. Current clinical landscape and global potential of bacteriophage therapy. Viruses 2023, 15, 1020. [Google Scholar] [CrossRef]
- Jones, J.D.; Trippett, C.; Suleman, M.; Clokie, M.R.J.; Clark, J.R. The future of clinical phage therapy in the United Kingdom. Viruses 2023, 15, 721. [Google Scholar] [CrossRef] [PubMed]
- Petrovic Fabijan, A.; Iredell, J.; Danis-Wlodarczyk, K.; Kebriaei, R.; Abedon, S.T. Translating phage therapy into the clinic: Recent accomplishments but continuing challenges. PLoS Biol. 2023, 21, e3002119. [Google Scholar] [CrossRef] [PubMed]
- Strathdee, S.A.; Hatfull, G.F.; Mutalik, V.K.; Schooley, R.T. Phage therapy: From biological mechanisms to future directions. Cell 2023, 186, 17–31. [Google Scholar] [CrossRef] [PubMed]
- García-Cruz, J.C.; Huelgas-Méndez, D.; Jiménez-Zúñiga, J.S.; Rebollar-Juárez, X.; Hernández-Garnica, M.; Fernández-Presas, A.M.; Husain, F.M.; Alenazy, R.; Alqasmi, M.; Albalawi, T.; et al. Myriad applications of bacteriophages beyond phage therapy. PeerJ 2023, 11, e15272. [Google Scholar] [CrossRef] [PubMed]
- Harper, D.R. Biological control by microorganisms. In eLS; John Wiley & Sons, Ltd: Chichester, UK, 2013. [Google Scholar] [CrossRef]
- Abedon, S.T. Further considerations on how to improve phage therapy experimentation, practice, and reporting: Pharmacodynamics perspectives. Phage 2022, 3, 98–111. [Google Scholar] [CrossRef]
- Stopar, D.; Abedon, S.T. Modeling bacteriophage population growth. In Bacteriophage Ecology; Abedon, S.T., Ed.; Cambridge University Press: Cambridge, UK, 2008; pp. 389–414. [Google Scholar]
- Abedon, S. Phage therapy pharmacology: Calculating phage dosing. Adv. Appl. Microbiol. 2011, 77, 1–40. [Google Scholar] [PubMed]
- Abedon, S.T. Bacteriophage adsorption: Likelihood of virion encounter with bacteria and other factors affecting rates. Antibiotics 2023, 12, 723. [Google Scholar] [CrossRef]
- Dulbecco, R. Appendix: On the reliability of the Poisson distribution as a distribution of the number of phage particles infecting individual bacteria in a population. Genetics 1949, 34, 122–125. [Google Scholar]
- Abedon, S.T. Phage therapy dosing: The problem(s) with multiplicity of infection (MOI). Bacteriophage 2016, 6, e1220348. [Google Scholar] [CrossRef] [PubMed]
- Kasman, L.M.; Kasman, A.; Westwater, C.; Dolan, J.; Schmidt, M.G.; Norris, J.S. Overcoming the phage replication threshold: A mathematical model with implications for phage therapy. J. Virol. 2002, 76, 5557–5564. [Google Scholar] [CrossRef]
- Benzer, S.; Hudson, W.; Weidel, W.; Delbrück, M.; Stent, G.S.; Weigle, J.J.; Dulbecco, R.; Watson, J.D.; Wollman, E.L. A syllabus on procedures, facts, and interpretations in phage. In Viruses 1950; Delbrück, M., Ed.; California Institute of Technology: Pasadena, CA, USA, 1950; pp. 100–147. [Google Scholar]
- Stent, G.S. Molecular Biology of Bacterial Viruses; WH Freeman and Co.: San Francisco, CA, USA, 1963. [Google Scholar]
- Bull, J.J.; Regoes, R.R. Pharmacodynamics of non-replicating viruses, bacteriocins and lysins. Proc. R. Soc. Lond. B Biol. Sci. 2006, 273, 2703–2712. [Google Scholar] [CrossRef]
- Abedon, S.T. Ecology and evolutionary biology of hindering phage therapy: The phage tolerance vs. phage resistance of bacterial biofilms. Antibiotics 2023, 12, 245. [Google Scholar] [CrossRef]
- Gallet, R.; Lenormand, T.; Wang, I.N. Phenotypic stochasticity protects lytic bacteriophage populations from extinction during the bacterial stationary phase. Evolution 2012, 66, 3485–3494. [Google Scholar] [CrossRef]
- Storms, Z.J.; Sauvageau, D. Evidence that the heterogeneity of a T4 population is the result of heritable traits. PLoS ONE 2014, 9, e116235. [Google Scholar] [CrossRef] [PubMed]
- Storms, Z.J.; Smith, L.; Sauvageau, D.; Cooper, D.G. Modeling bacteriophage attachment using adsorption efficiency. Biochem. Eng. J. 2012, 64, 22–29. [Google Scholar] [CrossRef]
- Storms, Z.J.; Arsenault, E.; Sauvageau, D.; Cooper, D.G. Bacteriophage adsorption efficiency and its effect on amplification. Bioprocess. Biosyst. Eng 2010, 33, 823–831. [Google Scholar] [CrossRef]
- Payne, R.J.H.; Jansen, V.A.A. Phage therapy: The peculiar kinetics of self-replicating pharmaceuticals. Clin. Pharmacol. Ther. 2000, 68, 225–230. [Google Scholar] [CrossRef] [PubMed]
- Payne, R.J.H.; Jansen, V.A.A. Understanding bacteriophage therapy as a density-dependent kinetic process. J. Theor. Biol. 2001, 208, 37–48. [Google Scholar] [CrossRef] [PubMed]
- Payne, R.J.H.; Jansen, V.A.A. Pharmacokinetic principles of bacteriophage therapy. Clin. Pharmacokinet. 2003, 42, 315–325. [Google Scholar] [CrossRef]
- Nang, S.C.; Lin, Y.W.; Petrovic Fabijan, A.; Chang, R.Y.K.; Rao, G.G.; Iredell, J.; Chan, H.K.; Li, J. Pharmacokinetics/pharmacodynamics of phage therapy: A major hurdle to clinical translation. Clin. Microbiol. Infect. 2023, 29, 702–709. [Google Scholar] [CrossRef] [PubMed]
- Hagens, S.; Habel, A.; von Ahsen, U.; von Gabain, A.; Bläsi, U. Therapy of experimental Pseudomonas infections with a nonreplicating genetically modified phage. Antimicrob. Agents Chemother. 2004, 48, 3817–3822. [Google Scholar] [CrossRef] [PubMed]
- Abedon, S.T. Active bacteriophage biocontrol and therapy on sub-millimeter scales towards removal of unwanted bacteria from foods and microbiomes. AIMS Microbiol. 2017, 3, 649–688. [Google Scholar] [CrossRef] [PubMed]
- Abedon, S.T. Spatial vulnerability: Bacterial arrangements, microcolonies, and biofilms as responses to low rather than high phage densities. Viruses 2012, 4, 663–687. [Google Scholar] [CrossRef]
- Eriksen, R.S.; Mitarai, N.; Sneppen, K. On phage adsorption to bacterial chains. Biophys. J. 2020, 119, 1896–1904. [Google Scholar] [CrossRef]
- Levin, B.R.; Stewart, F.M.; Chao, L. Resource limited growth, competition, and predation: A model and experimental studies with bacteria and bacteriophage. Am. Nat. 1977, 111, 3–24. [Google Scholar] [CrossRef]
- Bohannan, B.J.M.; Lenski, R.E. Effect of resource enrichment on a chemostat community of bacteria and bacteriophage. Ecology 1997, 78, 2303–2315. [Google Scholar] [CrossRef]
- Abedon, S.T.; Herschler, T.D.; Stopar, D. Bacteriophage latent-period evolution as a response to resource availability. Appl. Environ. Microbiol. 2001, 67, 4233–4241. [Google Scholar] [CrossRef] [PubMed]
- Campbell, A. Conditions for the existence of bacteriophages. Evolution 1961, 15, 153–165. [Google Scholar] [CrossRef]
- Storms, Z.J.; Teel, M.R.; Mercurio, K.; Sauvageau, D. The virulence index: A metric for quantitative analysis of phage virulence. Phage 2020, 1, 27–36. [Google Scholar] [CrossRef]
- Konopacki, M.; Grygorcewicz, B.; Dołęgowska, B.; Kordas, M.; Rakoczy, R. PhageScore: A simple method for comparative evaluation of bacteriophages lytic activity. Biochem. Eng. J. 2020, 161, 107652. [Google Scholar] [CrossRef]
- Grygorcewicz, B.; Roszak, M.; Rakoczy, R.; Augustyniak, A.; Konopacki, M.; Jabłońska, J.; Serwin, N.; Cecerska-Heryć, E.; Kordas, M.; Galant, K.; et al. PhageScore-based analysis of Acinetobacter baumannii infecting phages antibiotic interaction in liquid medium. Arch. Microbiol. 2022, 204, 421. [Google Scholar] [CrossRef] [PubMed]
- Gelman, D.; Yerushalmy, O.; Alkalay-Oren, S.; Rakov, C.; Ben-Porat, S.; Khalifa, L.; Adler, K.; Abdalrhman, M.; Coppenhagen-Glazer, S.; Aslam, S.; et al. Clinical phage microbiology: A suggested framework and recommendations for the in-vitro matching steps of phage therapy. Lancet Microbe 2021, 2, e555–e563. [Google Scholar] [CrossRef]
- Glonti, T.; Pirnay, J.-P. In vitro techniques and measurements of phage characteristics that are important for phage therapy success. Viruses 2022, 14, 1490. [Google Scholar] [CrossRef] [PubMed]
- Abedon, S.T. Look who’s talking: T-even phage lysis inhibition, the granddaddy of virus-virus intercellular communication research. Viruses 2019, 11, 951. [Google Scholar] [CrossRef]
- Hershey, A.D. Mutation of bacteriophage with respect to type of plaque. Genetics 1946, 31, 620–640. [Google Scholar] [CrossRef]
- Doermann, A.H. Lysis and lysis inhibition with Escherichia coli bacteriophage. J. Bacteriol. 1948, 55, 257–275. [Google Scholar] [CrossRef] [PubMed]
- Moussa, S.H.; Kuznetsov, V.; Tran, T.A.; Sacchettini, J.C.; Young, R. Protein determinants of phage T4 lysis inhibition. Protein Sci. 2012, 21, 571–582. [Google Scholar] [CrossRef]
- Gromkoua, R.H. T-related bacteriophage isolated from Shigella sonnei. J. Virol. 1968, 2, 692–694. [Google Scholar] [CrossRef] [PubMed]
- Schito, G.C. Dvelopment of coliphage N4: Ultrastructural studies. J. Virol. 1974, 13, 186–196. [Google Scholar] [CrossRef] [PubMed]
- Hays, S.G.; Seed, K.D. Dominant Vibrio cholerae phage exhibits lysis inhibition sensitive to disruption by a defensive phage satellite. eLife 2020, 9, e53200. [Google Scholar] [CrossRef]
- Danis-Wlodarczyk, K.M.; Cai, A.; Chen, A.; Gittrich, M.R.; Sullivan, M.B.; Wozniak, D.J.; Abedon, S.T. Friends or foes? Rapid determination of dissimilar colistin and ciprofloxacin antagonism of Pseudomonas aeruginosa phages. Pharmaceuticals 2021, 14, 1162. [Google Scholar] [CrossRef]
- Henry, M.; Lavigne, R.; Debarbieux, L. Predicting in vivo efficacy of therapeutic bacteriophages used to treat pulmonary infections. Antimicrob. Agents Chemother. 2013, 57, 5961–5968. [Google Scholar] [CrossRef]
- Lindberg, H.M.; McKean, K.A.; Wang, I.-N. Phage fitness may help predict phage therapy efficacy. Bacteriophage 2014, 4, e964081. [Google Scholar] [CrossRef]
- Bull, J.J.; Otto, G.; Molineux, I.J. In vivo growth rates are poorly correlated with phage therapy success in a mouse infection model. Antimicrob. Agents Chemother. 2012, 56, 949–954. [Google Scholar] [CrossRef]
- Kropinski, A.M. Practical advice on the one-step growth curve. Methods Mol. Biol. 2018, 1681, 41–47. [Google Scholar]
- Bull, J.J.; Gill, J.J. The habits of highly effective phages: Population dynamics as a framework for identifying therapeutic phages. Front. Microbiol. 2014, 5, 618. [Google Scholar] [CrossRef] [PubMed]
- Fungo, G.B.N.; Uy, J.C.W.; Porciuncula, K.L.J.; Candelario, C.M.A.; Chua, D.P.S.; Gutierrez, T.A.D.; Clokie, M.R.J.; Papa, D.M.D. “Two is better than one”: The multifactorial nature of phage-antibiotic combinatorial treatments against ESKAPE-induced infections. Phage 2023, 4, 55–67. [Google Scholar] [CrossRef]
- Marchi, J.; Zborowsky, S.; Debarbieux, L.; Weitz, J.S. The dynamic interplay of bacteriophage, bacteria and the mammalian host during phage therapy. iScience 2023, 26, 106004. [Google Scholar] [CrossRef] [PubMed]
- Kutter, E. Phage host range and efficiency of plating. Methods Mol. Biol. 2009, 501, 141–149. [Google Scholar] [PubMed]
- Khan Mirzaei, M.; Nilsson, A.S. Isolation of phages for phage therapy: A comparison of spot tests and efficiency of plating analyses for determination of host range and efficacy. PLoS ONE 2015, 10, e0118557. [Google Scholar] [CrossRef] [PubMed]
- Aggarwal, S.; Dhall, A.; Patiyal, S.; Choudhury, S.; Arora, A.; Raghava, G.P.S. An ensemble method for prediction of phage-based therapy against bacterial infections. Front. Microbiol. 2023, 14, 1148579. [Google Scholar] [CrossRef]
- Bajiya, N.; Dhall, A.; Aggarwal, S.; Raghava, G.P.S. Advances in the field of phage-based therapy with special emphasis on computational resources. Brief. Bioinform. 2023, 24, bbac574. [Google Scholar] [CrossRef]
- Schlesinger, M. Ueber die Bindung des Bakteriophagen an homologe Bakterien. II. Quantitative, Untersuchungen über die Bindungsgeschwindigkeit und die Sättigung. Berechnung der Teilchengröße des Bakteriophagen aus deren Ergebnissen. [Adsorption of bacteriophages to homologous bacteria. II Quantitative investigations of adsorption velocity and saturation. Estimation of particle size of the bacteriophage]. Z. Fur Hygenie Immunitiitsforschung 1932, 114, 149–160. [Google Scholar]
- Schlesinger, M. Adsorption of bacteriophages to homologous bacteria. II Quantitative investigations of adsorption velocity and saturation. Estimation of particle size of the bacteriophage [translation]. In Bacterial Viruses; Stent, G.S., Ed.; Little, Brown and Co.: Boston, MA, USA, 1960; pp. 26–36. [Google Scholar]
- Ellis, E.L.; Delbrück, M. The growth of bacteriophage. J. Gen. Physiol. 1939, 22, 365–384. [Google Scholar] [CrossRef]
- Levin, B.R.; Lenski, R.E. Coevolution in bacteria and their viruses and plasmids. In Coevolution; Futuyma, D.J., Slatkin, M., Eds.; Sinauer Associates, Inc.: Sunderland, MA, USA, 1983; pp. 99–127. [Google Scholar]
- Levin, B.R.; Lenski, R.E. Bacteria and phage: A model system for the study of the ecology and co-evolution of hosts and parasites. In Ecology and Genetics of Host-Parasite Interactions; Rollinson, D., Anderson, R.M., Eds.; Academic Press: London, UK, 1985; pp. 227–242. [Google Scholar]
- Levin, B.R.; Bull, J.J. Phage therapy revisited: The population biology of a bacterial infection and its treatment with bacteriophage and antibiotics. Am. Nat. 1996, 147, 881–898. [Google Scholar] [CrossRef]
- Levin, B.R.; Bull, J.J. Population and evolutionary dynamics of phage therapy. Nat. Rev. Microbiol. 2004, 2, 166–173. [Google Scholar] [CrossRef] [PubMed]
- Lenski, R.E. Dynamics of Interactions between Bacteria and Virulent Bacteriophage. In Advances in Microbial Ecology; Springer: Boston, MA, USA, 1988; Volume 10, pp. 1–44. [Google Scholar]
- Weld, R.J.; Butts, C.; Heinemann, J.A. Models of phage growth and their applicability to phage therapy. J. Theor. Biol. 2004, 227, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Kysela, D.T.; Turner, P.E. Optimal bacteriophage mutation rates for phage therapy. J. Theor. Biol. 2007, 249, 411–421. [Google Scholar] [CrossRef] [PubMed]
- Cairns, B.J.; Payne, R.J.H. Bacteriophage therapy and the mutant selection window. Antimicrob. Agents Chemother. 2008, 52, 4344–4350. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Gill, J.J. Modeling of bacteriophage therapy. In Bacteriophage Ecology; Abedon, S.T., Ed.; Cambridge University Press: Cambridge, UK, 2008; pp. 439–464. [Google Scholar]
- Cairns, B.J.; Timms, A.R.; Jansen, V.A.; Connerton, I.F.; Payne, R.J. Quantitative models of in vitro bacteriophage-host dynamics and their application to phage therapy. PLoS Pathog. 2009, 5, e1000253. [Google Scholar] [CrossRef]
- Schmerer, M.; Molineux, I.J.; Bull, J.J. Synergy as a rationale for phage therapy using phage cocktails. PeerJ 2014, 2, e590. [Google Scholar] [CrossRef]
- Leung, C.Y.J.; Weitz, J.S. Modeling the synergistic elimination of bacteria by phage and the innate immune system. J. Theor. Biol. 2017, 429, 241–252. [Google Scholar] [CrossRef]
- Roach, D.R.; Leung, C.Y.; Henry, M.; Morello, E.; Singh, D.; Di Santo, J.P.; Weitz, J.S.; Debarbieux, L. Synergy between the host immune system and bacteriophage is essential for successful phage therapy against an acute respiratory pathogen. Cell Host Microbe 2017, 22, 38–47. [Google Scholar] [CrossRef]
- Sinha, S.S.; Grewal, R.K.; Roy, S. Modeling bacteria–phage interactions and its implications for phage therapy. Adv. Appl. Microbiol. 2018, 103, 103–141. [Google Scholar]
- Li, G.; Leung, C.Y.; Wardi, Y.; Debarbieux, L.; Weitz, J.S. Optimizing the timing and composition of therapeutic phage cocktails: A control-theoretic approach. Bull. Math Biol. 2020, 82, 75. [Google Scholar] [CrossRef]
- Rodriguez-Gonzalez, R.A.; Leung, C.Y.; Chan, B.K.; Turner, P.E.; Weitz, J.S. Quantitative models of phage-antibiotic combination therapy. mSystems 2020, 5, e00756-19. [Google Scholar] [CrossRef] [PubMed]
- Styles, K.M.; Brown, A.T.; Sagona, A.P. A review of using mathematical modeling to improve our understanding of bacteriophage, bacteria, and eukaryotic interactions. Front. Microbiol. 2021, 12, 724767. [Google Scholar] [CrossRef] [PubMed]
- Kyaw, E.E.; Zheng, H.; Wang, J.; Hlaing, H.K. Stability analysis and persistence of a phage therapy model. Math. Biosci. Eng. 2021, 18, 5552–5572. [Google Scholar] [CrossRef]
- Abedon, S.T. Deconstructing chemostats towards greater phage-modeling precision. In Contemporary Trends in Bacteriophage Research; Adams, H.T., Ed.; Nova Science Publishers: Hauppauge, NY, USA, 2009; pp. 249–283. [Google Scholar]
- Abedon, S.T. Phage-antibiotic combination treatments: Antagonistic impacts of antibiotics on the pharmacodynamics of phage therapy? Antibiotics 2019, 8, 182. [Google Scholar] [CrossRef] [PubMed]
- Abedon, S.T.; Danis-Wlodarczyk, K.; Alves, D.R. Phage therapy in the 21st Century: Is there modern, clinical evidence of phage-mediated clinical efficacy? Pharmaceuticals 2021, 14, 1157. [Google Scholar] [CrossRef]
- Kebriaei, R.; Lehman, S.M.; Shah, R.M.; Stamper, K.C.; Kunz Coyne, A.J.; Holger, D.; El Ghali, A.; Rybak, M.J. Optimization of phage-antibiotic combinations against Staphylococcus aureus biofilms. Microbiol. Spectr. 2023, 11, e0491822. [Google Scholar] [CrossRef]
- Osman, A.H.; Kotey, F.C.N.; Odoom, A.; Darkwah, S.; Yeboah, R.K.; Dayie, N.T.K.D.; Donkor, E.S. The potential of bacteriophage-antibiotic combination therapy in treating infections with multidrug-resistant bacteria. Antibiotics 2023, 12, 1329. [Google Scholar] [CrossRef]
- Santamaría-Corral, G.; Senhaji-Kacha, A.; Broncano-Lavado, A.; Esteban, J.; García-Quintanilla, M. Bacteriophage-antibiotic combination therapy against Pseudomonas aeruginosa. Antibiotics 2023, 12, 1089. [Google Scholar] [CrossRef]
- Pons, B.J.; Dimitriu, T.; Westra, E.R.; van Houte, S. Antibiotics that affect translation can antagonize phage infectivity by interfering with the deployment of counter-defenses. Proc. Natl. Acad. Sci. USA 2023, 120, e2216084120. [Google Scholar] [CrossRef]
- Bhartiya, S.K.; Prasad, R.; Sharma, S.; Shukla, V.; Nath, G.; Kumar, R. Biological therapy on infected traumatic wounds: A case-control study. Int. J. Low. Extrem. Wounds 2022. [Google Scholar] [CrossRef]
- Blasco, L.; López-Hernández, I.; Rodríguez-Fernández, M.; Pérez-Florido, J.; Casimiro-Soriguer, C.S.; Djebara, S.; Merabishvili, M.; Pirnay, J.-P.; Rodríguez-Baño, J.; Tomás, M.; et al. Case report: Analysis of phage therapy failure in a patient with a Pseudomonas aeruginosa prosthetic vascular graft infection. Front Med. 2023, 10, 1199657. [Google Scholar] [CrossRef] [PubMed]
- Bradley, J.S.; Hajama, H.; Akong, K.; Jordan, M.; Stout, D.; Rowe, R.S.; Conrad, D.J.; Hingtgen, S.; Segall, A.M. Bacteriophage therapy of multidrug-resistant Achromobacter in an 11-year-old boy with cystic fibrosis assessed by metagenome analysis. Pediatr. Infect. Dis. J. 2023, 42, 754–759. [Google Scholar] [CrossRef] [PubMed]
- Cesta, N.; Pini, M.; Mulas, T.; Materazzi, A.; Ippolito, E.; Wagemans, J.; Kutateladze, M.; Fontana, C.; Sarmati, L.; Tavanti, A.; et al. Application of phage therapy in a case of a chronic hip-prosthetic joint infection due to Pseudomonas aeruginosa: An Italian real-life experience and in vitro analysis. Open Forum Infect. Dis. 2023, 10, ofad051. [Google Scholar] [CrossRef] [PubMed]
- Dedrick, R.M.; Smith, B.E.; Cristinziano, M.; Freeman, K.G.; Jacobs-Sera, D.; Belessis, Y.; Whitney, B.A.; Cohen, K.A.; Davidson, R.M.; Van, D.D.; et al. Phage therapy of Mycobacterium Infections: Compassionate-use of phages in twenty patients with drug-resistant mycobacterial disease. Clin. Infect. Dis. 2023, 76, 103–112. [Google Scholar] [CrossRef]
- Doub, J.B.; Chan, B.; Johnson, A.J. Salphage: Salvage bacteriophage therapy for a chronic Enterococcus faecalis prosthetic joint infection. IDCases 2023, 33, e01854. [Google Scholar] [CrossRef]
- Fedorov, E.; Samokhin, A.; Kozlova, Y.; Kretien, S.; Sheraliev, T.; Morozova, V.; Tikunova, N.; Kiselev, A.; Pavlov, V. Short-term outcomes of phage-antibiotic combination treatment in adult patients with periprosthetic hip joint infection. Viruses 2023, 15, 499. [Google Scholar] [CrossRef]
- Gainey, A.B.; Daniels, R.; Burch, A.K.; Hawn, J.; Fackler, J.; Biswas, B.; Brownstein, M.J. Recurrent ESBL Escherichia coli urosepsis in a pediatric renal transplant patient treated with antibiotics and bacteriophage therapy. Pediatr. Infect. Dis. J. 2023, 42, 43–46. [Google Scholar] [CrossRef]
- Green, S.I.; Clark, J.R.; Santos, H.H.; Weesner, K.E.; Salazar, K.C.; Aslam, S.; Campbell, W.J.; Doernberg, S.B.; Blodget, E.; Morris, M.I.; et al. A retrospective, observational study of 12 cases of expanded access customized phage therapy: Production, characteristics, and clinical outcomes. Clin. Infect. Dis. 2023, ciad335. [Google Scholar] [CrossRef]
- Hahn, A.; Sami, I.; Chaney, H.; Koumbourlis, A.C.; Del Valle, M.C.; Cochrane, C.; Chan, B.K.; Koff, J.L. Bacteriophage therapy for pan-drug-resistant Pseudomonas aeruginosa in two persons with cystic fibrosis. J. Investig. Med. High Impact. Case. Rep. 2023, 11, 23247096231188243. [Google Scholar] [CrossRef]
- Haidar, G.; Chan, B.K.; Cho, S.T.; Hughes, K.K.; Nordstrom, H.R.; Wallace, N.R.; Stellfox, M.E.; Holland, M.; Kline, E.G.; Kozar, J.M.; et al. Phage therapy in a lung transplant recipient with cystic fibrosis infected with multidrug-resistant Burkholderia multivorans. Transpl. Infect. Dis. 2023, 25, e14041. [Google Scholar] [CrossRef]
- Köhler, T.; Luscher, A.; Falconnet, L.; Resch, G.; McBride, R.; Mai, Q.A.; Simonin, J.L.; Chanson, M.; Maco, B.; Galiotto, R.; et al. Personalized aerosolised bacteriophage treatment of a chronic lung infection due to multidrug-resistant Pseudomonas aeruginosa. Nat. Commun. 2023, 14, 3629. [Google Scholar] [CrossRef] [PubMed]
- Le, T.; Nang, S.C.; Zhao, J.; Yu, H.H.; Li, J.; Gill, J.J.; Liu, M.; Aslam, S. Therapeutic potential of intravenous phage as standalone therapy for recurrent drug-resistant urinary tract infections. Antimicrob. Agents Chemother. 2023, 67, e0003723. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Zhong, Q.; Zhao, Y.; Bao, J.; Liu, B.; Zhong, Z.; Wang, J.; Yang, L.; Zhang, T.; Cheng, M.; et al. First-in-human application of double-stranded RNA bacteriophage in the treatment of pulmonary Pseudomonas aeruginosa infection. Microb. Biotechnol. 2023, 16, 862–867. [Google Scholar] [CrossRef] [PubMed]
- Onallah, H.; Hazan, R.; Nir-Paz, R.; Yerushalmy, O.; Rimon, A.; Braunstein, R.; Gelman, D.; Alkalay, S.; Abdalrhman, M.; Stuczynski, D.; et al. Compassionate use of bacteriophages for failed persistent infections during the first 5 years of the Israeli phage therapy center. Open Forum Infect. Dis. 2023, 10, ofad221. [Google Scholar] [CrossRef] [PubMed]
- Racenis, K.; Lacis, J.; Rezevska, D.; Mukane, L.; Vilde, A.; Putnins, I.; Djebara, S.; Merabishvili, M.; Pirnay, J.-P.; Kalnina, M.; et al. Successful bacteriophage-antibiotic combination therapy against multidrug-resistant Pseudomonas aeruginosa left ventricular assist device driveline infection. Viruses 2023, 15, 1210. [Google Scholar] [CrossRef]
- Samaee, H.R.; Eslami, G.; Rahimzadeh, G.; Saeedi, M.; Davoudi, B.A.; Asare-Addo, K.; Nokhodchi, A.; Roozbeh, F.; Moosazadeh, M.; Ghasemian, R.; et al. Inhalation phage therapy as a new approach to preventing secondary bacterial pneumonia in patients with moderate to severe COVID-19: A double-blind clinical trial study. J. Drug Deliv. Sci. Technol. 2023, 84, 104486. [Google Scholar] [CrossRef]
- Young, M.J.; Hall, L.M.L.; Merabishvilli, M.; Pirnay, J.-P.; Clark, J.R.; Jones, J.D. Phage therapy for diabetic foot infection: A case series. Clin. Ther. 2023, 45, 797–801. [Google Scholar] [CrossRef]
- Jean-Paul, P.; Sarah, D.; Griet, S.; Johann, G.; Christel, C.; Steven, D.S.; Tea, G.; An, S.; Emily, V.B.; Sabrina, G.; et al. Retrospective, observational analysis of the first one hundred consecutive cases of personalized bacteriophage therapy of difficult-to-treat infections facilitated by a Belgian consortium. medRxiv 2023. [Google Scholar] [CrossRef]
- Merril, C.R.; Biswas, B.; Carlton, R.; Jensen, N.C.; Creed, G.J.; Zullo, S.; Adhya, S. Long-circulating bacteriophage as antibacterial agents. Proc. Natl. Acad. Sci. USA 1996, 93, 3188–3192. [Google Scholar] [CrossRef]
- Vitiello, C.L.; Merril, C.R.; Adhya, S. An amino acid substitution in a capsid protein enhances phage survival in mouse circulatory system more than a 1000-fold. Virus Res. 2005, 114, 101–103. [Google Scholar] [CrossRef]
| Topic | Section | URL |
|---|---|---|
| Multiplicity of Infection | Section 2.1 | moi.phage.org |
| Phage Adsorptions | Section 2.2 | adsorptions.phage-therapy.org |
| Bacterial Half-Life | Section 2.3.1 | b-half-life.phage.org |
| Decimal Reduction Time | Section 2.3.2 | decimal.phage-therapy.org |
| Phage Half-Life | Section 2.3.4 | p-half-life.phage.org |
| Inundative Phage Quantities | Section 2.4 | inundative.phage-therapy.org |
| Poisson Frequencies | Section 2.5 | Poisson.phage.org |
| Killing Titers | Section 2.5.2 | killingtiter.phage-therapy.org |
| Active Phage Therapy | Section 2.6 | active.phage-therapy.org |
| Abbreviation | Description | Comments |
|---|---|---|
| Ac | Bacterial probability of being adsorbed | Likelihood of an individual bacterial cell being adsorbed per unit time, e.g., 1 min; the “c” stands for “cell” |
| At | Adsorptions over time | Number of phage adsorptions that occur over some interval of time, t |
| B | Burst size | Number of virions produced per phage infection; might range from 10 to well in excess of 100 |
| e | Base of the natural logarithm | =2.718… (a non-repeating decimal) |
| IP, IN | Decay rate | Rates of loss of free phages (IP) or bacteria (IN) that occur for reasons that are independent of phage adsorption |
| IPDmin | Inundative phage density | Minimum phage titer required to reduce a bacterial population from some starting number to some ending number over some specified interval of time, not assuming 100% phage adsorption |
| IPNmin | Inundative phage number | Minimum phage titer to achieve the same as IPDmin except here assuming 100% phage adsorption |
| k | Adsorption rate constant | Probability that one virion will adsorb one bacterium as suspended in a unit volume of fluid (e.g., 1 mL) over the course of some unit time (e.g., 1 min), hence, e.g., mL−1 min−1 units, though often expressed instead as mL min−1 |
| L | Latent period | Measure of the length of infection by a phage a bacterium |
| ln | Natural logarithm | For example, ln(2) = 0.69 = −ln(0.5) = −ln(1/2); ln(e) = 1 |
| MOIactual, n | Actual multiplicity of infection | Number of adsorbed phages divided by the number of adsorbable bacteria; equivalent to n as used in Poisson calculations |
| MOIinput or MOIaddition | Input multiplicity of infection | Number of phages added to targeted bacteria divided by the number of those bacteria |
| Μ | Malthusian parameter | A measure of bacterial population growth rate in per time units |
| N, N0, Nt | Bacterial concentrations | Subscript 0 refers to initial concentrations, though in many cases this is implied so the subscript is not always present; subscript t refers to the concentration of unadsorbed bacteria following a previous time interval, t |
| NF, NT | Bacterial numbers | Subscript F refers to a “Final” number of unadsorbed bacteria; subscript T refers to “Total” and is used instead of N0 to distinguish starting bacterial concentration (N0) from starting bacterial numbers (NT) |
| p | Probability | This is lower-case “p” without italicization |
| P, P0, PF, Pt | Phage titer | Subscripts are equivalent to those of N0, NF, Nt, with P in all cases referring to phage concentrations, i.e., phage titers |
| Padsorbed | Prior titer of adsorbed virions | Number of previously free phages that have now adsorbed, divided by volume, as to be distinguished from P0 |
| PK | Killing titer | Titer of phages required to reduce a bacterial population from a given starting number to a given ending number, assuming 100% adsorption |
| r | Poisson category | Here, e.g., 0 phages adsorbed, 1 phage adsorbed, etc., all per bacterium |
| r! | r factorial | For example, 3! = 1 × 2 × 3; 2! = 1 × 2; 1! = 1; 0! = 1 |
| t | Time | Generally, here, this is an interval over which adsorption occurs |
| t0.1, t0.01 | Decimal reduction time(s) | Time it takes for 90% of unadsorbed bacteria to become adsorbed (t0.1) or 99% (t0.01) |
| t0.5 | Bacterial half-life | Time it takes for one-half of unadsorbed bacteria to become adsorbed |
| tMFT | Mean free time | Average length of time it takes for a bacterium to become phage-adsorbed |
| V | Volume | Volume that targeted bacteria and targeting phages are suspended in during phage treatments |
| x | Fraction bacteria | As surviving following phage exposure (=NF/NT) |
| NT → | 1010 | 109 | 108 | 107 | 106 | 105 | 104 | 103 | 102 | |
|---|---|---|---|---|---|---|---|---|---|---|
| VNT → | 1012 | 1011 | 1010 | 109 | 108 | 107 | 106 | 105 | 104 | |
| NF ↓ | ||||||||||
| 10−3 | 2.3 × 108 | 2.1 × 108 | 2.0 × 108 | 1.8 × 108 | 1.7 × 108 | 1.5 × 108 | 1.4 × 108 | 1.2 × 108 | 1.1 × 108 | Eq. (15) |
| 10−3 | 3.5 × 1011 | 3.2 × 1010 | 3.0 × 109 | 3.6 × 108 | 1.8 × 108 | 1.5 × 108 | 1.4 × 108 | 1.2 × 108 | 1.1 × 108 | Eq. (16) |
| 10−3 | 3.0 × 1011 | 2.8 × 1010 | 2.5 × 109 | 2.3 × 108 | 2.1 × 107 | 1.8 × 106 | 1.6 × 105 | 1.4 × 104 | 1.2 × 103 | Eq. (18) |
| 10−2 | 2.1 × 108 | 2.0 × 108 | 1.8 × 108 | 1.7 × 108 | 1.5 × 108 | 1.4 × 108 | 1.2 × 108 | 1.1 × 108 | 9.2 × 107 | Eq. (15) |
| 10−2 | 3.2 × 1011 | 3.0 × 1010 | 2.8 × 109 | 3.3 × 108 | 1.7 × 108 | 1.4 × 108 | 1.2 × 108 | 1.1 × 108 | 9.2 × 107 | Eq. (16) |
| 10−2 | 2.8 × 1011 | 2.5 × 1010 | 2.3 × 109 | 2.1 × 108 | 1.8 × 107 | 1.6 × 106 | 1.4 × 105 | 1.2 × 104 | 9.2 × 102 | Eq. (18) |
| 10−1 | 2.0 × 108 | 1.8 × 108 | 1.7 × 108 | 1.5 × 108 | 1.4 × 108 | 1.2 × 108 | 1.1 × 108 | 9.2 × 107 | 7.7 × 107 | Eq. (15) |
| 10−1 | 3.0 × 1011 | 2.8 × 1010 | 2.5 × 109 | 3.0 × 108 | 1.5 × 108 | 1.2 × 108 | 1.1 × 108 | 9.2 × 107 | 7.7 × 107 | Eq. (16) |
| 10−1 | 2.5 × 1011 | 2.3 × 1010 | 2.1 × 109 | 1.8 × 108 | 1.6 × 107 | 1.4 × 106 | 1.2 × 105 | 9.2 × 103 | 6.9 × 102 | Eq. (18) |
| 100 | 1.8 × 108 | 1.7 × 108 | 1.5 × 108 | 1.4 × 108 | 1.2 × 108 | 1.1 × 108 | 9.2 × 107 | 7.7 × 107 | 6.1 × 107 | Eq. (15) |
| 100 | 2.8 × 1011 | 2.5 × 1010 | 2.3 × 109 | 2.7 × 108 | 1.3 × 108 | 1.1 × 108 | 9.2 × 107 | 7.7 × 107 | 6.1 × 107 | Eq. (16) |
| 100 | 2.3 × 1011 | 2.1 × 1010 | 1.8 × 109 | 1.6 × 108 | 1.4 × 107 | 1.2 × 106 | 9.2 × 104 | 6.9 × 103 | 4.6 × 102 | Eq. (18) |
| 101 | 1.7 × 108 | 1.5 × 108 | 1.4 × 108 | 1.2 × 108 | 1.1 × 108 | 9.2 × 107 | 7.7 × 107 | 6.1 × 107 | 4.6 × 107 | Eq. (15) |
| 101 | 2.5 × 1011 | 2.3 × 1010 | 2.1 × 109 | 2.4 × 108 | 1.2 × 108 | 9.3 × 107 | 7.7 × 107 | 6.1 × 107 | 4.6 × 107 | Eq. (16) |
| 101 | 2.1 × 1011 | 1.8 × 1010 | 1.6 × 109 | 1.4 × 108 | 1.2 × 107 | 9.2 × 105 | 6.9 × 104 | 4.6 × 103 | 2.3 × 102 | Eq. (18) |
| 102 | 1.5 × 108 | 1.4 × 108 | 1.2 × 108 | 1.1 × 108 | 9.2 × 107 | 7.7 × 107 | 6.1 × 107 | 4.6 × 107 | 3.1 × 107 | Eq. (15) |
| 102 | 2.3 × 1011 | 2.1 × 1010 | 1.8 × 109 | 2.1 × 108 | 9.9 × 107 | 7.7 × 107 | 6.1 × 107 | 4.6 × 107 | 3.1 × 107 | Eq. (16) |
| 102 | 1.8 × 1011 | 1.6 × 1010 | 1.4 × 109 | 1.2 × 108 | 9.2 × 106 | 6.9 × 105 | 4.6 × 104 | 2.3 × 103 | Eq. (18) | |
| 103 | 1.4 × 108 | 1.2 × 108 | 1.1 × 108 | 9.2 × 107 | 7.7 × 107 | 6.1 × 107 | 4.6 × 107 | 3.1 × 107 | 1.5 × 107 | Eq. (15) |
| 103 | 2.1 × 1011 | 1.8 × 1010 | 1.6 × 109 | 1.8 × 108 | 8.3 × 107 | 6.2 × 107 | 4.6 × 107 | 3.1 × 107 | 1.5 × 107 | Eq. (16) |
| 103 | 1.6 × 1011 | 1.4 × 1010 | 1.2 × 109 | 9.2 × 107 | 6.9 × 106 | 4.6 × 105 | 2.3 × 104 | Eq. (18) | ||
| 104 | 1.2 × 108 | 1.1 × 108 | 9.2 × 107 | 7.7 × 107 | 6.1 × 107 | 4.6 × 107 | 3.1 × 107 | 1.5 × 107 | Eq. (15) | |
| 104 | 1.8 × 1011 | 1.6 × 1010 | 1.4 × 109 | 1.5 × 108 | 6.6 × 107 | 4.6 × 107 | 3.1 × 107 | 1.5 × 107 | Eq. (16) | |
| 104 | 1.4 × 1011 | 1.2 × 1010 | 9.2 × 108 | 6.9 × 107 | 4.6 × 106 | 2.3 × 105 | Eq. (18) | |||
| 105 | 1.1 × 108 | 9.2 × 107 | 7.7 × 107 | 6.1 × 107 | 4.6 × 107 | 3.1 × 107 | 1.5 × 107 | Eq. (15) | ||
| 105 | 1.6 × 1011 | 1.4 × 1010 | 1.2 × 109 | 1.2 × 108 | 5.0 × 107 | 3.1 × 107 | 1.5 × 107 | Eq. (16) | ||
| 105 | 1.2 × 1011 | 9.2 × 109 | 6.9 × 108 | 4.6 × 107 | 2.3 × 106 | Eq. (18) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abedon, S.T. Automating Predictive Phage Therapy Pharmacology. Antibiotics 2023, 12, 1423. https://doi.org/10.3390/antibiotics12091423
Abedon ST. Automating Predictive Phage Therapy Pharmacology. Antibiotics. 2023; 12(9):1423. https://doi.org/10.3390/antibiotics12091423
Chicago/Turabian StyleAbedon, Stephen T. 2023. "Automating Predictive Phage Therapy Pharmacology" Antibiotics 12, no. 9: 1423. https://doi.org/10.3390/antibiotics12091423
APA StyleAbedon, S. T. (2023). Automating Predictive Phage Therapy Pharmacology. Antibiotics, 12(9), 1423. https://doi.org/10.3390/antibiotics12091423






