Analysis of Cellular Damage Resulting from Exposure of Bacteria to Graphene Oxide and Hybrids Using Fourier Transform Infrared Spectroscopy
Abstract
1. Introduction
2. Results
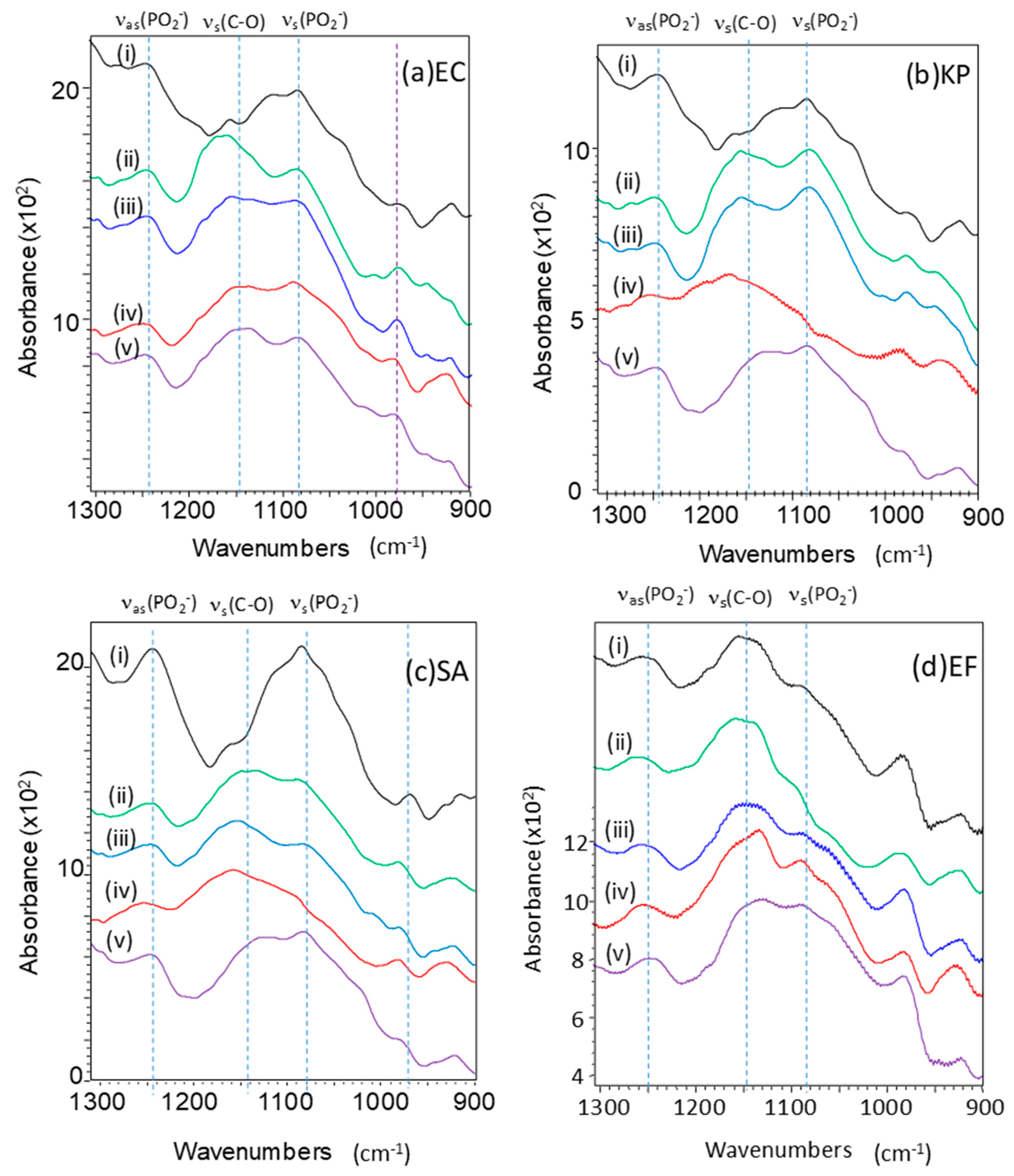
2.1. FTIR Spectra of Pristine Bacteria
2.2. Effect of GO Hybrids in FTIR Spectra of Bacteria
2.3. Minimum Bactericidal Concentrations and Peak Changes
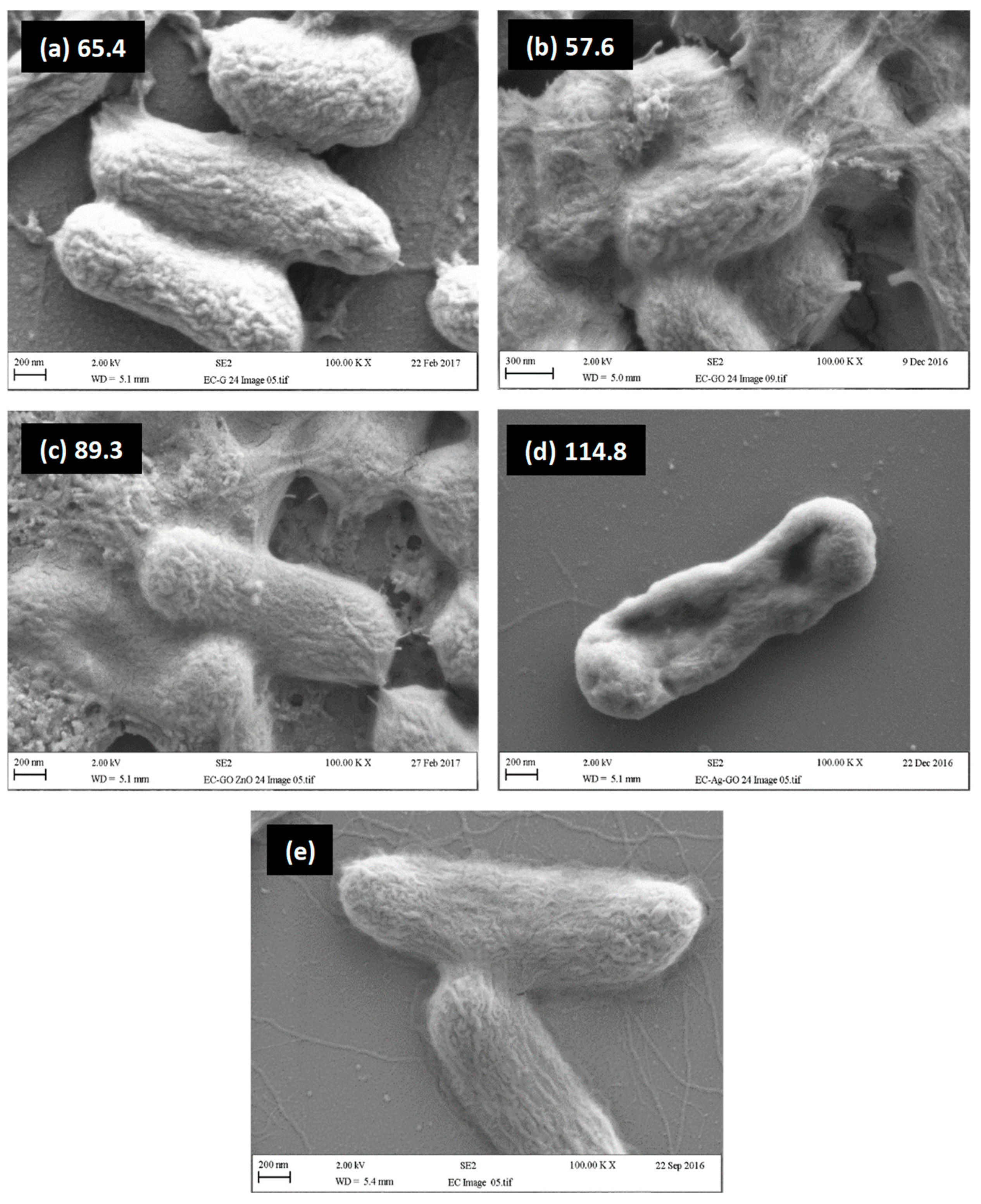
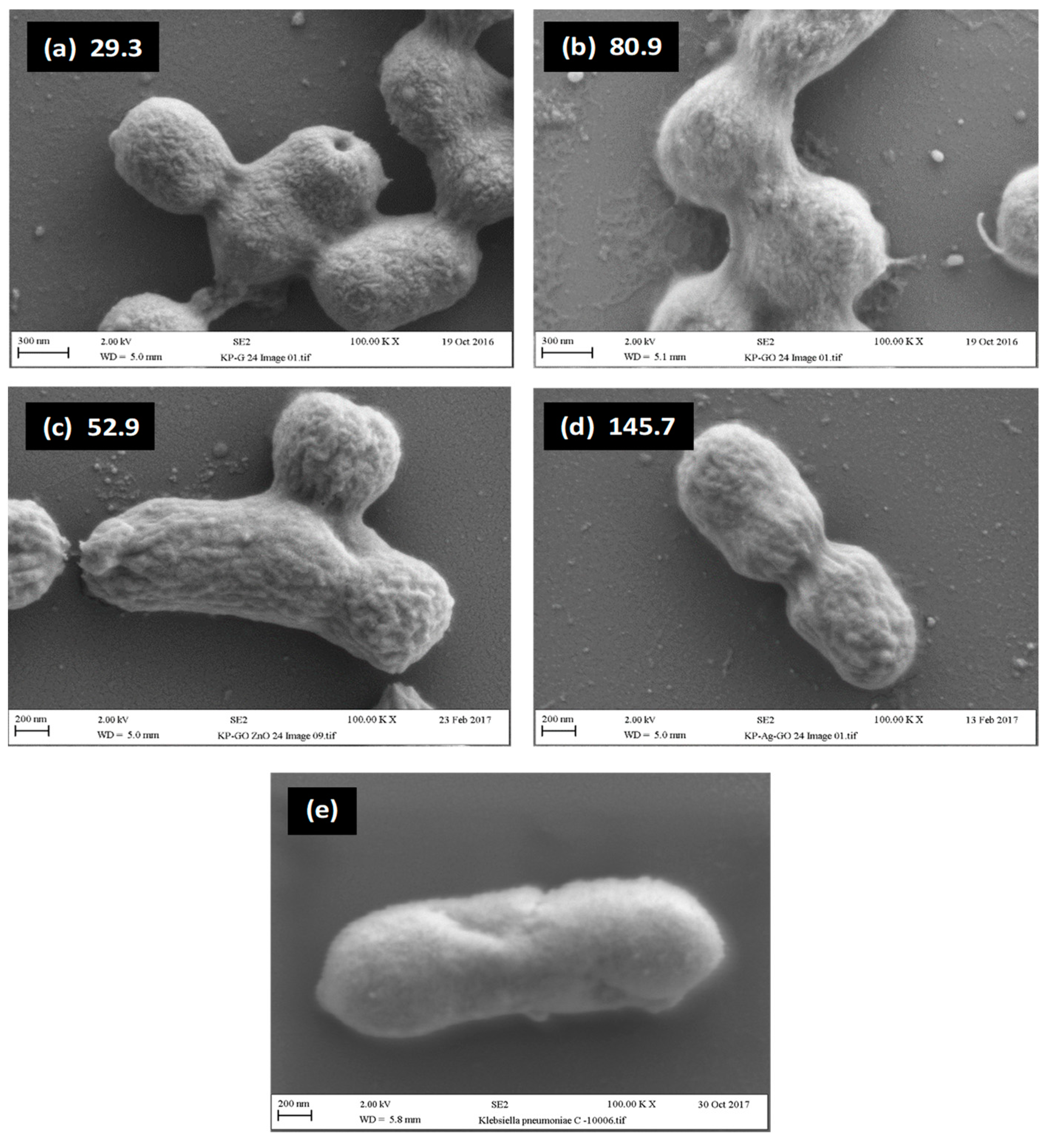
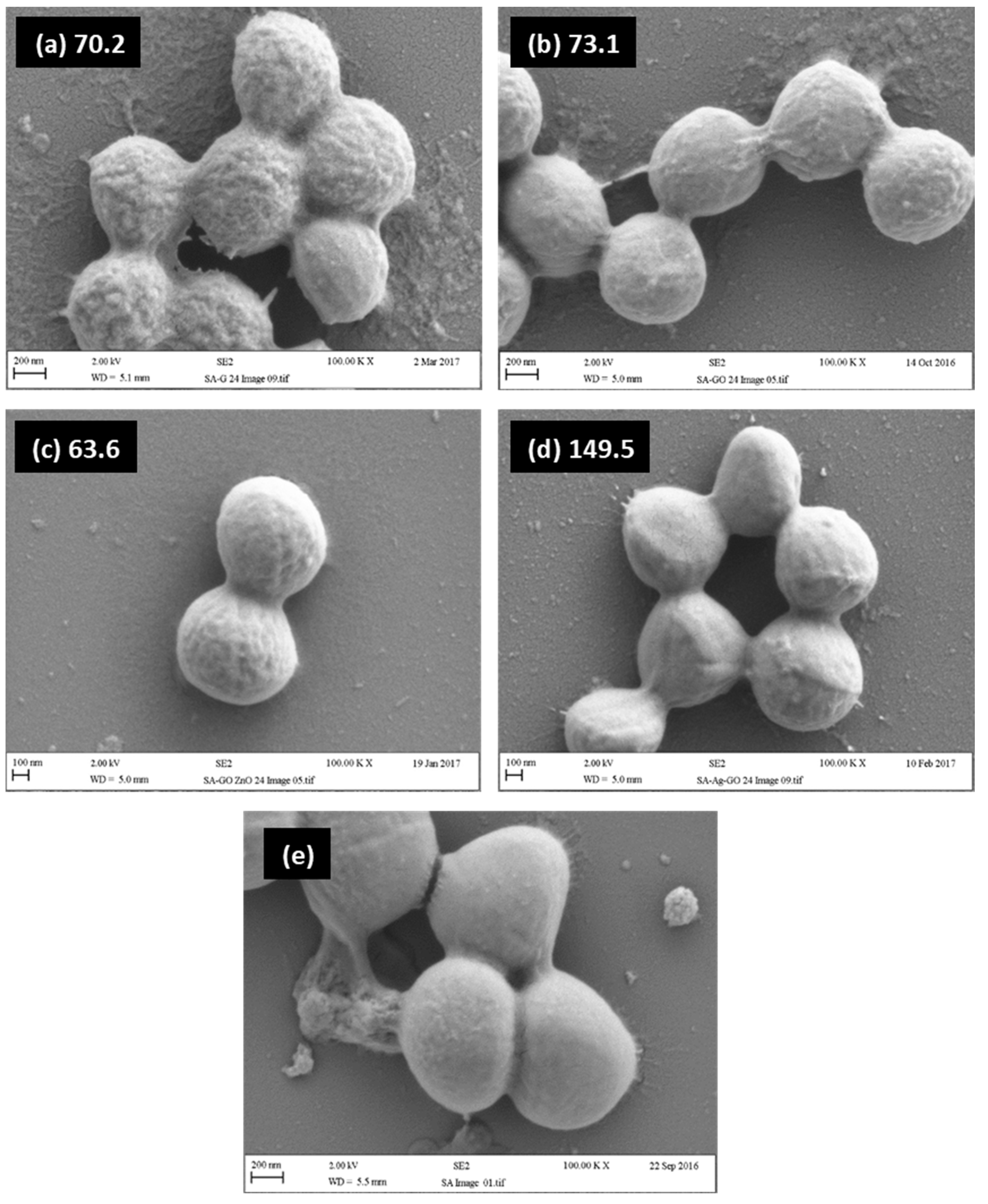
2.4. Relationship between Visual Assessment of Cell Damage (SEM), FTIR Metrics and MBC Values
3. Discussion
4. Conclusions
5. Material and Methods
5.1. Stock Cultures of Bacteria
5.2. Preparation of Microbiological Culture
5.3. Synthesis of Compounds and Characterisation
5.4. Preparation of Silicon Wafers
5.5. Minimal Bactericidal Concentrations (MBC)
5.6. Fourier Transform Infrared Spectroscopy
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Llor, C.; Bjerrum, L. Antimicrobial resistance: Risk associated with antibiotic overuse and initiatives to reduce the problem. Ther. Adv. Drug Saf. 2014, 5, 229–241. [Google Scholar] [CrossRef]
- Cosgrove, S.E. The Relationship between Antimicrobial Resistance and Patient Outcomes: Mortality, Length of Hospital Stay, and Health Care Costs. Clin. Infect. Dis. 2006, 42 (Suppl. S2), S82–S89. [Google Scholar] [CrossRef]
- Fraise, A.P. Biocide abuse and antimicrobial resistance—A cause for concern? J. Antimicrob. Chemother. 2002, 49, 11–12. [Google Scholar] [CrossRef] [PubMed]
- Pal, C.; Bengtsson-Palme, J.; Kristiansson, E.; Larsson, D.G.J. Co-occurrence of resistance genes to antibiotics, biocides and metals reveals novel insights into their co-selection potential. BMC Genom. 2015, 16, 964. [Google Scholar] [CrossRef] [PubMed]
- Webber, M.A.; Whitehead, R.N.; Mount, M.; Loman, N.J.; Pallen, M.J.; Piddock, L.J.V. Parallel evolutionary pathways to antibiotic resistance selected by biocide exposure. J. Antimicrob. Chemother. 2015, 70, 2241–2248. [Google Scholar] [CrossRef]
- Eriksson, J.; Gilek, M.; Rudén, C. Regulating Chemical Risks: European and Global Challenges; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2010. [Google Scholar]
- Lemire, J.A.; Harrison, J.J.; Turner, R.J. Antimicrobial activity of metals: Mechanisms, molecular targets and applications. Nat. Rev. Microbiol. 2013, 11, 371–384. [Google Scholar] [CrossRef]
- Borkow, G.; Gabbay, J. Copper, an ancient remedy returning to fight microbial, fungal and viral infections. Curr. Chem. Biol. 2009, 3, 272–278. [Google Scholar]
- Pricker, S.P. Medical uses of gold compounds: Past, present and future. Gold Bull. 1996, 29, 53–60. [Google Scholar] [CrossRef]
- Alexander, J.W.; Chen, Y.; Deng, Y.; Pu, Y.; Tang, B.; Su, Y.; Tang, J.; Wildt, B.E.; Celedon, A.; Maurer, E.I.; et al. History of the Medical Use of Silver. Surg. Infect. 2009, 10, 289–292. [Google Scholar] [CrossRef]
- Muller, M.; MacDougall, C.; Lim, M.; Armstrong, I.; Bialachowski, A.; Callery, S.; Ciccotelli, W.; Cividino, M.; Dennis, J.; Hota, S.; et al. Antimicrobial surfaces to prevent healthcare-associated infections: A systematic review. J. Hosp. Infect. 2016, 92, 7–13. [Google Scholar] [CrossRef]
- Rai, M.; Yadav, A.; Gade, A. Silver nanoparticles as a new generation of antimicrobials. Biotechnol. Adv. 2009, 27, 76–83. [Google Scholar] [CrossRef] [PubMed]
- Page, K.; Wilson, M.; Parkin, I.P. Antimicrobial surfaces and their potential in reducing the role of the inanimate environment in the incidence of hospital-acquired infections. J. Mater. Chem. 2009, 19, 3819–3831. [Google Scholar] [CrossRef]
- Woodward, B. Final Analysis: Palladium in Temporary and Permanently Implantable Medical Devices. Platin. Met. Rev. 2012, 56, 213–217. [Google Scholar] [CrossRef]
- Baltzer, N.; Copponnex, T. Precious Metals for Biomedical Applications; Elsevier Science & Technology: Amsterdam, The Netherlands, 2014. [Google Scholar]
- Cowley, A.; Woodward, B. A Healthy Future: Platinum in Medical Applications. Platin. Met. Rev. 2011, 55, 98–107. [Google Scholar] [CrossRef]
- Abdulkareem, E.H.; Memarzadeh, K.; Allaker, R.; Huang, J.; Pratten, J.; Spratt, D. Anti-biofilm activity of zinc oxide and hydroxyapatite nanoparticles as dental implant coating materials. J. Dent. 2015, 43, 1462–1469. [Google Scholar] [CrossRef]
- Sirelkhatim, A.; Mahmud, S.; Seeni, A.; Kaus, N.H.M.; Ann, L.C.; Bakhori, S.K.M.; Hasan, H.; Mohamad, D. Review on Zinc Oxide Nanoparticles: Antibacterial Activity and Toxicity Mechanism. Nano-Micro Lett. 2015, 7, 219–242. [Google Scholar] [CrossRef]
- Shahverdi, A.R.; Fakhimi, A.; Shahverdi, H.R.; Minaian, S. Synthesis and effect of silver nanoparticles on the antibacterial activity of different antibiotics against Staphylococcus aureus and Escherichia coli. Nanomed. Nanotechnol. Biol. Med. 2007, 3, 168–171. [Google Scholar] [CrossRef]
- Zhang, J.; Chen, Y.P.; Miller, K.P.; Ganewatta, M.; Bam, M.; Yan, Y.; Nagarkatti, M.; Decho, A.W.; Tang, C. Antimicrobial Metallopolymers and Their Bioconjugates with Conventional Antibiotics against Multidrug-Resistant Bacteria. J. Am. Chem. Soc. 2014, 136, 4873–4876. [Google Scholar] [CrossRef] [PubMed]
- Vaidya, M.Y.; McBain, A.J.; Butler, J.A.; Banks, C.E.; Whitehead, K.A. Antimicrobial Efficacy and Synergy of Metal Ions against Enterococcus faecium, Klebsiella pneumoniae and Acinetobacter baumannii in Planktonic and Biofilm Phenotypes. Sci. Rep. 2017, 7, 5911. [Google Scholar] [CrossRef] [PubMed]
- Akhavan, O.; Ghaderi, E. Toxicity of graphene and graphene oxide nanowalls against bacteria. ACS Nano 2010, 4, 5731–5736. [Google Scholar] [CrossRef] [PubMed]
- Feng, L.; Liu, Z. Graphene in biomedicine: Opportunities and challenges. Nanomedicine 2011, 6, 317–324. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Zeng, T.H.; Hofmann, M.; Burcombe, E.; Wei, J.; Jiang, R.; Kong, J.; Chen, Y. Antibacterial Activity of Graphite, Graphite Oxide, Graphene Oxide, and Reduced Graphene Oxide: Membrane and Oxidative Stress. ACS Nano 2011, 5, 6971–6980. [Google Scholar] [CrossRef]
- Slate, A.; Karaky, N.; Whitehead, K. Antimicrobial Properties of Modified Graphene and other Advanced 2D Material Coated Surfaces. In Advanced 2D Material–Characterisation, Production and Applications; Banks, C., Brownson, D., Eds.; CRC Press: Boca Raton, FL, USA, 2017. [Google Scholar]
- Wang, J.; Huang, N.; Yang, P.; Leng, Y.; Sun, H.; Liu, Z.; Chu, P. The effects of amorphous carbon films deposited on polyethylene terephthalate on bacterial adhesion. Biomaterials 2004, 25, 3163–3170. [Google Scholar] [CrossRef] [PubMed]
- Erridge, C.; Bennett-Guerrero, E.; Poxton, I.R. Structure and function of lipopolysaccharides. Microbes Infect. 2002, 4, 837–851. [Google Scholar] [CrossRef]
- Kim, S.J.; Chang, J.; Singh, M. Peptidoglycan architecture of Gram-positive bacteria by solid-state NMR. Biochim. Biophys. Acta (BBA)-Biomembr. 2015, 1848, 350–362. [Google Scholar] [CrossRef]
- Shockman, G.D.; Barrett, J.F. Structure, function, and assembly of cell walls of gram-positive bacteria. Annu. Rev. Microbiol. 1983, 37, 501–527. [Google Scholar] [CrossRef] [PubMed]
- Singleton, P. Bacteria in Biology, Biotechnology and Medicine; John Wiley & Sons: Hoboken, NJ, USA, 2004. [Google Scholar]
- Johnson, J.R.; Stamm, W.E. Urinary tract infections in women: Diagnosis and treatment. Ann. Intern. Med. 1989, 111, 906–917. [Google Scholar] [CrossRef] [PubMed]
- Foxman, B.; Barlow, R.; D’Arcy, H.; Gillespie, B.; Sobel, J.D. Urinary Tract Infection: Self-Reported Incidence and Associated Costs. Ann. Epidemiol. 2000, 10, 509–515. [Google Scholar] [CrossRef]
- Ryan, K.J.; Ray, C.G. Medical Microbiology; McGraw Hill: New York, NY, USA, 2004; Volume 4, p. 370. [Google Scholar]
- Weinstein, R.A.; Gaynes, R.; Edwards, J.R.; System, N.N.I.S. Overview of Nosocomial Infections Caused by Gram-Negative Bacilli. Clin. Infect. Dis. 2005, 41, 848–854. [Google Scholar] [CrossRef]
- Flores-Mireles, A.L.; Walker, J.N.; Caparon, M.; Hultgren, S.J. Urinary tract infections: Epidemiology, mechanisms of infection and treatment options. Nat. Rev. Microbiol. 2015, 13, 269–284. [Google Scholar] [CrossRef] [PubMed]
- Top, J.; Willems, R.; Bonten, M. Emergence of CC17 Enterococcus faecium: From commensal to hospital-adapted pathogen. FEMS Immunol. Med. Microbiol. 2008, 52, 297–308. [Google Scholar] [CrossRef] [PubMed]
- Arias, C.A.; Murray, B.E. The rise of the Enterococcus: Beyond vancomycin resistance. Nat. Rev. Microbiol. 2012, 10, 266–278. [Google Scholar] [CrossRef]
- Arias, C.A.; Murray, B.E. Emergence and management of drug-resistant enterococcal infections. Expert Rev. Anti-Infect. Ther. 2008, 6, 637–655. [Google Scholar] [CrossRef] [PubMed]
- Grice, E.A.; Kong, H.H.; Conlan, S.; Deming, C.B.; Davis, J.; Young, A.C.; NISC Comparative Sequencing Program; Bouffard, G.G.; Blakesley, R.W.; Murray, P.R.; et al. Topographical and Temporal Diversity of the Human Skin Microbiome. Science 2009, 324, 1190–1192. [Google Scholar] [CrossRef]
- Schenck, L.P.; Surette, M.G.; Bowdish, D.M.E. Composition and immunological significance of the upper respiratory tract microbiota. FEBS Lett. 2016, 590, 3705–3720. [Google Scholar] [CrossRef]
- Tong, S.Y.C.; Davis, J.S.; Eichenberger, E.; Holland, T.L.; Fowler, V.G., Jr. Staphylococcus aureus Infections: Epidemiology, Pathophysiology, Clinical Manifestations, and Management. Clin. Microbiol. Rev. 2015, 28, 603–661. [Google Scholar] [CrossRef]
- Li, W.-R.; Xie, X.-B.; Shi, Q.-S.; Zeng, H.-Y.; Ou-Yang, Y.-S.; Chen, Y.-B. Antibacterial activity and mechanism of silver nanoparticles on Escherichia coli. Appl. Microbiol. Biotechnol. 2009, 85, 1115–1122. [Google Scholar] [CrossRef]
- Sondi, I.; Salopek-Sondi, B. Silver nanoparticles as antimicrobial agent: A case study on E. coli as a model for Gram-negative bacteria. J. Colloid Interface Sci. 2004, 275, 177–182. [Google Scholar] [CrossRef] [PubMed]
- Jung, W.K.; Koo, H.C.; Kim, K.W.; Shin, S.; Kim, S.H.; Park, Y.H. Antibacterial Activity and Mechanism of Action of the Silver Ion in Staphylococcus aureus and Escherichia coli. Appl. Environ. Microbiol. 2008, 74, 2171–2178. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.S.; Kuk, E.; Yu, K.N.; Kim, J.-H.; Park, S.J.; Lee, H.J.; Kim, S.H.; Park, Y.K.; Park, Y.H.; Hwang, C.-Y.; et al. Antimicrobial effects of silver nanoparticles. Nanomed. Nanotechnol. Boil. Med. 2007, 3, 95–101, Erratum in Nanomed. Nanotechnol. Biol. Med. 2014, 10, e1119. [Google Scholar] [CrossRef] [PubMed]
- Bragg, P.D.; Rainnie, D.J. The effect of silver ions on the respiratory chain of Escherichia coli. Can. J. Microbiol. 1974, 20, 883–889. [Google Scholar] [CrossRef]
- Webster, T.J.; Seil, J.T. Antimicrobial applications of nanotechnology: Methods and literature. Int. J. Nanomed. 2012, 7, 2767–2781. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; He, Y.; Irwin, P.L.; Jin, T.; Shi, X. Antibacterial Activity and Mechanism of Action of Zinc Oxide Nanoparticles against Campylobacter jejuni. Appl. Environ. Microbiol. 2011, 77, 2325–2331. [Google Scholar] [CrossRef]
- Nair, S.; Sasidharan, A.; Rani, V.V.D.; Menon, D.; Nair, S.; Manzoor, K.; Raina, S. Role of size scale of ZnO nanoparticles and microparticles on toxicity toward bacteria and osteoblast cancer cells. J. Mater. Sci. Mater. Med. 2008, 20, 235–241. [Google Scholar] [CrossRef]
- Padmavathy, N.; Vijayaraghavan, R. Enhanced bioactivity of ZnO nanoparticles—An antimicrobial study. Sci. Technol. Adv. Mater. 2008, 9, 035004. [Google Scholar] [CrossRef]
- Chen, J.; Peng, H.; Wang, X.; Shao, F.; Yuan, Z.; Han, H. Graphene oxide exhibits broad-spectrum antimicrobial activity against bacterial phytopathogens and fungal conidia by intertwining and membrane perturbation. Nanoscale 2013, 6, 1879–1889. [Google Scholar] [CrossRef]
- Perreault, F.; de Faria, A.F.; Nejati, S.; Elimelech, M. Antimicrobial Properties of Graphene Oxide Nanosheets: Why Size Matters. ACS Nano 2015, 9, 7226–7236. [Google Scholar] [CrossRef]
- Tu, Y.; Lv, M.; Xiu, P.; Huynh, T.; Zhang, M.; Castelli, M.; Liu, Z.; Huang, Q.; Fan, C.; Fang, H.; et al. Destructive extraction of phospholipids from Escherichia coli membranes by graphene nanosheets. Nat. Nanotechnol. 2013, 8, 594–601. [Google Scholar] [CrossRef] [PubMed]
- Krishnamoorthy, K.; Veerapandian, M.; Zhang, L.-H.; Yun, K.; Kim, S.J. Antibacterial Efficiency of Graphene Nanosheets against Pathogenic Bacteria via Lipid Peroxidation. J. Phys. Chem. C 2012, 116, 17280–17287. [Google Scholar] [CrossRef]
- Rietschel, E.T.; Kirikae, T.; Schade, F.U.; Mamat, U.; Schmidt, G.; Loppnow, H.; Ulmer, A.J.; Zähringer, U.; Seydel, U.; Di Padova, F.; et al. Bacterial endotoxin: Molecular relationships of structure to activity and function. FASEB J. 1994, 8, 217–225. [Google Scholar] [CrossRef]
- Silhavy, T.J.; Kahne, D.; Walker, S. The bacterial cell envelope. Cold Spring Harb. Perspect. Biol. 2010, 2, a000414. [Google Scholar] [CrossRef] [PubMed]
- Meroueh, S.O.; Bencze, K.Z.; Hesek, D.; Lee, M.; Fisher, J.F.; Stemmler, T.L.; Mobashery, S. Three-dimensional structure of the bacterial cell wall peptidoglycan. Proc. Natl. Acad. Sci. USA 2006, 103, 4404–4409. [Google Scholar] [CrossRef] [PubMed]
- Vollmer, W.; Blanot, D.; De Pedro, M.A. Peptidoglycan structure and architecture. FEMS Microbiol. Rev. 2008, 32, 149–167. [Google Scholar] [CrossRef] [PubMed]
- Schirmer, T.; Keller, T.A.; Wang, Y.-F.; Rosenbusch, J.P. Structural Basis for Sugar Translocation Through Maltoporin Channels at 3.1 Å Resolution. Science 1995, 267, 512–514. [Google Scholar] [CrossRef]
- Macnab, R.M. How Bacteria Assemble Flagella. Annu. Rev. Microbiol. 2003, 57, 77–100. [Google Scholar] [CrossRef]
- Symmons, M.F.; Bokma, E.; Koronakis, E.; Hughes, C.; Koronakis, V. The assembled structure of a complete tripartite bacterial multidrug efflux pump. Proc. Natl. Acad. Sci. USA 2009, 106, 7173–7178. [Google Scholar] [CrossRef]
- Navarre, W.W.; Schneewind, O. Surface Proteins of Gram-Positive Bacteria and Mechanisms of Their Targeting to the Cell Wall Envelope. Microbiol. Mol. Biol. Rev. 1999, 63, 174–229. [Google Scholar] [CrossRef]
- Bellais, S.; Arthur, M.; Dubost, L.; Hugonnet, J.-E.; Gutmann, L.; van Heijenoort, J.; Legrand, R.; Brouard, J.-P.; Rice, L.; Mainardi, J.-L. Aslfm, the D-Aspartate Ligase Responsible for the Addition of D-Aspartic Acid onto the Peptidoglycan Precursor of Enterococcus faecium. J. Biol. Chem. 2006, 281, 11586–11594. [Google Scholar] [CrossRef]
- Tong, G.; Pan, Y.; Dong, H.; Pryor, R.; Wilson, G.E.; Schaefer, J. Structure and Dynamics of Pentaglycyl Bridges in the Cell Walls of Staphylococcus aureus by 13C− 15N REDOR NMR. Biochemistry 1997, 36, 9859–9866. [Google Scholar] [CrossRef]
- Leung, Y.H.; Xu, X.; Ma, A.P.Y.; Liu, F.; Ng, A.M.C.; Shen, Z.; Gethings, L.A.; Guo, M.Y.; Djurišić, A.B.; Lee, P.K.H.; et al. Toxicity of ZnO and TiO2 to Escherichia coli cells. Sci. Rep. 2016, 6, 35243. [Google Scholar] [CrossRef]
- Ansari, M.A.; Khan, H.M.; Khan, A.A.; Cameotra, S.S.; Saquib, Q.; Musarrat, J. Interaction of Al2O3 nanoparticles with Escherichia coli and their cell envelope biomolecules. J. Appl. Microbiol. 2014, 116, 772–783. [Google Scholar] [CrossRef]
- Whitehead, K.; Vaidya, M.; Liauw, C.; Brownson, D.; Ramalingam, P.; Kamieniak, J.; Rowley-Neale, S.; Tetlow, L.; Wilson-Nieuwenhuis, J.; Brown, D.; et al. Antimicrobial activity of graphene oxide-metal hybrids. Int. Biodeterior. Biodegrad. 2017, 123, 182–190. [Google Scholar] [CrossRef]
- Hummers, W.S., Jr.; Offeman, R.E. Preparation of graphitic oxide. J. Am. Chem. Soc. 1958, 80, 1339. [Google Scholar] [CrossRef]
- Anandan, S.; Muthukumaran, S. Microstructural, crystallographic and optical characterizations of Cu-doped ZnO nanoparticles co-doped with Ni. J. Mater. Sci. Mater. Electron. 2015, 26, 4298–4307. [Google Scholar] [CrossRef]
- Liu, Q.; Yao, X.; Zhou, X.; Qin, Z.; Liu, Z. Varistor effect in Ag–graphene/epoxy resin nanocomposites. Scr. Mater. 2012, 66, 113–116. [Google Scholar] [CrossRef]
- Rajab, F.H.; Liauw, C.M.; Benson, P.S.; Li, L.; Whitehead, K.A. Production of hybrid macro/micro/nano surface structures on Ti6Al4V surfaces by picosecond laser surface texturing and their antifouling characteristics. Colloids Surf. B Biointerfaces 2017, 160, 688–696. [Google Scholar] [CrossRef] [PubMed]
- Miles, A.A.; Misra, S.S.; Irwin, J.O. The estimation of the bactericidal power of the blood. Epidemiol. Infect. 1938, 38, 732–749. [Google Scholar] [CrossRef]



| Metric Number | Metric | Abbreviation | Wave Number Range (cm−1) | Comments |
|---|---|---|---|---|
| 1 | Amide 1 and ester shift | ∆ν(C=O)Amide1+ester | 1700–1680 | Interaction with amide I |
| 2 | Absorbance ratio (amide I + C=O) to (C-N from R-NH2 and R-NH3+) | AbsC=O/AbsC-N | 1670–1615 | Interaction of amine groups with candidate antimicrobials |
| 3 | a-methylene and/or C-O (deprotonated carboxylate) shift | ∆(δα > CH-H) + νs(C-O) | 1400–1430 | Indicative of changes in the order of LPS or peptidoglycan structures |
| 4 | Change in hydrogen bonded OH/NH peak width | ∆PW(OH + NH) | 3600–2800 | Indicative of changes in the order of LPS or peptidoglycan structures |
| 5 | Alkenic C-H stretch shift | ∆ν(H-CH=) | 3060–3090 | Interaction with amides |
| 6 | Methyl asymmetric C-H stretch shift | ∆νas(H-CH2-) | 2955–2980 | Indicative of changes in the order of LPS or peptidoglycan structures |
| 7 | Methylene asymmetric C-H stretch shift | ∆νas(H-CH<) | 2930–2942 | Indicative of changes in the order of LPS or peptidoglycan structures |
| 8 | Methyl symmetric C-H stretch shift | ∆ns(H-CH2-) | 2873–2883 | Indicative of changes in the order of LPS or peptidoglycan structures |
| 9 | Perturbation index | PI | Sum of metrics 1 to 8 * | Provides a single figure indicating the amount of spectral perturbation that may be related to cell damage |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liauw, C.M.; Vaidya, M.; Slate, A.J.; Hickey, N.A.; Ryder, S.; Martínez-Periñán, E.; McBain, A.J.; Banks, C.E.; Whitehead, K.A. Analysis of Cellular Damage Resulting from Exposure of Bacteria to Graphene Oxide and Hybrids Using Fourier Transform Infrared Spectroscopy. Antibiotics 2023, 12, 776. https://doi.org/10.3390/antibiotics12040776
Liauw CM, Vaidya M, Slate AJ, Hickey NA, Ryder S, Martínez-Periñán E, McBain AJ, Banks CE, Whitehead KA. Analysis of Cellular Damage Resulting from Exposure of Bacteria to Graphene Oxide and Hybrids Using Fourier Transform Infrared Spectroscopy. Antibiotics. 2023; 12(4):776. https://doi.org/10.3390/antibiotics12040776
Chicago/Turabian StyleLiauw, Christopher M., Misha Vaidya, Anthony J. Slate, Niall A. Hickey, Steven Ryder, Emiliano Martínez-Periñán, Andrew J. McBain, Craig E. Banks, and Kathryn A. Whitehead. 2023. "Analysis of Cellular Damage Resulting from Exposure of Bacteria to Graphene Oxide and Hybrids Using Fourier Transform Infrared Spectroscopy" Antibiotics 12, no. 4: 776. https://doi.org/10.3390/antibiotics12040776
APA StyleLiauw, C. M., Vaidya, M., Slate, A. J., Hickey, N. A., Ryder, S., Martínez-Periñán, E., McBain, A. J., Banks, C. E., & Whitehead, K. A. (2023). Analysis of Cellular Damage Resulting from Exposure of Bacteria to Graphene Oxide and Hybrids Using Fourier Transform Infrared Spectroscopy. Antibiotics, 12(4), 776. https://doi.org/10.3390/antibiotics12040776






