Antimicrobial Susceptibility of Bacteria Isolated from Freshwater Mussels in the Wildcat Creek Watershed, Indiana, United States
Abstract
:1. Introduction
2. Results
2.1. Bacteria Isolated
2.2. Antimicrobial Susceptibility
3. Discussion
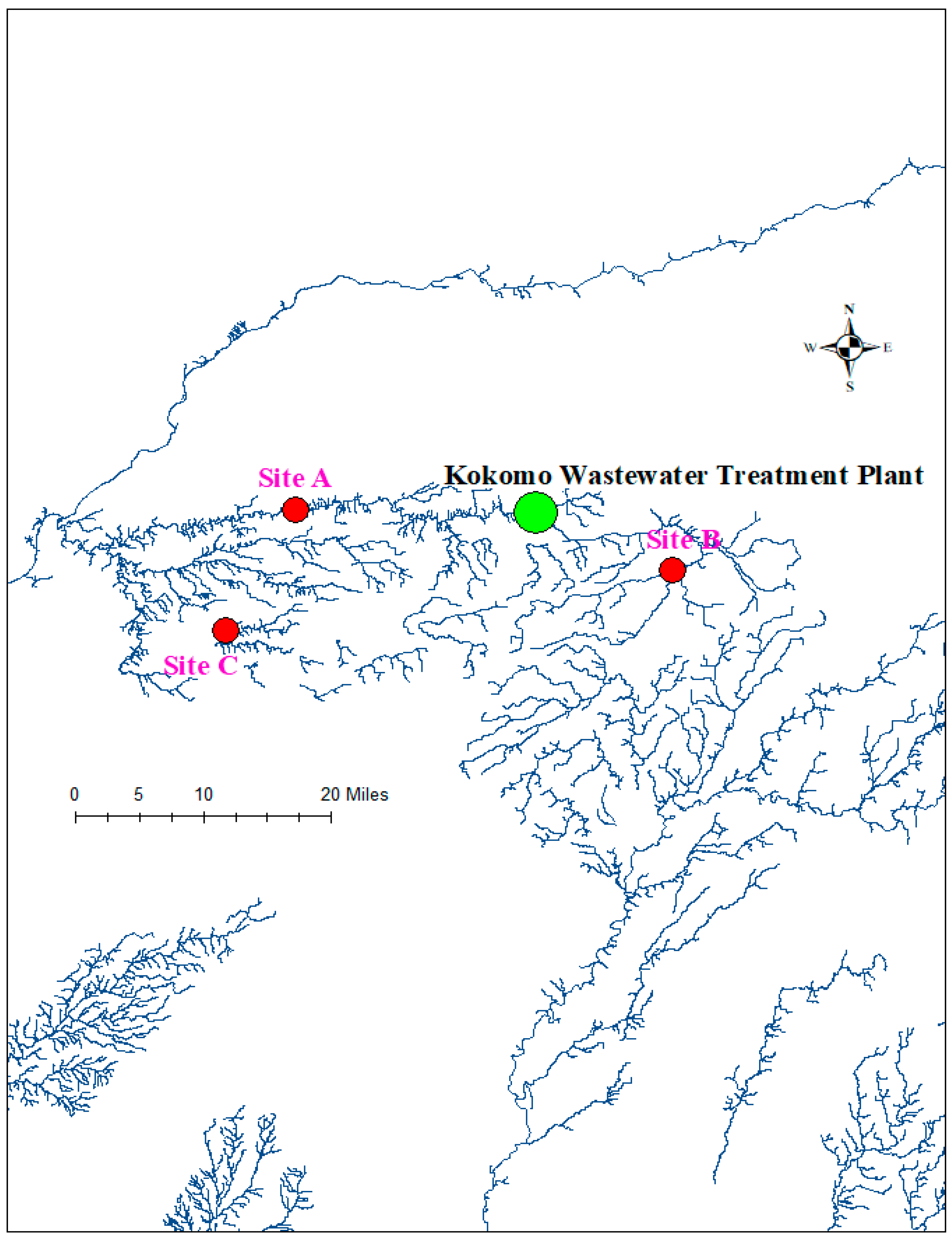
4. Materials and Methods
4.1. Test Organisms and Tissue Preparation
4.2. Bacterial Culture
4.2.1. Aerobic Culture
4.2.2. Campylobacter Culture
4.2.3. Salmonella Culture
4.2.4. Bacterial Isolate Identification
4.3. Antimicrobial Susceptibility Testing
4.4. Data Analyses
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Köck, R.; Kreienbrock, L.; van Duijkeren, E.; Schwarz, S. Antimicrobial Resistance at the Interface of Human and Veterinary Medicine. Vet. Microbiol. 2017, 200, 1–5. [Google Scholar] [CrossRef]
- Aslam, B.; Wang, W.; Arshad, M.I.; Khurshid, M.; Muzammil, S.; Rasool, M.H.; Nisar, M.A.; Alvi, R.F.; Aslam, M.A.; Qamar, M.U.; et al. Antibiotic Resistance: A Rundown of a Global Crisis. Infect. Drug Resist. 2018, 11, 1645–1658. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Christaki, E.; Marcou, M.; Tofarides, A. Antimicrobial Resistance in Bacteria: Mechanisms, Evolution, and Persistence. J. Mol. Evol. 2020, 88, 26–40. [Google Scholar] [CrossRef]
- D’Costa, V.M.; King, C.E.; Kalan, L.; Morar, M.; Sung, W.W.L.; Schwarz, C.; Froese, D.; Zazula, G.; Calmels, F.; Debruyne, R.; et al. Antibiotic Resistance Is Ancient. Nature 2011, 477, 457–461. [Google Scholar] [CrossRef] [PubMed]
- Holmes, A.H.; Moore, L.S.P.; Sundsfjord, A.; Steinbakk, M.; Regmi, S.; Karkey, A.; Guerin, P.J.; Piddock, L.J.V. Understanding the Mechanisms and Drivers of Antimicrobial Resistance. Lancet 2016, 387, 176–187. [Google Scholar] [CrossRef] [PubMed]
- Prestinaci, F.; Pezzotti, P.; Pantosti, A. Antimicrobial Resistance: A Global Multifaceted Phenomenon. Pathog. Glob. Health 2015, 109, 309–318. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ten Health Issues WHO Will Tackle This Year. Available online: https://www.who.int/news-room/spotlight/ten-threats-to-global-health-in-2019 (accessed on 27 January 2023).
- Klein, E.Y.; Van Boeckel, T.P.; Martinez, E.M.; Pant, S.; Gandra, S.; Levin, S.A.; Goossens, H.; Laxminarayan, R. Global Increase and Geographic Convergence in Antibiotic Consumption between 2000 and 2015. Proc. Natl. Acad. Sci. USA 2018, 115, E3463–E3470. [Google Scholar] [CrossRef] [Green Version]
- Grundmann, H.; Klugman, K.P.; Walsh, T.; Ramon-Pardo, P.; Sigauque, B.; Khan, W.; Laxminarayan, R.; Heddini, A.; Stelling, J. A Framework for Global Surveillance of Antibiotic Resistance. Drug Resist. Updat. 2011, 14, 79–87. [Google Scholar] [CrossRef]
- Kusi, J.; Ojewole, C.O.; Ojewole, A.E.; Nwi-Mozu, I. Antimicrobial Resistance Development Pathways in Surface Waters and Public Health Implications. Antibiotics 2022, 11, 821. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.; Singh, A.P.; Kumar, S.; Giri, B.S.; Kim, K.-H. Antibiotic Resistance in Major Rivers in the World: A Systematic Review on Occurrence, Emergence, and Management Strategies. J. Clean. Prod. 2019, 234, 1484–1505. [Google Scholar] [CrossRef]
- Xu, J.; Xu, Y.; Wang, H.; Guo, C.; Qiu, H.; He, Y.; Zhang, Y.; Li, X.; Meng, W. Occurrence of Antibiotics and Antibiotic Resistance Genes in a Sewage Treatment Plant and Its Effluent-Receiving River. Chemosphere 2015, 119, 1379–1385. [Google Scholar] [CrossRef]
- Rodriguez-Mozaz, S.; Chamorro, S.; Marti, E.; Huerta, B.; Gros, M.; Sànchez-Melsió, A.; Borrego, C.M.; Barceló, D.; Balcázar, J.L. Occurrence of Antibiotics and Antibiotic Resistance Genes in Hospital and Urban Wastewaters and Their Impact on the Receiving River. Water Res. 2015, 69, 234–242. [Google Scholar] [CrossRef]
- Pruden, A.; Vikesland, P.J.; Davis, B.C.; de Roda Husman, A.M. Seizing the Moment: Now Is the Time for Integrated Global Surveillance of Antimicrobial Resistance in Wastewater Environments. Curr. Opin. Microbiol. 2021, 64, 91–99. [Google Scholar] [CrossRef]
- Huijbers, P.M.C.; Blaak, H.; de Jong, M.C.M.; Graat, E.A.M.; Vandenbroucke-Grauls, C.M.J.E.; de Roda Husman, A.M. Role of the Environment in the Transmission of Antimicrobial Resistance to Humans: A Review. Environ. Sci. Technol. 2015, 49, 11993–12004. [Google Scholar] [CrossRef] [PubMed]
- White, A.; Hughes, J.M. Critical Importance of a One Health Approach to Antimicrobial Resistance. EcoHealth 2019, 16, 404–409. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Environmental Sampling|Background|Environmental Guidelines|Guidelines Library|Infection Control|CDC. Available online: https://www.cdc.gov/infectioncontrol/guidelines/environmental/background/sampling.html (accessed on 27 January 2023).
- Vaughn, C.C. Ecosystem Services Provided by Freshwater Mussels. Hydrobiologia 2018, 810, 15–27. [Google Scholar] [CrossRef]
- Chiesa, L.M.; Nobile, M.; Malandra, R.; Panseri, S.; Arioli, F. Occurrence of Antibiotics in Mussels and Clams from Various FAO Areas. Food Chem. 2018, 240, 16–23. [Google Scholar] [CrossRef] [Green Version]
- Boufafa, M.; Kadri, S.; Redder, P.; Bensouilah, M. Occurrence and Distribution of Fecal Indicators and Pathogenic Bacteria in Seawater and Perna Perna Mussel in the Gulf of Annaba (Southern Mediterranean). Env. Sci. Pollut. Res. 2021, 28, 46035–46052. [Google Scholar] [CrossRef] [PubMed]
- Ilic, N.; Velebit, B.; Teodorovic, V.; Djordjevic, V.; Karabasil, N.; Vasilev, D.; Djuric, S.; Adzic, B.; Dimitrijevic, M. Influence of Environmental Conditions on Norovirus Presence in Mussels Harvested in Montenegro. Food Env. Virol. 2017, 9, 406–414. [Google Scholar] [CrossRef] [PubMed]
- Leis, E.; Erickson, S.; Waller, D.; Richard, J.; Goldberg, T. A Comparison of Bacteria Cultured from Unionid Mussel Hemolymph between Stable Populations in the Upper Mississippi River Basin and Populations Affected by a Mortality Event in the Clinch River. FMBC 2019, 22, 70–80. [Google Scholar] [CrossRef]
- Vaughn, C.C.; Nichols, S.J.; Spooner, D.E. Community and Foodweb Ecology of Freshwater Mussels. J. North Am. Benthol. Soc. 2008, 27, 409–423. [Google Scholar] [CrossRef]
- Strubbia, S.; Lyons, B.P.; Lee, R.J. Geographical and Temporal Variation of E. Coli and Norovirus in Mussels. Mar. Pollut. Bull. 2016, 107, 66–70. [Google Scholar] [CrossRef] [PubMed]
- Bighiu, M.A.; Norman Haldén, A.; Goedkoop, W.; Ottoson, J. Assessing Microbial Contamination and Antibiotic Resistant Bacteria Using Zebra Mussels (Dreissena Polymorpha). Sci. Total Env. 2019, 650, 2141–2149. [Google Scholar] [CrossRef] [PubMed]
- Cooke, M.D. Antibiotic Resistance among Coliform and Fecal Coliform Bacteria Isolated from the Freshwater Mussel Hydridella Menziesii. Antimicrob. Agents Chemother. 1976, 9, 885–888. [Google Scholar] [CrossRef] [Green Version]
- Saavedra, M.J.; Fernandes, C.; Teixeira, A.; Álvarez, X.; Varandas, S. Multiresistant Bacteria: Invisible Enemies of Freshwater Mussels. Environ. Pollut. 2022, 295, 118671. [Google Scholar] [CrossRef]
- Zacharias, N.; Löckener, I.; Essert, S.M.; Sib, E.; Bierbaum, G.; Kistemann, T.; Schreiber, C. Antibiotic-Resistant Bacteria in Clams—A Study on Mussels in the River Rhine. Antibiotics 2021, 10, 571. [Google Scholar] [CrossRef]
- Fincher, L.M.; Parker, C.D.; Chauret, C.P. Occurrence and Antibiotic Resistance of Escherichia Coli O157:H7 in a Watershed in North-Central Indiana. J. Env. Qual. 2009, 38, 997–1004. [Google Scholar] [CrossRef]
- Indiana Department of Environmental Management Wildcat Creek Upper. Available online: https://www.in.gov/idem/nps/resources/total-maximum-daily-load-reports/wildcat-creek-upper/ (accessed on 31 January 2023).
- Mukherjee, M.; Laird, E.; Gentry, T.J.; Brooks, J.P.; Karthikeyan, R. Increased Antimicrobial and Multidrug Resistance Downstream of Wastewater Treatment Plants in an Urban Watershed. Front. Microbiol. 2021, 12, 657353. [Google Scholar] [CrossRef]
- Khan, G.A.; Berglund, B.; Khan, K.M.; Lindgren, P.-E.; Fick, J. Occurrence and Abundance of Antibiotics and Resistance Genes in Rivers, Canal and near Drug Formulation Facilities—A Study in Pakistan. PLoS ONE 2013, 8, e62712. [Google Scholar] [CrossRef]
- Sabri, N.A.; Schmitt, H.; Van der Zaan, B.; Gerritsen, H.W.; Zuidema, T.; Rijnaarts, H.H.M.; Langenhoff, A.A.M. Prevalence of Antibiotics and Antibiotic Resistance Genes in a Wastewater Effluent-Receiving River in the Netherlands. J. Environ. Chem. Eng. 2020, 8, 102245. [Google Scholar] [CrossRef]
- Iwane, T.; Urase, T.; Yamamoto, K. Possible Impact of Treated Wastewater Discharge on Incidence of Antibiotic Resistant Bacteria in River Water. Water Sci. Technol. 2001, 43, 91–99. [Google Scholar] [CrossRef]
- Weingarten, E.A.; Atkinson, C.L.; Jackson, C.R. The Gut Microbiome of Freshwater Unionidae Mussels Is Determined by Host Species and Is Selectively Retained from Filtered Seston. PLoS ONE 2019, 14, e0224796. [Google Scholar] [CrossRef]
- Gill, S.P.; Learman, D.R.; Annis, M.L.; Woolnough, D.A. Freshwater Mussels and Host Fish Gut Microbe Community Composition Shifts after Agricultural Contaminant Exposure. J. Appl. Microbiol. 2022, 133, 3645–3658. [Google Scholar] [CrossRef] [PubMed]
- Poirel, L.; Madec, J.-Y.; Lupo, A.; Schink, A.-K.; Kieffer, N.; Nordmann, P.; Schwarz, S. Antimicrobial Resistance in Escherichia Coli. Microbiol. Spectr. 2018, 6, 4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- World Health Organization. WHO Guidelines on Use of Medically Important Antimicrobials in Food-Producing Animals; World Health Organization: Geneva, Switzerland, 2017; ISBN 978-92-4-155013-0. [Google Scholar]
- Wright, G.D. Antibiotic Resistance in the Environment: A Link to the Clinic? Curr. Opin. Microbiol. 2010, 13, 589–594. [Google Scholar] [CrossRef] [PubMed]
- Bengtsson-Palme, J.; Kristiansson, E.; Larsson, D.G.J. Environmental Factors Influencing the Development and Spread of Antibiotic Resistance. FEMS Microbiol. Rev. 2018, 42, fux053. [Google Scholar] [CrossRef]
- Bouza, E.; Cercenado, E. Klebsiella and Enterobacter: Antibiotic Resistance and Treatment Implications. Semin. Respir. Infect. 2002, 17, 215–230. [Google Scholar] [CrossRef]
- Lee, J.-H. Perspectives towards Antibiotic Resistance: From Molecules to Population. J. Microbiol. 2019, 57, 181–184. [Google Scholar] [CrossRef] [Green Version]
- Leclercq, R.; Cantón, R.; Brown, D.F.J.; Giske, C.G.; Heisig, P.; MacGowan, A.P.; Mouton, J.W.; Nordmann, P.; Rodloff, A.C.; Rossolini, G.M.; et al. EUCAST Expert Rules in Antimicrobial Susceptibility Testing. Clin. Microbiol. Infect. 2013, 19, 141–160. [Google Scholar] [CrossRef] [Green Version]
- Boerlin, P.; White, D.G. Antimicrobial Resistance and Its Epidemiology. In Antimicrobial Therapy in Veterinary Medicine; Giguère, S., Prescott, J.F., Dowling, P.M., Eds.; Wiley: Hoboken, NJ, USA, 2013; pp. 21–40. ISBN 978-0-470-96302-9. [Google Scholar]
- Tamma, P.D.; Doi, Y.; Bonomo, R.A.; Johnson, J.K.; Simner, P.J. Antibacterial Resistance Leadership Group A Primer on AmpC β-Lactamases: Necessary Knowledge for an Increasingly Multidrug-Resistant World. Clin. Infect. Dis. 2019, 69, 1446–1455. [Google Scholar] [CrossRef] [Green Version]
- Wyres, K.L.; Holt, K.E. Klebsiella Pneumoniae as a Key Trafficker of Drug Resistance Genes from Environmental to Clinically Important Bacteria. Curr. Opin. Microbiol. 2018, 45, 131–139. [Google Scholar] [CrossRef]
- Moya, C.; Maicas, S. Antimicrobial Resistance in Klebsiella Pneumoniae Strains: Mechanisms and Outbreaks. Proceedings 2020, 66, 11. [Google Scholar] [CrossRef]
- Glen, K.A.; Lamont, I.L. β-Lactam Resistance in Pseudomonas Aeruginosa: Current Status, Future Prospects. Pathogens 2021, 10, 1638. [Google Scholar] [CrossRef] [PubMed]
- Angeletti, S. Matrix Assisted Laser Desorption Time of Flight Mass Spectrometry (MALDI-TOF MS) in Clinical Microbiology. J. Microbiol. Methods 2017, 138, 20–29. [Google Scholar] [CrossRef] [PubMed]
- Veterinary Microbiology Standards Documents—CLSI Shop. Available online: https://clsi.org/standards/products/veterinary-medicine/documents/ (accessed on 27 January 2023).
- Sweeney, M.T.; Lubbers, B.V.; Schwarz, S.; Watts, J.L. Applying Definitions for Multidrug Resistance, Extensive Drug Resistance and Pandrug Resistance to Clinically Significant Livestock and Companion Animal Bacterial Pathogens-Authors’ Response. J. Antimicrob. Chemother. 2019, 74, 536–537. [Google Scholar] [CrossRef]
- Magiorakos, A.-P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-Resistant, Extensively Drug-Resistant and Pandrug-Resistant Bacteria: An International Expert Proposal for Interim Standard Definitions for Acquired Resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| Bacterial Organisms (Number of Isolates) | Antimicrobial Agents | Dilution Range (µg/mL) | M.I.C. (µg/mL) | Number of Susceptible Isolates |
|---|---|---|---|---|
| Escherichia coli (5) | Ampicillin | 0.25–16 | 2–4 * | 5 |
| Penicillin | 0.12–8 | >8 | 0 | |
| Ceftiofur | 0.25–8 | ≤0.25–0.5 * | 5 | |
| Gentamycin | 1–16 | <1 | 5 | |
| Neomycin | 4–32 | ≤4 | 5 | |
| Spectinomycin | 8–64 | 16–32 * | 0 | |
| Tilmicosin | 2–16 | >16 | 0 | |
| Clindamycin | 0.25–16 | >16 | 0 | |
| Sulphadimethoxine | 256 | ≤256–>256 * | 4 | |
| Tetracycline | 0.5–8 | 1–2 * | 5 | |
| Tiamulin | 0.5–32 | >32 | 0 | |
| Florfenicol | 0.25–8 | 2–8 * | 1 | |
| Enterobacter cloacae (1) | Ampicillin | 0.25–16 | 16 | 0 |
| Penicillin | 0.12–8 | >8 | 0 | |
| Ceftiofur | 0.25–8 | 1 | 1 | |
| Gentamycin | 1–16 | ≤1 | 1 | |
| Neomycin | 4–32 | ≤4 | 1 | |
| Spectinomycin | 8–64 | 16 | 0 | |
| Tilmicosin | 2–16 | >16 | 0 | |
| Clindamycin | 0.25–16 | >16 | 0 | |
| Sulphadimethoxine | 256 | ≤256 | 1 | |
| Tetracycline | 0.5–8 | 2 | 1 | |
| Tiamulin | 0.5–32 | >32 | 0 | |
| Florfenicol | 0.25–8 | 4 | 0 | |
| Klebsiella pneumoniae (1) | Ampicillin | 0.25–16 | - | 0 |
| Penicillin | 0.12–8 | >8 | 0 | |
| Ceftiofur | 0.25–8 | 1 | 1 | |
| Gentamycin | 1–16 | ≤1 | 1 | |
| Neomycin | 4–32 | ≤4 | 1 | |
| Spectinomycin | 8–64 | 16 | 0 | |
| Tilmicosin | 2–16 | >16 | 0 | |
| Clindamycin | 0.25–16 | >16 | 0 | |
| Sulphadimethoxine | 256 | >256 | 0 | |
| Tetracycline | 0.5–8 | 4 | 1 | |
| Tiamulin | 0.5–32 | >32 | 0 | |
| Florfenicol | 0.25–8 | 4 | 0 | |
| Pseudomonas aeruginosa (1) | Ampicillin | 0.25–16 | >16 | 0 |
| Penicillin | 0.12–8 | >8 | 0 | |
| Ceftiofur | 0.25–8 | >8 | 0 | |
| Gentamycin | 1–16 | ≤1 | 1 | |
| Neomycin | 4–32 | ≤4 | 1 | |
| Spectinomycin | 8–64 | >64 | 0 | |
| Tilmicosin | 2–16 | >16 | 0 | |
| Clindamycin | 0.25–16 | >16 | 0 | |
| Sulphadimethoxine | 256 | >256 | 0 | |
| Tiamulin | 0.5–32 | >32 | 0 | |
| Florfenicol | 0.25–8 | >8 | 0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ekakoro, J.E.; Guptill, L.F.; Hendrix, G.K.; Dorsey, L.; Ruple, A. Antimicrobial Susceptibility of Bacteria Isolated from Freshwater Mussels in the Wildcat Creek Watershed, Indiana, United States. Antibiotics 2023, 12, 728. https://doi.org/10.3390/antibiotics12040728
Ekakoro JE, Guptill LF, Hendrix GK, Dorsey L, Ruple A. Antimicrobial Susceptibility of Bacteria Isolated from Freshwater Mussels in the Wildcat Creek Watershed, Indiana, United States. Antibiotics. 2023; 12(4):728. https://doi.org/10.3390/antibiotics12040728
Chicago/Turabian StyleEkakoro, John E., Lynn F. Guptill, G. Kenitra Hendrix, Lauren Dorsey, and Audrey Ruple. 2023. "Antimicrobial Susceptibility of Bacteria Isolated from Freshwater Mussels in the Wildcat Creek Watershed, Indiana, United States" Antibiotics 12, no. 4: 728. https://doi.org/10.3390/antibiotics12040728
APA StyleEkakoro, J. E., Guptill, L. F., Hendrix, G. K., Dorsey, L., & Ruple, A. (2023). Antimicrobial Susceptibility of Bacteria Isolated from Freshwater Mussels in the Wildcat Creek Watershed, Indiana, United States. Antibiotics, 12(4), 728. https://doi.org/10.3390/antibiotics12040728






