Abstract
Animals have been identified as potential reservoirs and vectors of resistance genes, with studies showing that Gram-negative bacteria can acquire resistance through the horizontal transmission of resistance genes on plasmids. It is important to understand the distribution of antimicrobial-resistant bacteria and their drug-resistant genes in animals. Previous review articles mostly focused on a single bacterium or a single animal. Our objective is to compile all ESBL-producing bacteria isolated from various animals in recent years and provide a comprehensive viewpoint. Using a thorough PubMed literature search spanning from 1 January 2020 to 30 June 2022, studies exploring extended-spectrum beta-lactamase (ESBL) producing bacteria in animals were included. ESBL-producing bacteria are present in animals from various countries around the world. The most common sources of these bacteria were farm animals, and the most frequently isolated bacteria were Escherichia coli and Klebsiella pneumoniae. The most detected ESBL genes were blaTEM, blaSHV, and blaCTX-M. The presence of ESBL-producing bacteria in animals highlights the importance of the One Health approach to address the issue of antibiotic resistance. Further research is needed to better understand the epidemiology and mechanisms of the spread of ESBL-producing bacteria in animal populations and their potential impact on human and animal health.
1. Introduction
Antibiotics are important weapons for humans in fighting microbial infections and reducing overall mortality from infectious diseases. However, the increasing prevalence of antimicrobial-resistant bacteria (AMRs) in recent decades is a great challenge [1]. Studies suggest that animals are potential reservoirs and vectors of resistance genes [2]. Gram-negative bacteria (GNB), especially Enterobacterales strains, can acquire resistance through the plasmid-mediated horizontal transmission of resistance genes [3]. Increased use of antibiotics in livestock has been identified as a potential contributor to antimicrobial resistance in humans [4]. Therefore, it is also important to understand the distribution of antimicrobial-resistant bacteria and their drug-resistant genes in animals.
Bacteria that produce extended-spectrum β-lactamases (ESBLs) are considered one of the critical priority pathogens by the World Health Organization (WHO). ESBLs are a type of β-lactamase that can hydrolyze penicillins, first, second, and third-generation cephalosporins, and aztreonam but are unable to break down cephamycins or carbapenems [5]. The ESBL-encoding genes can be grouped into several families: blaTEM, blaSHV, blaCTX-M, etc. In the past, TEM and SHV-type ESBLs were the mainstream of ESBLs. However, CTX-M-type enzymes are much more commonly found in the ESBL type in recent research [5].
The widespread use of antibiotics is a contributing factor in the rise of antimicrobial resistance, particularly in the case of ESBL-producing bacteria. Previous studies have shown that the use of antibiotics within the past three months and monotherapy with specific drug classes (cephalosporins, tetracycline, macrolide, and cotrimoxazole) are associated with the prevalence of these bacteria [6,7]. The plasmids responsible for ESBL production often carry genes encoding resistance to other drug classes, such as aminoglycosides, trimethoprim, and fluoroquinolones [8]. This makes the treatment of infections caused by ESBL-producing bacteria more challenging, as the presence of these plasmids exacerbates the problem of antibiotic resistance and limits therapeutic options. In short, the overuse of antibiotics creates a favorable environment for the spread of plasmids responsible for ESBL production, which in turn contributes to the rise of antibiotic resistance.
Animals have been identified as potential reservoirs and vectors of resistance genes. The widespread use of antibiotics in animals has been linked to the increasing prevalence of antimicrobial-resistant bacteria in humans, highlighting the importance of understanding the distribution of antimicrobial-resistant bacteria and their drug-resistant genes in both humans and animals.
There are many articles on the analysis of ESBL-producing bacteria and their drug-resistance genes in particular animals. While some review articles have attempted to summarize these studies, most of them have focused on a specific type of bacteria (such as Escherichia coli or Klebsiella pneumoniae) or only reviewed one type of animal, lacking a comprehensive review. Our objective is to compile all ESBL-producing bacteria isolated from various animals in recent years, providing a comprehensive understanding of the distribution of ESBL-producing bacteria and genes in animals worldwide. This review attempts to underscore the role of animals in the rising incidence of ESBL-producing bacteria and the need for a coordinated effort to address this growing threat.
2. Results
The general findings of the reviewed articles are summarized in Table 1. Samples of ESBL-producing bacteria were mostly obtained from farms in Africa (Egypt, Kenya, Tunisia, Nigeria, and Algeria), Asia (Pakistan, India, Qatar, Iran, Malaysia, China, Saudi Arabia, Bangladesh, and Thailand), Europe (Finland, Portugal, Spain, Netherlands, Germany, France, and Switzerland), North American (USA), and South America (Brazil, Guadeloupe, and Peru). Other sampling locations included the airport, animal clinics, animal shelters, hunting grounds, petting zoos, slaughterhouses, research facilities, universities, and wild colonies. Most samples were obtained from rectal swabs and fresh feces of animals. However, other samples including raw milk, blood and visceral samples, cloacal swabs, uterine swabs, external surface and gut homogenates, urine, pus, and respiratory pathological specimens were also included. The most reported bacteria were Escherichia coli and Klebsiella pneumoniae. Other Enterobacterales were also in abundance while Pseudomonas aeruginosa was only found on the uterine swabs of farm cows, camels, and mares in one study from Saudi Arabia.

Table 1.
Studies of ESBL-producing bacteria in animals.
Our review included 23 articles on domestic animals, 6 articles on wild animals, and 1 article on both. Other than farm animals, pets, zoo animals, vampire bats, and cockroaches were sampled. Four studies emphasized that the specimens were sourced from diseased animals, including diseased companion animals, diseased horses, diseased cows, camels, mares, and diseased pigs. The compilation of animals screened across different countries is presented in Figure 1a.
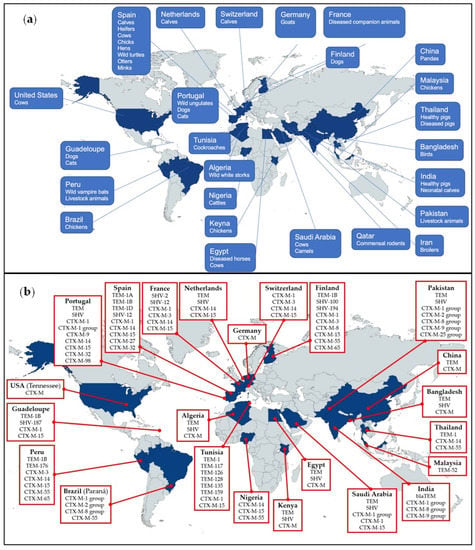
Figure 1.
(a) Countries that reported the presence of ESBL-producing bacteria in animals. (b) Countries that reported the presence of ESBL-subtype distribution in animals.
Table 2 summarizes the details of the ESBL genes. Most samples were grown using MacConkey agar. Fifteen articles included in our review utilized selective media supplemented with third-generation cephalosporins for initial ESBL screening. Most targeted bacteria were identified by Matrix-Assisted Laser Desorption Ionization–Time-of-Flight (MALDI-TOF) and polymerase chain reaction (PCR). The prevalence of ESBL in the samples varied widely from 0 to 100%. Double-disc synergy test was mostly used for identifying ESBL-producing bacteria. The most detected ESBL genes were blaTEM, blaSHV, and blaCTX-M. Subtype distribution around the world can be found in Figure 1b. Primers used in the reviewed articles are listed in Table 3. No standardized primer was used for each target gene. However, Woodford et al. (2006) was the most highly cited article for primers targeting specific groups of blaCTX-M.

Table 2.
Characteristics of ESBL genes in animals in the review.

Table 3.
Primers used for detecting ESBL-encoding genes in the review.
3. Discussion
The results of the literature review provide a comprehensive comparison with previous studies on ESBL-producing bacteria in animals. While most previous studies have focused on limited geographic regions and animal populations, the current literature review offers a broader perspective on the highly diverse nature of ESBL-producing bacteria. This review provides insights into the distribution and occurrence of ESBL-producing bacteria in different regions and animal populations, helping to fill gaps in our understanding of this important issue.
Although previous studies have established the spread of ESBL-producing Enterobacterales in food-producing animals and companion pets around the world [67,68], this review highlights the presence of ESBL-producing bacteria in wild vampire bats, wild ungulates, and cockroaches. These findings suggest that the spread of ESBL-producing bacteria is not limited to domesticated animals, but can also occur in wild animal populations. The presence of ESBL-producing bacteria in wild animals can have significant implications for their health, as well as for the health of other animal populations and humans that may encounter them. It is important to consider the potential sources of ESBL-producing bacteria in wild animals, including exposure to contaminated food and water sources, contact with domesticated animals and their environment, or exposure to antibiotics in the environment [69,70]. The spread of ESBL-producing bacteria in wild animal populations can also have ecological consequences, such as altering the balance of microbial communities and affecting the health of the animals and their habitat. It is crucial to continue monitoring the presence of ESBL-producing bacteria in wild animal populations and to implement strategies to reduce their spread. The One Health approach, which recognizes the interconnections between human, animal, and environmental health, is crucial in addressing the issue of ESBL-producing bacteria in wild animals. In the past, WHO had launched an integrated global surveillance on ESBL-producing E. coli. with the same approach [71]. Similar programs covering a wider range of ESBL-producing bacteria may be considered.
Samples in this review were mostly obtained from Enterobacterales-rich areas such as the rectum and fresh feces. However, ESBL-producing bacteria were also found in raw milk and blood and visceral samples of animals in this review [15,17,20]. Unlike most other samples where ESBL-producing genes were found in Enterobacterales, one study reviewed showed the presence of Pseudomonas aeruginosa in uterine swabs of farm animals [28]. The potential for animals to act as reservoirs and vectors of resistance genes is therefore not limited to Enterobacterales found in food-producing animals and pets in contact with humans and antibiotics. One study in Kenya was conducted to collect cloacal swabs from the chickens and fecal samples from the farms. Out of the 544 cloacal isolates of Enterobacterales, 30 were found to contain ESBL genes. Among these, 14 isolates had blaTEM, 5 had blaSHV, and 11 had blaCTX-M. In contrast, among the 47 human isolates, 3 were found to contain ESBL genes, including 2 with blaTEM and 1 with blaCTX-M [25].
There are many kinds of ESBL-encoding genes, including blaTEM, blaSHV, blaCTX-M, blaGES, blaVEB, blaIRT, blaCMT, blaBEL, blaTLA, and blaPER [5]. However, most studies reviewed that investigated bacteria from animals only screened for blaTEM, blaSHV, and blaCTX-M. Genes including blaGES, blaVEB, blaIRT, blaCMT, blaBEL, blaTLA, and blaPER were rarely described and did not occur as frequently as the former three. This is possibly due to the genes being encoded on the chromosomes and not plasmids [8,72]. Furthermore, researchers used various primers for the detection and sequencing of the target genes. The results of this review draw attention to the need for standardized and comprehensive surveillance of ESBL-producing bacteria in animal populations. The limited screening for only a few types of ESBL-encoding genes, such as blaTEM, blaSHV, and blaCTX-M, may not fully capture the diversity and distribution of ESBL-producing bacteria in animal populations. Standardized surveillance covering a wider range of animals and regions will be necessary to better understand the spread of ESBL-producing bacteria and the potential impact on human and animal health. In addition, the use of different primers for the detection and sequencing of ESBL-encoding genes can lead to variability in the results and limit the comparability of studies [73]. Standardized protocols for the detection and sequencing of ESBL-encoding genes are necessary to ensure consistent and accurate results, which is the cornerstone of a better understanding of the spread of these bacteria and their potential impact on human and animal health.
In recent years, there has been a growing concern over the emergence of blaESBL-harboring plasmids in animal isolates. These plasmids are capable of transmitting blaESBL genes among different bacterial species and even among different hosts, including animals and humans [74,75]. Studies have shown that blaESBL-harboring plasmids can be found in various animal isolates, including those from bovine, camels, dogs, cats, goats, and poultry [10,14,76,77,78]. These plasmids can spread rapidly across and within bacterial populations, leading to the dissemination of antibiotic-resistance genes and the emergence of multidrug-resistant bacteria [79]. Overall, the occurrence of blaESBL-harboring plasmids in animal isolates highlights the need for effective surveillance and control measures to limit the spread of antibiotic resistance in both animal and human populations.
PCR with oligonucleotide primers that are specific for a β-lactamase gene was the most common and simplest molecular method used to identify the presence of a β-lactamase enzyme belonging to a specific family. The chosen primers were designed to anneal to regions where no point mutations were known to occur [8,72]. However, in some cases, specific primers may have had special restrictions. For example, the blaCTX-M primer Forward (5′-CGCTTTGCGATGTGCAG-3′) and Reverse (5′-ACCGCGATATCGTTGGT-3′) should not be used to detect CTX-M enzymes in Klebsiella oxytoca as it would result in amplifying chromosomal blaoxy genes [80]. However, the direct comparison of the sensitivity and specificity of different primers in detecting specific ESBL genes is lacking. Therefore, exploring the impact of different primers on blaESBL epidemiology may be a potential research direction.
Though the presence of ESBL-producing bacteria could be seen around the world from this review, regions such as Australia, Canada, and Russia were not covered. This may be due to the fewer numbers of literature reviewed. The prevalence of the ESBL-producing bacteria ranged from 0–100% in this review. Results varied widely among different species and regions. Further research in different regions and animal populations is needed to gain a more comprehensive understanding of the distribution of ESBL-producing bacteria in animals. To limit the spread of ESBL-producing bacteria in animals, the development of new strategies, including the use of alternative treatments, improved animal husbandry practices, and increased public awareness are also vital.
Our review is subject to significant limitations. Specifically, our use of only one database (PubMed) and a limited set of keywords, as well as our exclusion of non-English literature. These may have resulted in some relevant publications being overlooked.
4. Materials and Methods
4.1. Literature Search Strategy
Using a thorough PubMed literature search from 1 January 2020 to 30 June 2022, studies that investigated bacteria from animals (whether wild or domestic) with details of ESBLs were included along with current contents and references from relevant articles. We combined the medical MeSH terminology with free-text terms to conduct a systematic literature search. These were the four keyword combinations we used to search: [(Animals) AND (extended-spectrum beta-lactamase)], [(Animals) AND ESBL], [“Animals” [Mesh] AND (extended-spectrum beta-lactamase)], and [“Animals” [Mesh] AND ESBL]. The bibliographic search was carried out by two researchers. The review protocol is provided as Supplementary Materials File S1.
4.2. Inclusion and Exclusion Criteria
The inclusion criteria include: (1) cultivation of bacteria from animal specimens, whether the animal is healthy or sick; (2) conducting ESBL testing on the bacteria, whether it is phenotype or genotype; and (3) the language of publication was English. The exclusion criteria were as follows: (1) specimens from humans or the surrounding environment of animals were excluded; (2) specimens from animals, humans, and the environment, with no clear distinction between them, were also excluded; and (3) specimens that may be contaminated by environmental or human factors were excluded, such as dairy products, supermarket-packaged raw meat, poultry litter, and pooled feces. However, raw milk and fresh feces are acceptable specimens, as we believe the probability of bacteria cultured from these two types of specimens being contaminated by environmental or human factors is low.
4.3. Study Selection
Our search uncovered 2430 bibliographic references to articles published between 1 January 2021 and 30 June 2022. Thereafter, 1187 duplicate records were removed. Finally, 1243 references remained for screening. The PRISMA 2020 flow diagram [81] for literature screening can be viewed in Figure 2. After the screening of the 1243 records, 1123 records did not match the type of articles we wanted to include. Hence, 120 pieces of full-text literature were assessed for eligibility. Sixty-five of them were excluded because the samples were from the surrounding environment of animals, not from animals themselves. Forty-three of them were excluded because the samples were not only from animals but also from humans or the environment, which could not be distinguished based on the content of the article. Twelve articles were excluded because no denominator (whether the number of animals, number of samples, or number of cultured bacteria) was provided. Finally, 30 documents were selected for further review and analysis.
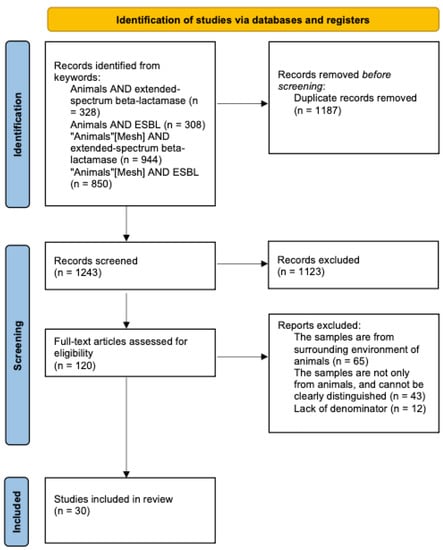
Figure 2.
Flow diagram of the study selection process.
4.4. Data Extraction
We extracted data from all selected literature using a standardized table. The data were grouped as follows: author, date of publication, countries, sampling date and location, sample type, animal species, targeted bacteria, selective media, methods for target identification, the number of denominators, the number of ESBL target, methods for detecting ESBL, methods for detecting ESBL genes, and the number of particular ESBL genes. The collected data were entered into standardized data extraction sheets using Microsoft Excel 2019 (Microsoft Corp, Seattle, WA, USA) for data extraction.
5. Conclusion
The results of this systematic literature review show that ESBL-producing bacteria are present in animals from various countries around the world. We focused on articles where samples were obtained from animals, excluding data from the environment or humans. The most common sources of these bacteria were farm animals, and the most frequently isolated bacteria were E. coli and K. pneumoniae. The prevalence of ESBL in the samples varied widely and the most commonly detected ESBL genes were blaTEM, blaSHV, and blaCTX-M.
The presence of ESBL-producing bacteria in animals highlights the importance of the One Health approach to address the issue of antibiotic resistance. Further research is needed to better understand the epidemiology and mechanisms of the spread of ESBL-producing bacteria in animal populations and their potential impact on human and animal health.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/antibiotics12040661/s1, File S1: Review Protocol. Reference [82] is cited in the supplementary materials.
Author Contributions
Conceptualization: C.-H.T. and P.-Y.L.; Methodology: C.-H.T.; Formal analysis: C.-H.T., C.-W.L. and P.-Y.L.; Investigation: C.-H.T. and P.-Y.L.; Data Curation: C.-H.T., C.-W.L. and P.-Y.L.; Writing—Original Draft: C.-H.T. and C.-W.L.; Writing—Review and Editing: P.-Y.L.; Funding acquisition: P.-Y.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Science and Technology Council, grant number 110-2314-B-075A-011, and Taichung Veterans General Hospital, grant numbers TCVGH-1113901D and TCVGH-1113901C.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Antimicrobial Resistance Collaborators. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef] [PubMed]
- Darwich, L.; Vidal, A.; Seminati, C.; Albamonte, A.; Casado, A.; Lopez, F.; Molina-Lopez, R.A.; Migura-Garcia, L. High prevalence and diversity of extended-spectrum beta-lactamase and emergence of OXA-48 producing Enterobacterales in wildlife in Catalonia. PLoS ONE 2019, 14, e0210686. [Google Scholar] [CrossRef] [PubMed]
- Dandachi, I.; Sokhn, E.S.; Dahdouh, E.A.; Azar, E.; El-Bazzal, B.; Rolain, J.M.; Daoud, Z. Prevalence and Characterization of Multi-Drug-Resistant Gram-Negative Bacilli Isolated from Lebanese Poultry: A Nationwide Study. Front. Microbiol. 2018, 9, 550. [Google Scholar] [CrossRef]
- Tang, K.L.; Caffrey, N.P.; Nobrega, D.B.; Cork, S.C.; Ronksley, P.E.; Barkema, H.W.; Polachek, A.J.; Ganshorn, H.; Sharma, N.; Kellner, J.D.; et al. Restricting the use of antibiotics in food-producing animals and its associations with antibiotic resistance in food-producing animals and human beings: A systematic review and meta-analysis. Lancet Planet Health 2017, 1, e316–e327. [Google Scholar] [CrossRef] [PubMed]
- Castanheira, M.; Simner, P.J.; Bradford, P.A. Extended-spectrum beta-lactamases: An update on their characteristics, epidemiology and detection. JAC Antimicrob. Resist. 2021, 3, dlab092. [Google Scholar] [CrossRef]
- Zhu, F.H.; Rodado, M.P.; Asmar, B.I.; Salimnia, H.; Thomas, R.; Abdel-Haq, N. Risk factors for community acquired urinary tract infections caused by extended spectrum beta-lactamase (ESBL) producing Escherichia coli in children: A case control study. Infect. Dis. 2019, 51, 802–809. [Google Scholar] [CrossRef]
- Tacconelli, E.; Gorska, A.; De Angelis, G.; Lammens, C.; Restuccia, G.; Schrenzel, J.; Huson, D.H.; Carevic, B.; Preotescu, L.; Carmeli, Y.; et al. Estimating the association between antibiotic exposure and colonization with extended-spectrum beta-lactamase-producing Gram-negative bacteria using machine learning methods: A multicentre, prospective cohort study. Clin. Microbiol. Infect. 2020, 26, 87–94. [Google Scholar] [CrossRef]
- Paterson, D.L.; Bonomo, R.A. Extended-spectrum beta-lactamases: A clinical update. Clin. Microbiol. Rev. 2005, 18, 657–686. [Google Scholar] [CrossRef]
- Johansson, V.; Nykasenoja, S.; Myllyniemi, A.L.; Rossow, H.; Heikinheimo, A. Genomic characterization of ESBL/AmpC-producing and high-risk clonal lineages of Escherichia coli and Klebsiella pneumoniae in imported dogs with shelter and stray background. J. Glob. Antimicrob. Resist. 2022, 30, 183–190. [Google Scholar] [CrossRef]
- Shafiq, M.; Rahman, S.U.; Bilal, H.; Ullah, A.; Noman, S.M.; Zeng, M.; Yuan, Y.; Xie, Q.; Li, X.; Jiao, X. Incidence and molecular characterization of ESBL-producing and colistin-resistant Escherichia coli isolates recovered from healthy food-producing animals in Pakistan. J. Appl. Microbiol. 2022, 133, 1169–1182. [Google Scholar] [CrossRef]
- Torres, R.T.; Cunha, M.V.; Araujo, D.; Ferreira, H.; Fonseca, C.; Palmeira, J.D. A walk on the wild side: Wild ungulates as potential reservoirs of multi-drug resistant bacteria and genes, including Escherichia coli harbouring CTX-M beta-lactamases. Environ. Pollut. 2022, 306, 119367. [Google Scholar] [CrossRef] [PubMed]
- Tello, M.; Ocejo, M.; Oporto, B.; Lavin, J.L.; Hurtado, A. Within-farm dynamics of ESBL-producing Escherichia coli in dairy cattle: Resistance profiles and molecular characterization by long-read whole-genome sequencing. Front. Microbiol. 2022, 13, 936843. [Google Scholar] [CrossRef] [PubMed]
- Mengistu, T.S.; Garcias, B.; Castellanos, G.; Seminati, C.; Molina-Lopez, R.A.; Darwich, L. Occurrence of multidrug resistant Gram-negative bacteria and resistance genes in semi-aquatic wildlife—Trachemys scripta, Neovison vison and Lutra lutra—As sentinels of environmental health. Sci. Total Environ. 2022, 830, 154814. [Google Scholar] [CrossRef]
- Aldea, I.; Gibello, A.; Hernandez, M.; Leekitcharoenphon, P.; Bortolaia, V.; Moreno, M.A. Clonal and plasmid-mediated flow of ESBL/AmpC genes in Escherichia coli in a commercial laying hen farm. Vet. Microbiol. 2022, 270, 109453. [Google Scholar] [CrossRef] [PubMed]
- Elkenany, R.; Eltaysh, R.; Elsayed, M.; Abdel-Daim, M.; Shata, R. Characterization of multi-resistant Shigella species isolated from raw cow milk and milk products. J. Vet. Med. Sci. 2022, 84, 890–897. [Google Scholar] [CrossRef] [PubMed]
- Bello Gonzalez, T.D.J.; Kant, A.; Dijkstra, Q.; Marcato, F.; van Reenen, K.; Veldman, K.T.; Brouwer, M.S.M. Changes in Fecal Carriage of Extended-Spectrum Beta-Lactamase Producing Enterobacterales in Dutch Veal Calves by Clonal Spread of Klebsiella pneumoniae. Front. Microbiol. 2022, 13, 866674. [Google Scholar] [CrossRef]
- Gelalcha, B.D.; Ensermu, D.B.; Agga, G.E.; Vancuren, M.; Gillespie, B.E.; D’Souza, D.H.; Okafor, C.C.; Kerro Dego, O. Prevalence of Antimicrobial Resistant and Extended-Spectrum Beta-Lactamase-producing Escherichia coli in Dairy Cattle Farms in East Tennessee. Foodborne Pathog. Dis. 2022, 19, 408–416. [Google Scholar] [CrossRef]
- Gottling, J.; Heckel, J.O.; Hotzel, H.; Fruth, A.; Pfeifer, Y.; Henning, K.; Kopp, P.; Mertens-Scholz, K.; Rietschel, W.; Pfeffer, M. Zoonotic bacteria in clinically healthy goats in petting zoo settings of zoological gardens in Germany. Zoonoses Public Health 2022, 69, 333–343. [Google Scholar] [CrossRef]
- Karabasanavar, N.; Sivaraman, G.K.; S, P.S.; Nair, A.S.; Vijayan, A.; Rajan, V.; Girish, P.S. Non-diarrhoeic pigs as source of highly virulent and multidrug-resistant non-typhoidal Salmonella. Braz. J. Microbiol. 2022, 53, 1039–1049. [Google Scholar] [CrossRef]
- Islam, M.M.; Farag, E.; Hassan, M.M.; Enan, K.A.; Mohammad Sabeel, K.V.; Alhaddad, M.M.; Smatti, M.K.; Al-Marri, A.M.; Al-Zeyara, A.A.; Al-Romaihi, H.; et al. Diversity of bacterial pathogens and their antimicrobial resistance profile among commensal rodents in Qatar. Vet. Res. Commun. 2022, 46, 487–498. [Google Scholar] [CrossRef]
- Khawaskar, D.P.; Sinha, D.K.; Lalrinzuala, M.V.; Athira, V.; Kumar, M.; Chhakchhuak, L.; Mohanapriya, K.; Sophia, I.; Abhishek; Kumar, O.R.V.; et al. Pathotyping and antimicrobial susceptibility testing of Escherichia coli isolates from neonatal calves. Vet. Res. Commun. 2022, 46, 353–362. [Google Scholar] [CrossRef] [PubMed]
- Haeili, M.; Salehzeinali, H.; Mirzaei, S.; Pishnian, Z.; Ahmadi, A. Molecular characterization of quinolone resistance and antimicrobial resistance profiles of Klebsiella pneumoniae and Escherichia coli isolated from human and broiler chickens. Int. J. Environ. Health Res. 2022, 32, 1382–1392. [Google Scholar] [CrossRef] [PubMed]
- Aklilu, E.; Harun, A.; Singh, K.K.B. Molecular characterization of bla(NDM), bla(OXA-48), mcr-1 and bla(TEM-52) positive and concurrently carbapenem and colistin resistant and extended spectrum beta-lactamase producing Escherichia coli in chicken in Malaysia. BMC Vet. Res. 2022, 18, 190. [Google Scholar] [CrossRef]
- Menck-Costa, M.F.; Baptista, A.A.S.; Gazal, L.E.S.; Justino, L.; Sanches, M.S.; de Souza, M.; Nishio, E.K.; Queiroz Dos Santos, B.; Cruz, V.D.; Berbert, J.V.M.; et al. High-Frequency Detection of fosA3 and bla (CTX-M-55) Genes in Escherichia coli From Longitudinal Monitoring in Broiler Chicken Farms. Front. Microbiol. 2022, 13, 846116. [Google Scholar] [CrossRef] [PubMed]
- Ndukui, J.G.; Gikunju, J.K.; Aboge, G.O.; Mwaniki, J.K.; Maina, J.N.; Mbaria, J.M. Molecular Characterization of ESBLs and QnrS Producers from Selected Enterobacteriaceae Strains Isolated from Commercial Poultry Production Systems in Kiambu County, Kenya. Microbiol. Insights 2022, 15, 11786361211063619. [Google Scholar] [CrossRef] [PubMed]
- Su, X.; Yan, X.; Li, Y.; Zhang, D.; Li, L.; Geng, Y.; Su, F.; Yue, C.; Hou, R.; Liu, S. Identification of extended-spectrum beta-lactamase (CTX-M)-producing Klebsiella pneumoniae belonging to ST37, ST290, and ST2640 in captive giant pandas. BMC Vet. Res. 2022, 18, 186. [Google Scholar] [CrossRef] [PubMed]
- Gruel, G.; Couvin, D.; Guyomard-Rabenirina, S.; Arlet, G.; Bambou, J.C.; Pot, M.; Roy, X.; Talarmin, A.; Tressieres, B.; Ferdinand, S.; et al. High Prevalence of bla (CTXM-1)/IncI1-Igamma/ST3 Plasmids in Extended-Spectrum Beta-Lactamase-Producing Escherichia coli Isolates Collected from Domestic Animals in Guadeloupe (French West Indies). Front. Microbiol. 2022, 13, 882422. [Google Scholar] [CrossRef]
- Mahmoud, S.F.; Fayez, M.; Swelum, A.A.; Alswat, A.S.; Alkafafy, M.; Alzahrani, O.M.; Alsunaini, S.J.; Almuslem, A.; Al Amer, A.S.; Yusuf, S. Genetic Diversity, Biofilm Formation, and Antibiotic Resistance of Pseudomonas aeruginosa Isolated from Cow, Camel, and Mare with Clinical Endometritis. Vet. Sci. 2022, 9, 239. [Google Scholar] [CrossRef]
- Islam, M.S.; Sobur, M.A.; Rahman, S.; Ballah, F.M.; Ievy, S.; Siddique, M.P.; Rahman, M.; Kafi, M.A.; Rahman, M.T. Detection of bla(TEM), bla(CTX-M), bla(CMY), and bla(SHV) Genes Among Extended-Spectrum Beta-Lactamase-Producing Escherichia coli Isolated from Migratory Birds Travelling to Bangladesh. Microb. Ecol. 2022, 83, 942–950. [Google Scholar] [CrossRef]
- Landolsi, S.; Selmi, R.; Hadjadj, L.; Ben Haj Yahia, A.; Ben Romdhane, K.; Messadi, L.; Rolain, J.M. First Report of Extended-Spectrum beta-Lactamase (bla(CTX-M1)) and Colistin Resistance Gene mcr-1 in E. coli of Lineage ST648 from Cockroaches in Tunisia. Microbiol. Spectr. 2022, 10, e0003621. [Google Scholar] [CrossRef]
- Garcia-Fierro, R.; Drapeau, A.; Dazas, M.; Saras, E.; Rodrigues, C.; Brisse, S.; Madec, J.Y.; Haenni, M. Comparative phylogenomics of ESBL-, AmpC- and carbapenemase-producing Klebsiella pneumoniae originating from companion animals and humans. J. Antimicrob. Chemother. 2022, 77, 1263–1271. [Google Scholar] [CrossRef]
- Belas, A.; Marques, C.; Menezes, J.; da Gama, L.T.; Cavaco-Silva, P.; Pomba, C. ESBL/pAmpC-Producing Escherichia coli Causing Urinary Tract Infections in Non-Related Companion Animals and Humans. Antibiotics 2022, 11, 559. [Google Scholar] [CrossRef] [PubMed]
- Aworh, M.K.; Ekeng, E.; Nilsson, P.; Egyir, B.; Owusu-Nyantakyi, C.; Hendriksen, R.S. Extended-Spectrum ss-Lactamase-Producing Escherichia coli among Humans, Beef Cattle, and Abattoir Environments in Nigeria. Front. Cell Infect. Microbiol. 2022, 12, 869314. [Google Scholar] [CrossRef]
- Nuesch-Inderbinen, M.; Hanni, C.; Zurfluh, K.; Hartnack, S.; Stephan, R. Antimicrobial resistance profiles of Escherichia coli and prevalence of extended-spectrum beta-lactamase-producing Enterobacteriaceae in calves from organic and conventional dairy farms in Switzerland. Microbiologyopen 2022, 11, e1269. [Google Scholar] [CrossRef]
- Samir, A.; Abdel-Moein, K.A.; Zaher, H.M. The Public Health Burden of Virulent Extended-Spectrum Beta-Lactamase-Producing Klebsiella pneumoniae Strains Isolated from Diseased Horses. Vector-Borne Zoonotic Dis. 2022, 22, 217–224. [Google Scholar] [CrossRef]
- Loucif, L.; Chelaghma, W.; Cherak, Z.; Bendjama, E.; Beroual, F.; Rolain, J.M. Detection of NDM-5 and MCR-1 antibiotic resistance encoding genes in Enterobacterales in long-distance migratory bird species Ciconia ciconia, Algeria. Sci. Total Environ. 2022, 814, 152861. [Google Scholar] [CrossRef] [PubMed]
- Benavides, J.A.; Godreuil, S.; Opazo-Capurro, A.; Mahamat, O.O.; Falcon, N.; Oravcova, K.; Streicker, D.G.; Shiva, C. Long-term maintenance of multidrug-resistant Escherichia coli carried by vampire bats and shared with livestock in Peru. Sci. Total Environ. 2022, 810, 152045. [Google Scholar] [CrossRef]
- Trongjit, S.; Assavacheep, P.; Samngamnim, S.; My, T.H.; An, V.T.T.; Simjee, S.; Chuanchuen, R. Plasmid-mediated colistin resistance and ESBL production in Escherichia coli from clinically healthy and sick pigs. Sci. Rep. 2022, 12, 2466. [Google Scholar] [CrossRef] [PubMed]
- Dallenne, C.; Da Costa, A.; Decre, D.; Favier, C.; Arlet, G. Development of a set of multiplex PCR assays for the detection of genes encoding important beta-lactamases in Enterobacteriaceae. J. Antimicrob. Chemother. 2010, 65, 490–495. [Google Scholar] [CrossRef]
- Randall, L.P.; Cooles, S.W.; Osborn, M.K.; Piddock, L.J.; Woodward, M.J. Antibiotic resistance genes, integrons and multiple antibiotic resistance in thirty-five serotypes of Salmonella enterica isolated from humans and animals in the UK. J. Antimicrob. Chemother. 2004, 53, 208–216. [Google Scholar] [CrossRef]
- Strauss, L.M.; Dahms, C.; Becker, K.; Kramer, A.; Kaase, M.; Mellmann, A. Development and evaluation of a novel universal beta-lactamase gene subtyping assay for blaSHV, blaTEM and blaCTX-M using clinical and livestock-associated Escherichia coli. J. Antimicrob. Chemother. 2015, 70, 710–715. [Google Scholar] [CrossRef][Green Version]
- Gootz, T.D.; Lescoe, M.K.; Dib-Hajj, F.; Dougherty, B.A.; He, W.; Della-Latta, P.; Huard, R.C. Genetic organization of transposase regions surrounding blaKPC carbapenemase genes on plasmids from Klebsiella strains isolated in a New York City hospital. Antimicrob. Agents Chemother. 2009, 53, 1998–2004. [Google Scholar] [CrossRef]
- Bhattacharjee, A.; Sen, M.R.; Anupurba, S.; Prakash, P.; Nath, G. Detection of OXA-2 group extended-spectrum-beta-lactamase-producing clinical isolates of Escherichia coli from India. J. Antimicrob. Chemother. 2007, 60, 703–704. [Google Scholar] [CrossRef]
- Christophy, R.; Osman, M.; Mallat, H.; Achkar, M.; Ziedeh, A.; Moukaddem, W.; Dabboussi, F.; Hamze, M. Prevalence, antibiotic susceptibility and characterization of antibiotic resistant genes among carbapenem-resistant Gram-negative bacilli and yeast in intestinal flora of cancer patients in North Lebanon. J. Infect. Public Health 2017, 10, 716–720. [Google Scholar] [CrossRef]
- Grobner, S.; Linke, D.; Schutz, W.; Fladerer, C.; Madlung, J.; Autenrieth, I.B.; Witte, W.; Pfeifer, Y. Emergence of carbapenem-non-susceptible extended-spectrum beta-lactamase-producing Klebsiella pneumoniae isolates at the university hospital of Tubingen, Germany. J. Med. Microbiol. 2009, 58, 912–922. [Google Scholar] [CrossRef]
- Ly, T.D.A.; Hadjadj, L.; Hoang, V.T.; Louni, M.; Dao, T.L.; Badiaga, S.; Tissot-Dupont, H.; Raoult, D.; Rolain, J.M.; Gautret, P. Low prevalence of resistance genes in sheltered homeless population in Marseille, France, 2014–2018. Infect. Drug Resist. 2019, 12, 1139–1151. [Google Scholar] [CrossRef] [PubMed]
- Hosu, M.C.; Vasaikar, S.D.; Okuthe, G.E.; Apalata, T. Detection of extended spectrum beta-lactamase genes in Pseudomonas aeruginosa isolated from patients in rural Eastern Cape Province, South Africa. Sci. Rep. 2021, 11, 7110. [Google Scholar] [CrossRef]
- Pitout, J.D.; Thomson, K.S.; Hanson, N.D.; Ehrhardt, A.F.; Moland, E.S.; Sanders, C.C. beta-Lactamases responsible for resistance to expanded-spectrum cephalosporins in Klebsiella pneumoniae, Escherichia coli, and Proteus mirabilis isolates recovered in South Africa. Antimicrob. Agents Chemother. 1998, 42, 1350–1354. [Google Scholar] [CrossRef]
- Fang, H.; Ataker, F.; Hedin, G.; Dornbusch, K. Molecular epidemiology of extended-spectrum beta-lactamases among Escherichia coli isolates collected in a Swedish hospital and its associated health care facilities from 2001 to 2006. J. Clin. Microbiol. 2008, 46, 707–712. [Google Scholar] [CrossRef] [PubMed]
- Hasman, H.; Mevius, D.; Veldman, K.; Olesen, I.; Aarestrup, F.M. Beta-Lactamases among extended-spectrum beta-lactamase (ESBL)-resistant Salmonella from poultry, poultry products and human patients in The Netherlands. J. Antimicrob. Chemother. 2005, 56, 115–121. [Google Scholar] [CrossRef] [PubMed]
- Ali, T.; Rahman, S.U.; Zhang, L.; Shahid, M.; Han, D.; Gao, J.; Zhang, S.; Ruegg, P.L.; Saddique, U.; Han, B. Characteristics and genetic diversity of multi-drug resistant extended-spectrum beta-lactamase (ESBL)-producing Escherichia coli isolated from bovine mastitis. Oncotarget 2017, 8, 90144–90163. [Google Scholar] [CrossRef] [PubMed]
- Weill, F.X.; Demartin, M.; Tande, D.; Espie, E.; Rakotoarivony, I.; Grimont, P.A. SHV-12-like extended-spectrum-beta-lactamase-producing strains of Salmonella enterica serotypes Babelsberg and Enteritidis isolated in France among infants adopted from Mali. J. Clin. Microbiol. 2004, 42, 2432–2437. [Google Scholar] [CrossRef] [PubMed]
- Colom, K.; Perez, J.; Alonso, R.; Fernandez-Aranguiz, A.; Larino, E.; Cisterna, R. Simple and reliable multiplex PCR assay for detection of blaTEM, bla(SHV) and blaOXA-1 genes in Enterobacteriaceae. FEMS Microbiol. Lett. 2003, 223, 147–151. [Google Scholar] [CrossRef]
- Rayamajhi, N.; Kang, S.G.; Lee, D.Y.; Kang, M.L.; Lee, S.I.; Park, K.Y.; Lee, H.S.; Yoo, H.S. Characterization of TEM-, SHV- and AmpC-type beta-lactamases from cephalosporin-resistant Enterobacteriaceae isolated from swine. Int. J. Food Microbiol. 2008, 124, 183–187. [Google Scholar] [CrossRef] [PubMed]
- Lob, S.H.; Kazmierczak, K.M.; Badal, R.E.; Hackel, M.A.; Bouchillon, S.K.; Biedenbach, D.J.; Sahm, D.F. Trends in susceptibility of Escherichia coli from intra-abdominal infections to ertapenem and comparators in the United States according to data from the SMART program, 2009 to 2013. Antimicrob. Agents Chemother. 2015, 59, 3606–3610. [Google Scholar] [CrossRef]
- El-Shazly, S.; Dashti, A.; Vali, L.; Bolaris, M.; Ibrahim, A.S. Molecular epidemiology and characterization of multiple drug-resistant (MDR) clinical isolates of Acinetobacter baumannii. Int. J. Infect. Dis. 2015, 41, 42–49. [Google Scholar] [CrossRef]
- Van, T.T.; Chin, J.; Chapman, T.; Tran, L.T.; Coloe, P.J. Safety of raw meat and shellfish in Vietnam: An analysis of Escherichia coli isolations for antibiotic resistance and virulence genes. Int. J. Food Microbiol. 2008, 124, 217–223. [Google Scholar] [CrossRef]
- Archambault, M.; Petrov, P.; Hendriksen, R.S.; Asseva, G.; Bangtrakulnonth, A.; Hasman, H.; Aarestrup, F.M. Molecular characterization and occurrence of extended-spectrum beta-lactamase resistance genes among Salmonella enterica serovar Corvallis from Thailand, Bulgaria, and Denmark. Microb. Drug Resist. 2006, 12, 192–198. [Google Scholar] [CrossRef]
- Ahmed, O.I.; El-Hady, S.A.; Ahmed, T.M.; Ahmed, I.Z. Detection of bla SHV and bla CTX-M genes in ESBL producing Klebsiella pneumoniae isolated from Egyptian patients with suspected nosocomial infections. Egypt. J. Med. Hum. Genet. 2013, 14, 277–283. [Google Scholar] [CrossRef]
- Gundran, R.S.; Cardenio, P.A.; Villanueva, M.A.; Sison, F.B.; Benigno, C.C.; Kreausukon, K.; Pichpol, D.; Punyapornwithaya, V. Prevalence and distribution of bla(CTX-M), bla(SHV), bla(TEM) genes in extended- spectrum beta- lactamase- producing E. coli isolates from broiler farms in the Philippines. BMC Vet. Res. 2019, 15, 227. [Google Scholar] [CrossRef]
- Batchelor, M.; Hopkins, K.; Threlfall, E.J.; Clifton-Hadley, F.A.; Stallwood, A.D.; Davies, R.H.; Liebana, E. Bla(CTX-M) genes in clinical Salmonella isolates recovered from humans in England and Wales from 1992 to 2003. Antimicrob. Agents Chemother. 2005, 49, 1319–1322. [Google Scholar] [CrossRef]
- Edelstein, M.; Pimkin, M.; Palagin, I.; Edelstein, I.; Stratchounski, L. Prevalence and molecular epidemiology of CTX-M extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae in Russian hospitals. Antimicrob. Agents Chemother. 2003, 47, 3724–3732. [Google Scholar] [CrossRef] [PubMed]
- Horton, R.A.; Randall, L.P.; Snary, E.L.; Cockrem, H.; Lotz, S.; Wearing, H.; Duncan, D.; Rabie, A.; McLaren, I.; Watson, E.; et al. Fecal carriage and shedding density of CTX-M extended-spectrum beta-lactamase-producing Escherichia coli in cattle, chickens, and pigs: Implications for environmental contamination and food production. Appl. Environ. Microbiol. 2011, 77, 3715–3719. [Google Scholar] [CrossRef] [PubMed]
- Woodford, N.; Fagan, E.J.; Ellington, M.J. Multiplex PCR for rapid detection of genes encoding CTX-M extended-spectrum (beta)-lactamases. J. Antimicrob. Chemother. 2006, 57, 154–155. [Google Scholar] [CrossRef] [PubMed]
- Pfeifer, Y.; Matten, J.; Rabsch, W. Salmonella enterica serovar Typhi with CTX-M beta-lactamase, Germany. Emerg. Infect. Dis. 2009, 15, 1533–1535. [Google Scholar] [CrossRef]
- Wetzker, W.; Pfeifer, Y.; Wolke, S.; Haselbeck, A.; Leistner, R.; Kola, A.; Gastmeier, P.; Salm, F. Extended-Spectrum Beta-Lactamase (ESBL)-Producing Escherichia coli Isolated from Flies in the Urban Center of Berlin, Germany. Int. J. Environ. Res. Public Health 2019, 16, 1530. [Google Scholar] [CrossRef]
- Carattoli, A. Animal reservoirs for extended spectrum beta-lactamase producers. Clin. Microbiol. Infect. 2008, 14 (Suppl. S1), 117–123. [Google Scholar] [CrossRef]
- Dantas Palmeira, J.; Ferreira, H.M.N. Extended-spectrum beta-lactamase (ESBL)-producing Enterobacteriaceae in cattle production—A threat around the world. Heliyon 2020, 6, e03206. [Google Scholar] [CrossRef]
- Mesbah Zekar, F.; Granier, S.A.; Marault, M.; Yaici, L.; Gassilloud, B.; Manceau, C.; Touati, A.; Millemann, Y. From Farms to Markets: Gram-Negative Bacteria Resistant to Third-Generation Cephalosporins in Fruits and Vegetables in a Region of North Africa. Front. Microbiol. 2017, 8, 1569. [Google Scholar] [CrossRef]
- De Boeck, H.; Miwanda, B.; Lunguya-Metila, O.; Muyembe-Tamfum, J.J.; Stobberingh, E.; Glupczynski, Y.; Jacobs, J. ESBL-positive Enterobacteria isolates in drinking water. Emerg. Infect. Dis. 2012, 18, 1019–1020. [Google Scholar] [CrossRef]
- World Health Organization. WHO Integrated Global Surveillance on ESBL-Producing E. coli Using a “One Health” Approach: Implementation and Opportunities; World Health Organization: Geneva, Switzerland, 2021.
- Bradford, P.A. Extended-spectrum beta-lactamases in the 21st century: Characterization, epidemiology, and detection of this important resistance threat. Clin. Microbiol. Rev. 2001, 14, 933–951. [Google Scholar] [CrossRef] [PubMed]
- Brazelton de Cardenas, J.N.; Garner, C.D.; Su, Y.; Tang, L.; Hayden, R.T. Comparative Evaluation of Assays for Broad Detection of Molecular Resistance Mechanisms in Enterobacterales Isolates. J. Clin. Microbiol. 2021, 59, e0103321. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Stephan, R.; Karczmarczyk, M.; Yan, Q.; Hachler, H.; Fanning, S. Molecular characterization of bla ESBL-harboring conjugative plasmids identified in multi-drug resistant Escherichia coli isolated from food-producing animals and healthy humans. Front. Microbiol. 2013, 4, 188. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Stephan, R.; Power, K.; Yan, Q.; Hachler, H.; Fanning, S. Nucleotide sequences of 16 transmissible plasmids identified in nine multidrug-resistant Escherichia coli isolates expressing an ESBL phenotype isolated from food-producing animals and healthy humans. J. Antimicrob. Chemother. 2014, 69, 2658–2668. [Google Scholar] [CrossRef]
- Anes, J.; Nguyen, S.V.; Eshwar, A.K.; McCabe, E.; Macori, G.; Hurley, D.; Lehner, A.; Fanning, S. Molecular characterisation of multi-drug resistant Escherichia coli of bovine origin. Vet. Microbiol. 2020, 242, 108566. [Google Scholar] [CrossRef]
- Saidani, M.; Messadi, L.; Mefteh, J.; Chaouechi, A.; Soudani, A.; Selmi, R.; Daaloul-Jedidi, M.; Ben Chehida, F.; Mamlouk, A.; Jemli, M.H.; et al. Various Inc-type plasmids and lineages of Escherichia coli and Klebsiella pneumoniae spreading bla(CTX-M-15,)bla(CTX-M-1) and mcr-1 genes in camels in Tunisia. J. Glob. Antimicrob. Resist. 2019, 19, 280–283. [Google Scholar] [CrossRef]
- Pepin-Puget, L.; El Garch, F.; Bertrand, X.; Valot, B.; Hocquet, D. Genome analysis of enterobacteriaceae with non-wild type susceptibility to third-generation cephalosporins recovered from diseased dogs and cats in Europe. Vet. Microbiol. 2020, 242, 108601. [Google Scholar] [CrossRef]
- Rozwandowicz, M.; Brouwer, M.S.M.; Fischer, J.; Wagenaar, J.A.; Gonzalez-Zorn, B.; Guerra, B.; Mevius, D.J.; Hordijk, J. Plasmids carrying antimicrobial resistance genes in Enterobacteriaceae. J. Antimicrob. Chemother. 2018, 73, 1121–1137. [Google Scholar] [CrossRef]
- Wiegand, I.; Geiss, H.K.; Mack, D.; Sturenburg, E.; Seifert, H. Detection of extended-spectrum beta-lactamases among Enterobacteriaceae by use of semiautomated microbiology systems and manual detection procedures. J. Clin. Microbiol. 2007, 45, 1167–1174. [Google Scholar] [CrossRef]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 statement: An updated guideline for reporting systematic reviews. BMJ 2021, 372, n71. [Google Scholar] [CrossRef]
- University of Alabama. A Research Guide for Systematic Literature Reviews. University Libraries. Available online: https://guides.lib.ua.edu/c.php?g=1135040&p=8286355 (accessed on 24 May 2022).
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).