Cross-Talking of Pathway-Specific Regulators in Glycopeptide Antibiotics (Teicoplanin and A40926) Production
Abstract
1. Introduction
2. Results
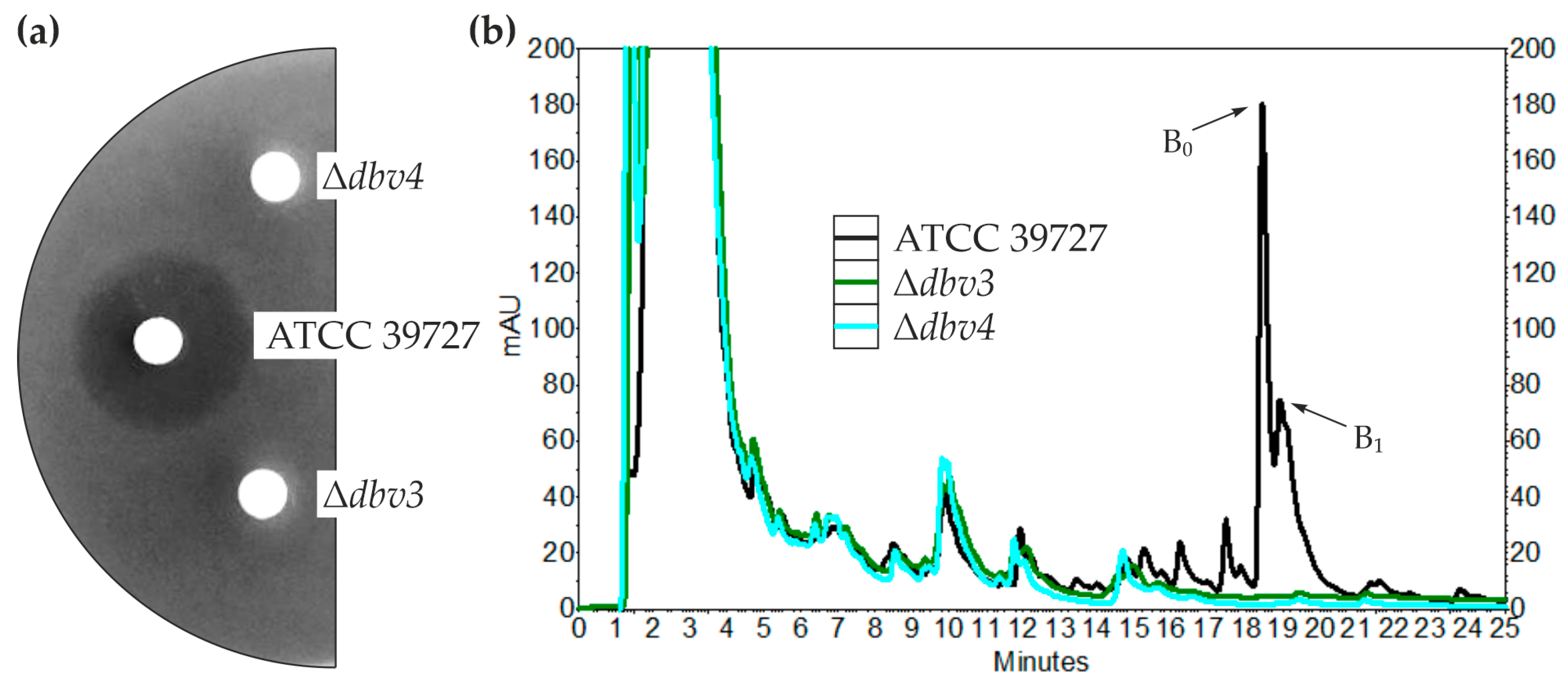
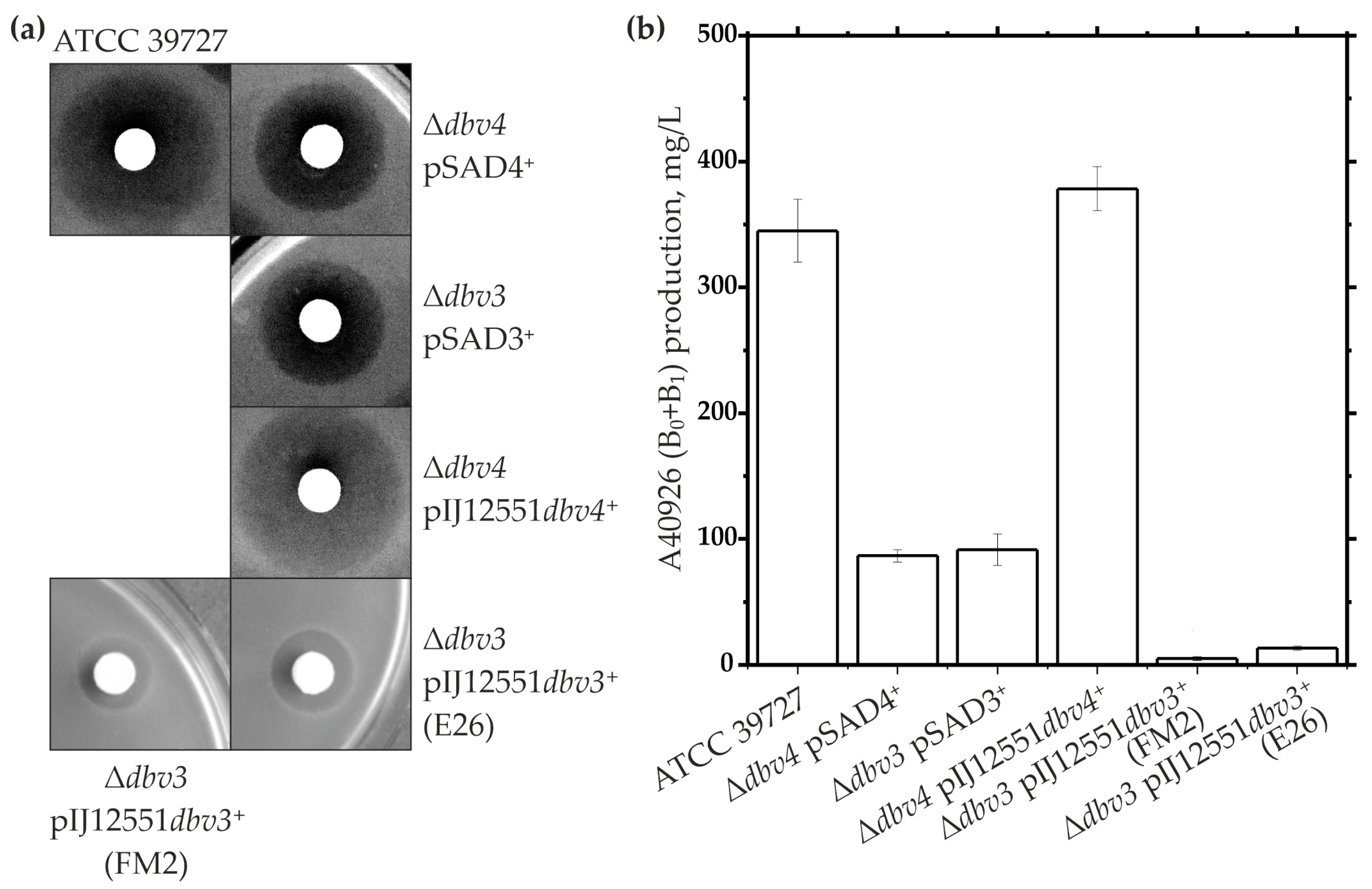
2.1. Generation and Complementation of dbv3 and dbv4 N. gerenzanensis Knockout Mutants
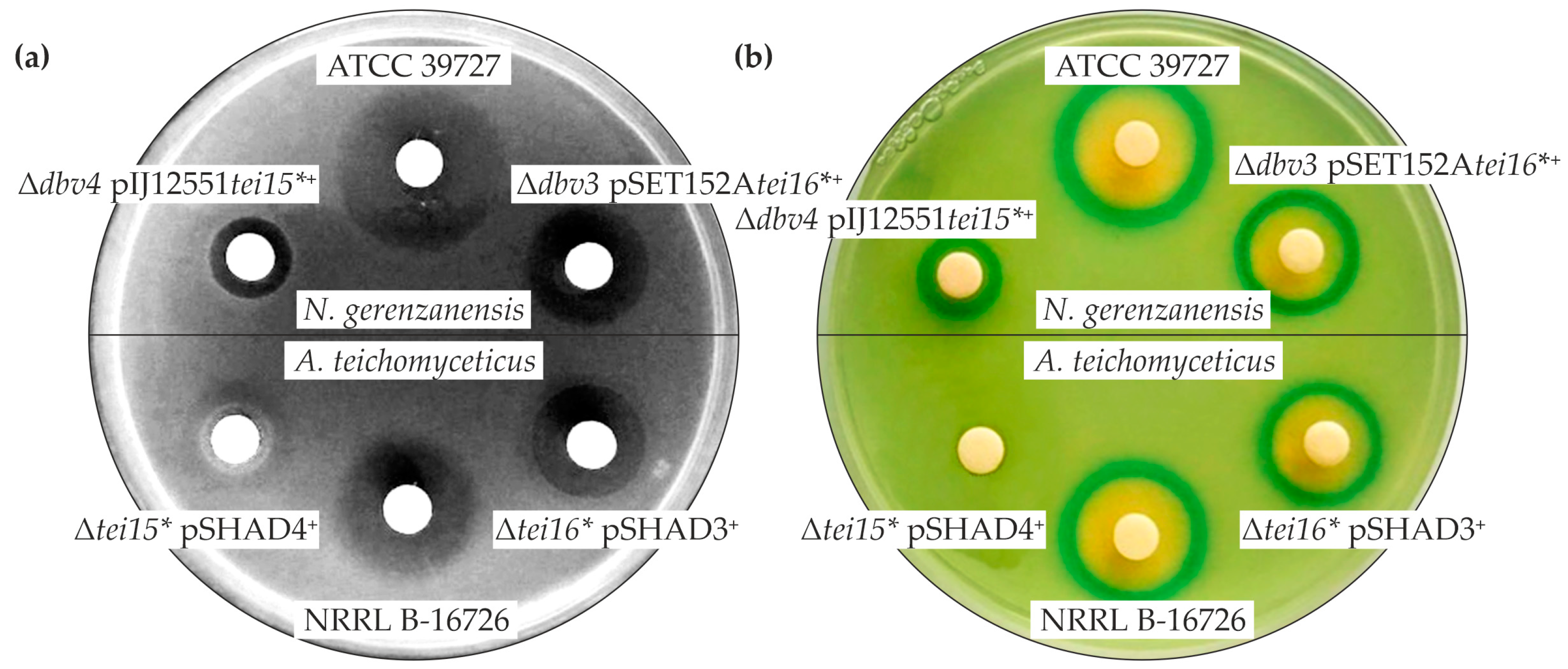
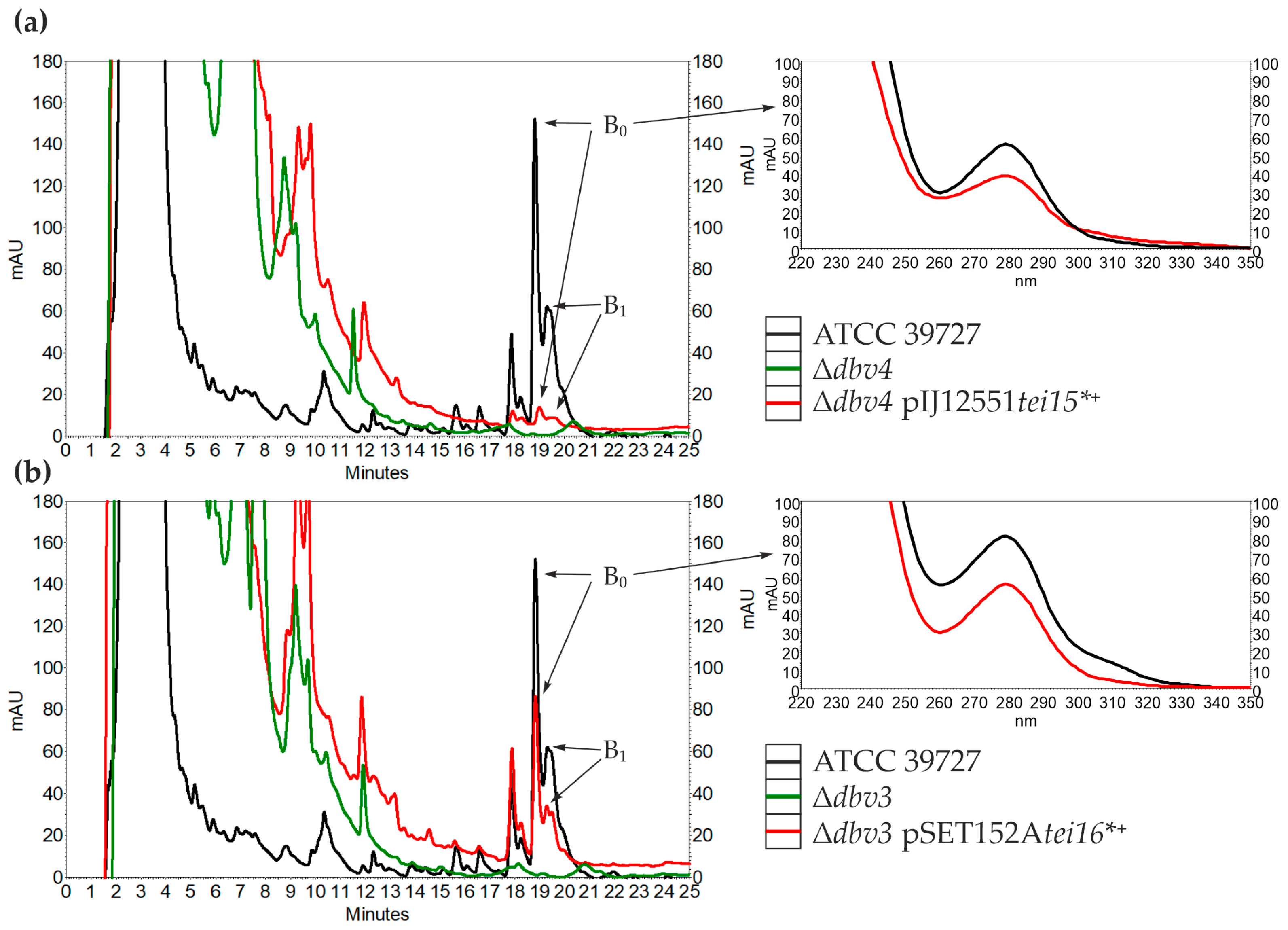
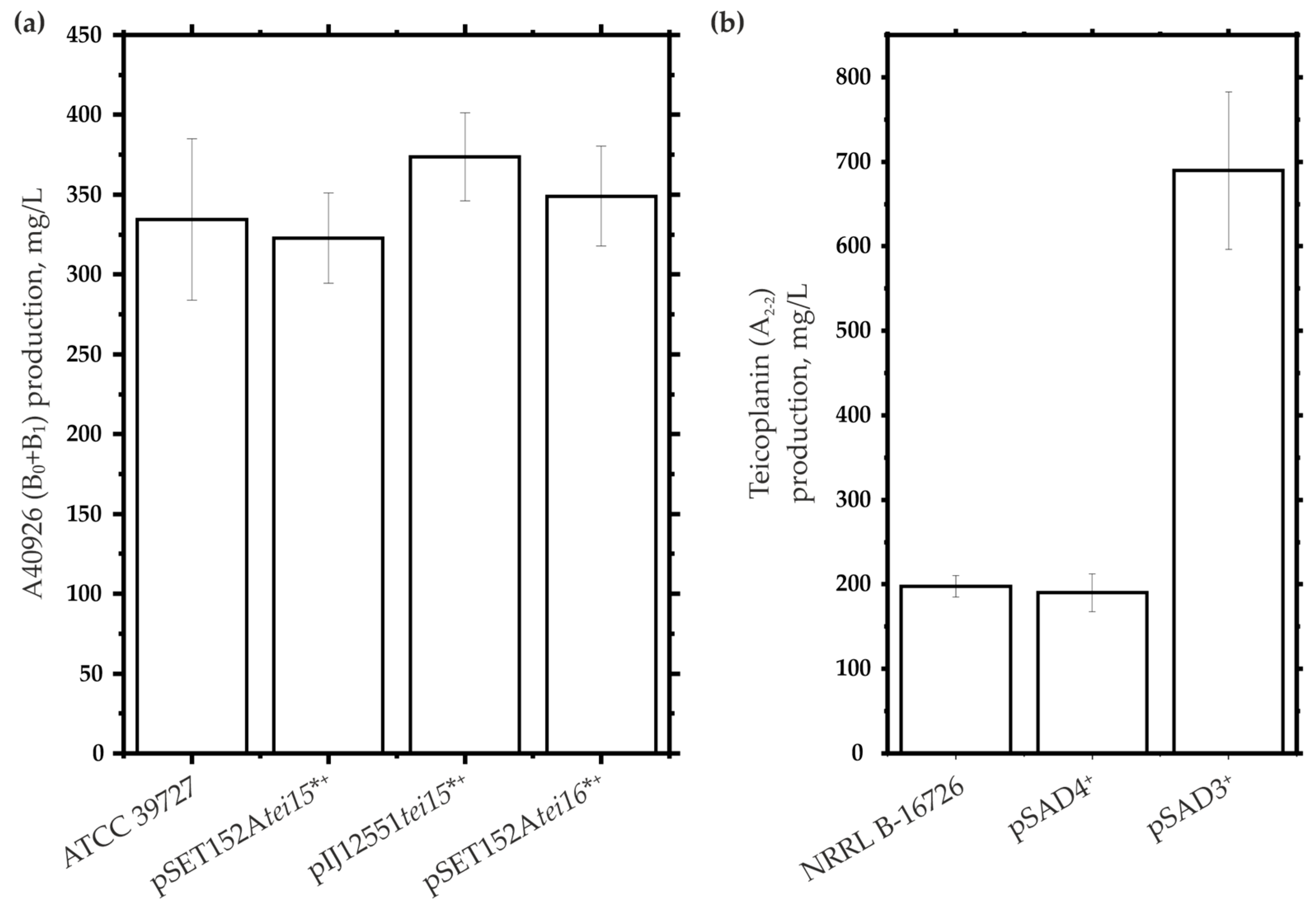
2.2. Cross-Complementation of N. gerenzanensis Δdbv4 and A. teichomyceticus Δtei15* with tei15* and dbv4
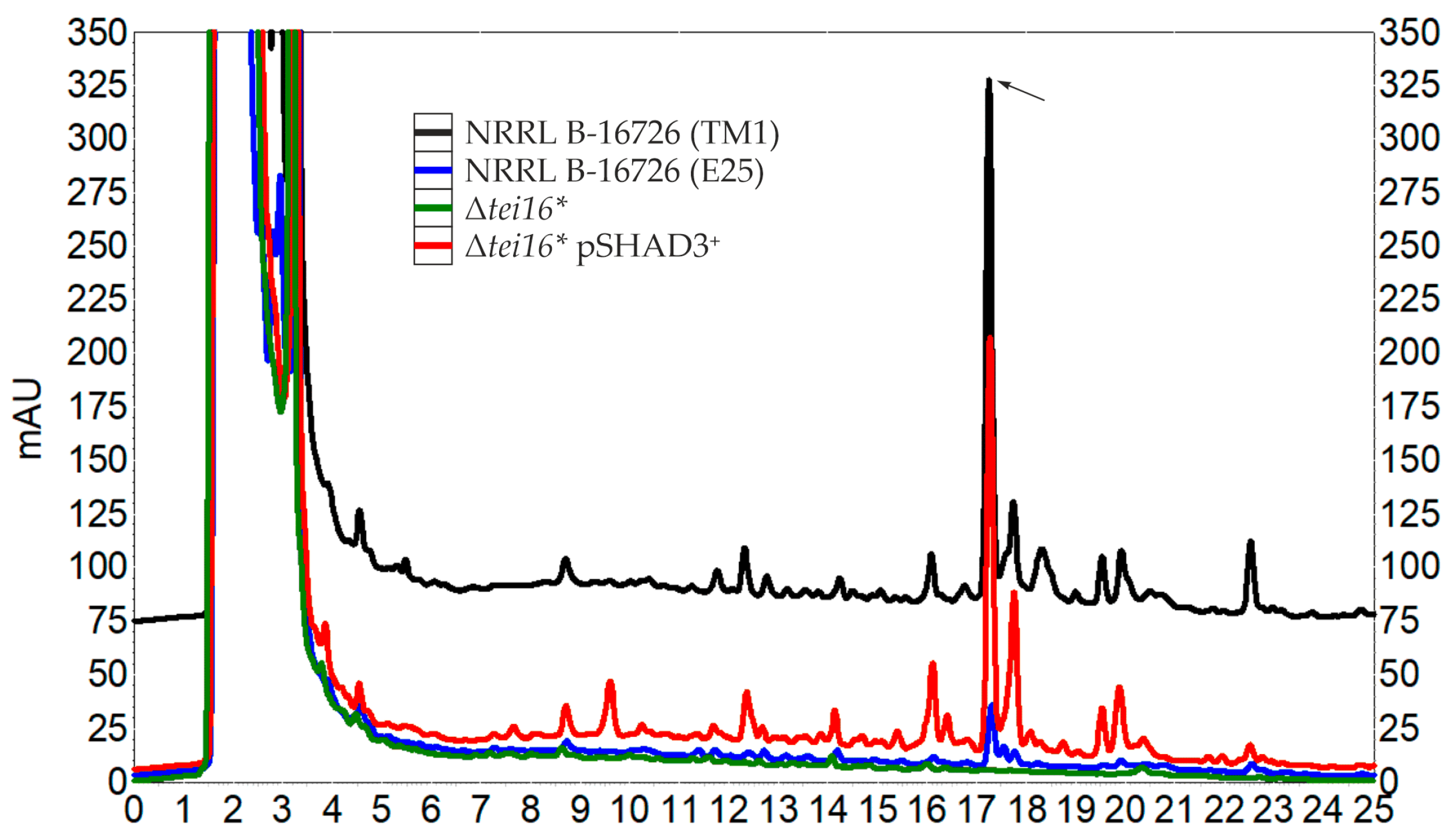
2.3. Cross-Complementation of N. gerenzanensis Δdbv3 and A. teichomyceticus Δtei16* with tei16* and dbv3
2.4. Expression of tei15* and tei16* in the Double dbv3 and dbv4 Knockout Mutant of N. gerenzanensis
2.5. Cross-Overexpression of dbv and tei CSRGs in N. gerenzanensis ATCC 39727 and A. teichomyceticus NRRL B-16726
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains and Growth Conditions
| Plasmids | Description | Source or Reference |
|---|---|---|
| pIJ778 | Template vector for the amplification of oriT-aadA cassette for PCR-targeted mutagenesis | [57] |
| A40Y | SuperCos1 derivative, including 22 kb fragment of dbv cluster (dbv1–dbv17) | [58] |
| A40dbv3::aadA | A40Y derivative with dbv3 replaced by oriT-aadA cassette | This work |
| A40dbv4::aadA | A40Y derivative with dbv4 replaced with oriT-aadA | This work |
| A40dbv3–4::aadA | A40Y derivative with dbv3 and dbv4 replaced with oriT-aadA cassette derived from plasmid pIJ778 | This work |
| pKC1132 | Suicide plasmid for gene knockouts, AmR | [59] |
| pKCKOD3 | pKC1132 derivative carrying dbv3 flanking regions surrounding oriT-aadA, AmR | This work |
| pKCKOD4 | pKC1132 derivative carrying dbv4 flanking regions surrounding oriT-aadA, AmR | This work |
| pKCKOD3–4 | pKC1132 derivative carrying dbv3–4 flanking regions surrounding oriT-aadA, AmR | This work |
| pSAD3 | pSET152A derivative carrying dbv3 under the control of aac(3)IVp, AmR | [38] |
| pSAD4 | pSET152A derivative carrying dbv4 under the control of aac(3)IVp, AmR | [38] |
| pIJ10700 | Template vector for the amplification of hygR | [43] |
| pSHAD3 | pSAD3 derivative, HgR | This work |
| pSHAD4 | pSAD4 derivative, HgR | This work |
| pIJ12551 | ϕC31-actinophage-based integrative vector for gene expression under the control of ermE* promoter, AmR | [46] |
| pIJ12551dbv4 | pIJ12551 derivative carrying dbv4 under the control of ermE*, AmR | This work |
| pIJ12551dbv3 | pIJ12551 derivative carrying dbv3 under the control of ermE*, AmR | This work |
| pSET152Atei15* | pSET152A derivative carrying tei15* under the control of aac(3)IVp, AmR | [35] |
| pSET152Atei16* | pSET152A derivative carrying tei16* under the control of aac(3)IVp, AmR | [35] |
| pIJ12551tei15* | pIJ12551 derivative carrying tei15* under the control of ermE* | This work |
| Bacterial Strains | ||
| N. gerenzanensis | Wild type, A40926 producer | ATCC 39727 |
| N. gerenzanensis Δdbv3 | Wild type derivative with dbv3 replaced by oriT-aadA cassette | This work |
| N. gerenzanensis Δdbv4 | Wild type derivative with dbv4 replaced by oriT-aadA cassette | This work |
| N. gerenzanensis Δdbv3–4 | Wild type derivative with dbv3–4 replaced by oriT-aadA cassette | This work |
| N. gerenzanensis Δdbv3 pSAD3+ | N. gerenzanensis Δdbv3 derivative carrying pSAD3 | This work |
| N. gerenzanensis Δdbv4 pSAD4+ | N. gerenzanensis Δdbv4 derivative carrying pSAD4 | This work |
| N. gerenzanensis Δdbv4 pIJ12551dbv4+ | N. gerenzanensis Δdbv4 derivative carrying pIJ12551dbv4 | This work |
| N. gerenzanensis pSET152Atei15*+ | Wild type derivative carrying pSET152Atei15* | This work |
| N. gerenzanensis pSET152Atei16*+ | Wild type derivative carrying pSET152Atei16* | This work |
| N. gerenzanensis pIJ12551tei15*+ | Wild type derivative carrying pIJ12551tei15* | This work |
| N. gerenzanensis Δdbv4 pSET152Atei15*+ | N. gerenzanensis Δdbv4 derivative carrying pSET152Atei15* | This work |
| N. gerenzanensis Δdbv3 pSET152Atei16*+ | N. gerenzanensis Δdbv3 derivative carrying pSET152Atei16* | This work |
| N. gerenzanensis Δdbv4 pIJ12551dbv4+ | N. gerenzanensis Δdbv4 derivative carrying pIJ12551dbv4 | This work |
| N. gerenzanensis Δdbv3–4 pIJ12551tei15*+ | N. gerenzanensis Δdbv3–4 derivative carrying pIJ12551tei15* | This work |
| N. gerenzanensis Δdbv3–4 pSET152Atei16*+ | N. gerenzanensis Δdbv3–4 derivative carrying pSET152Atei16* | This work |
| A. teichomyceticus | Wild type, teicoplanin producer | NRRL-B16726 |
| A. teichomyceticus pSAD3+ | Wild type derivative carrying pSAD3 | This work |
| A. teichomyceticus pSAD4+ | Wild type derivative carrying pSAD4 | This work |
| A. teichomyceticus Δtei15* | Wild type derivative with tei15* gene replaced by oriT-aac(3)IV cassette | [35] |
| A. teichomyceticus Δtei16* | Wild type derivative with tei16* gene replaced with oriT-aac(3)IV cassette | [35] |
| A. teichomyceticus Δtei15* pSHAD4+ | A. teichomyceticus Δtei15* derivative carrying pSHAD4 | This work |
| A. teichomyceticus Δtei16* pSHAD3+ | A. teichomyceticus Δtei16* derivative carrying pSHAD3 | This work |
| E. coli DH5α | General cloning host | MBI Fermentas, US |
| E. coli ET12567 pUZ8002+ | (dam-13::Tn9 dcm-6), pUZ8002+ (ΔoriT), used for conjugative transfer of DNA | [60] |
| E. coli BW25113 pIJ790+ | λ-Red-mediated recombineering host | [43] |
| B. subtilis HB0933 | CU1065 liaR::kan | [61] |
| B. subtilis HB0950 | CU1065 SPβ2Δ2::Tn917::Φ(PliaI-74-cat-lacZ) | [50] |
4.2. Extraction of Genomic DNA
4.3. Inactivation of dbv3 and dbv4 in N. gerenzanensis
4.4. Generation of the Recombinant Plasmids
4.5. Conjugative Transfer of Plasmids into N. gerenzanensis and A. teichomyceticus
4.6. Microbial Growth Inhibition Assays
4.7. HPLC Analysis of Teicoplanin and A40926
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- van der Meij, A.; Worsley, S.F.; Hutchings, M.I.; van Wezel, G.P. Chemical ecology of antibiotic production by actinomycetes. FEMS Microbiol. Rev. 2017, 41, 392–416. [Google Scholar] [CrossRef]
- van Bambeke, F. Lipoglycopeptide antibacterial agents in Gram-positive infections: A comparative review. Drugs 2015, 75, 2073–2095. [Google Scholar] [CrossRef]
- Hansen, M.H.; Stegmann, E.; Cryle, M.J. Beyond vancomycin: Recent advances in the modification, reengineering, production and discovery of improved glycopeptide antibiotics to tackle multidrug-resistant bacteria. Curr. Opin. Biotechnol. 2022, 77, 102767. [Google Scholar] [CrossRef]
- Nicolaou, K.C.; Boddy, C.N.C.; Bräse, S.; Winssinger, N. Chemistry, biology, and medicine of the glycopeptide antibiotics. Angew. Chem.-Int. Ed. 1999, 38, 2096–2152. [Google Scholar] [CrossRef]
- Parenti, F.; Cavalleri, B. Proposal to name the vancomycin-ristocetin like glycopeptides as dalbaheptides. J. Antibiot. 1989, 42, 1882–1883. [Google Scholar] [CrossRef] [PubMed]
- Butler, M.S.; Hansford, K.A.; Blaskovich, M.A.T.; Halai, R.; Cooper, M.A. Glycopeptide antibiotics: Back to the future. J. Antibiot. 2014, 67, 631–644. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Huang, H.; Wei, W.; Zhong, Y.; Tang, B.; Yuan, H.; Zhu, L.; Huang, W.; Ge, M.; Yang, S.; et al. Complete genome sequence and comparative genomic analyses of the vancomycin-producing Amycolatopsis orientalis. BMC Genom. 2014, 15, 363. [Google Scholar] [CrossRef] [PubMed]
- Borgh, A.; Coronelli, C.; Faniuolo, L.; Allievi, G.; Pallanza, R.; Gallo, G.G. Teichomycins, new antibiotics from Actinoplanes teichomyceticus nov. sp.: IV. Separation and characterization of the components of teichomycin (teicoplanin). J. Antibiot. 1984, 37, 615–620. [Google Scholar] [CrossRef]
- Taurino, C.; Frattini, L.; Marcone, G.L.; Gastaldo, L.; Marinelli, F. Actinoplanes teichomyceticus ATCC 31121 as a cell factory for producing teicoplanin. Microb. Cell Fact. 2011, 10, 82. [Google Scholar] [CrossRef]
- Gunnarsson, N.; Bruheim, P.; Nielsen, J. Production of the glycopeptide antibiotic A40926 by Nonomuraea sp. ATCC 39727: Influence of medium composition in batch fermentation. J. Ind. Microbiol. Biotechnol. 2003, 30, 150–156. [Google Scholar] [CrossRef]
- Marcone, G.L.; Binda, E.; Carrano, L.; Bibb, M.; Marinelli, F. Relationship between glycopeptide production and resistance in the actinomycete Nonomuraea sp. ATCC 39727. Antimicrob. Agents Chemother. 2014, 58, 5191–5201. [Google Scholar] [CrossRef]
- Spohn, M.; Kirchner, N.; Kulik, A.; Jochim, A.; Wolf, F.; Muenzer, P.; Borst, O.; Gross, H.; Wohlleben, W.; Stegmann, E. Overproduction of ristomycin a by activation of a silent gene cluster in Amycolatopsis japonicum MG417-CF17. Antimicrob. Agents Chemother. 2014, 58, 6185–6196. [Google Scholar] [CrossRef]
- Liu, K.; Hu, X.R.; Zhao, L.X.; Wang, Y.; Deng, Z.; Taoa, M. Enhancing ristomycin a production by overexpression of ParB-like StrR family regulators controlling the biosynthesis genes. Appl. Environ. Microbiol. 2021, 87, e0106621. [Google Scholar] [CrossRef] [PubMed]
- Padma, P.N.; Rao, A.B.; Yadav, J.S.; Reddy, G. Optimization of fermentation conditions for production of glycopeptide antibiotic vancomycin by Amycolatopsis orientalis. Appl. Biochem. Biotechnol. 2002, 102–103, 395–405. [Google Scholar] [CrossRef]
- Levine, D.P. Vancomycin: A history. Clin. Infect. Dis. 2006, 42, 5–12. [Google Scholar] [CrossRef] [PubMed]
- Babul, N.; Pasko, M. Teicoplanin: A new glycopeptide antibiotic complex. Drug Intell. Clin. Pharm. 1988, 22, 218–226. [Google Scholar] [CrossRef] [PubMed]
- Glupczynski, Y.; Lagast, H.; Van der Auwera, P.; Thys, J.P.; Crokaert, F.; Yourassowsky, E.; Meunier-Carpentier, F.; Klastersky, J.; Kains, J.P.; Serruys-Schoutens, E. Clinical evaluation of teicoplanin for therapy of severe infections caused by Gram-positive bacteria. Antimicrob. Agents Chemother. 1986, 29, 52–57. [Google Scholar] [CrossRef]
- Klinker, K.P.; Borgert, S.J. Beyond vancomycin: The tail of the lipoglycopeptides. Clin. Ther. 2015, 37, 2619–2636. [Google Scholar] [CrossRef] [PubMed]
- Steiert, M.; Schmitz, F.-J. Dalbavancin (Biosearch Italia/Versicor). Curr. Opin. Investig. Drugs 2002, 3, 229–233. [Google Scholar]
- Goldstein, B.P.; Selva, E.; Gastaldo, L.; Berti, M.; Pallanza, R.; Ripamonti, F.; Ferrari, P.; Denaro, M.; Arioli, V.; Cassani, G. A40926, a new glycopeptide antibiotic with anti-Neisseria activity. Antimicrob. Agents Chemother. 1987, 31, 1961–1966. [Google Scholar] [CrossRef]
- Yim, G.; Thaker, M.N.; Koteva, K.; Wright, G. Glycopeptide antibiotic biosynthesis. J. Antibiot. 2014, 67, 31–41. [Google Scholar] [CrossRef] [PubMed]
- Yim, G.; Wang, W.; Thaker, M.N.; Tan, S.; Wright, G.D. How to make a glycopeptide: A synthetic biology approach to expand antibiotic chemical diversity. ACS Infect. Dis. 2016, 2, 642–650. [Google Scholar] [CrossRef] [PubMed]
- van Groesen, E.; Innocenti, P.; Martin, N.I. Recent advances in the development of semisynthetic glycopeptide antibiotics: 2014–2022. ACS Infect. Dis. 2022, 8, 1381–1407. [Google Scholar] [CrossRef] [PubMed]
- Acharya, Y.; Bhattacharyya, S.; Dhanda, G.; Haldar, J. Emerging roles of glycopeptide antibiotics: Moving beyond Gram-positive bacteria. ACS Infect. Dis. 2022, 8, 1–28. [Google Scholar] [CrossRef]
- Tian, L.; Shi, S.; Zhang, X.; Han, F.; Dong, H. Newest perspectives of glycopeptide antibiotics: Biosynthetic cascades, novel derivatives, and new appealing antimicrobial applications. World J. Microbiol. Biotechnol. 2023, 39, 67. [Google Scholar] [CrossRef]
- van der Heul, H.U.; Bilyk, B.L.; McDowall, K.J.; Seipke, R.F.; Van Wezel, G.P. Regulation of antibiotic production in Actinobacteria: New perspectives from the post-genomic era. Nat. Prod. Rep. 2018, 35, 575–604. [Google Scholar] [CrossRef]
- Ostash, B.; Yushchuk, O.; Tistechok, S.; Mutenko, H.; Horbal, L.; Muryn, A.; Dacyuk, Y.; Kalinowski, J.; Luzhetskyy, A.; Fedorenko, V. The adpA-like regulatory gene from Actinoplanes teichomyceticus: In silico analysis and heterologous expression. World J. Microbiol. Biotechnol. 2015, 31, 1297–1301. [Google Scholar] [CrossRef]
- Alduina, R.; Sosio, M.; Donadio, S. Complex regulatory networks governing production of the glycopeptide A40926. Antibiotics 2018, 7, 30. [Google Scholar] [CrossRef]
- Retzlaff, L.; Distler, J. The regulator of streptomycin gene expression, StrR, of Streptomyces griseus is a DNA binding activator protein with multiple recognition sites. Mol. Microbiol. 1995, 18, 151–162. [Google Scholar] [CrossRef]
- Yushchuk, O.; Horbal, L.; Ostash, B.; Marinelli, F.; Wohlleben, W.; Stegmann, E.; Fedorenko, V. Regulation of teicoplanin biosynthesis: Refining the roles of tei cluster-situated regulatory genes. Appl. Microbiol. Biotechnol. 2019, 103, 4089–4102. [Google Scholar] [CrossRef]
- Santos, C.L.; Correia-Neves, M.; Moradas-Ferreira, P.; Mendes, M.V. A Walk into the LuxR regulators of Actinobacteria: Phylogenomic distribution and functional diversity. PLoS ONE 2012, 7, e46758. [Google Scholar] [CrossRef] [PubMed]
- De Schrijver, A.; De Mot, R. A subfamily of MalT-related ATP-dependent regulators in the LuxR family. Microbiology 1999, 145, 1287–1288. [Google Scholar] [CrossRef]
- Shawky, R.M.; Puk, O.; Wietzorrek, A.; Pelzer, S.; Takano, E.; Wohlleben, W.; Stegmann, E. The border sequence of the balhimycin biosynthesis gene cluster from Amycolatopsis balhimycina contains bbr, encoding a StrR-like pathway-specific regulator. J. Mol. Microbiol. Biotechnol. 2007, 13, 76–88. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhang, C.; Zhao, Y.; Lei, X.; Jiang, Z.; Zhang, X.; Zheng, Z.; Si, S.; Wang, L.; Hong, B. Comparative genomics and transcriptomics analyses provide insights into the high yield and regulatory mechanism of norvancomycin biosynthesis in Amycolatopsis orientalis NCPC 2-48. Microb. Cell Fact. 2021, 20, 28. [Google Scholar] [CrossRef] [PubMed]
- Horbal, L.; Kobylyanskyy, A.; Truman, A.W.; Zaburranyi, N.; Ostash, B.; Luzhetskyy, A.; Marinelli, F.; Fedorenko, V. The pathway-specific regulatory genes, tei15* and tei16*, are the master switches of teicoplanin production in Actinoplanes teichomyceticus. Appl. Microbiol. Biotechnol. 2014, 98, 9295–9309. [Google Scholar] [CrossRef]
- Alduina, R.; Lo Piccolo, L.; D’Alia, D.; Ferraro, C.; Gunnarsson, N.; Donadio, S.; Puglia, A.M. Phosphate-controlled regulator for the biosynthesis of the dalbavancin precursor A40926. J. Bacteriol. 2007, 189, 8120–8129. [Google Scholar] [CrossRef]
- Lo Grasso, L.; Maffioli, S.; Sosio, M.; Bib, M.; Puglia, A.M.; Alduina, R. Two master switch regulators trigger A40926 biosynthesis in Nonomuraea sp. strain ATCC 39727. J. Bacteriol. 2015, 197, 2536–2544. [Google Scholar] [CrossRef]
- Yushchuk, O.; Andreo-Vidal, A.; Marcone, G.L.; Bibb, M.; Marinelli, F.; Binda, E. New molecular tools for regulation and improvement of A40926 glycopeptide antibiotic production in Nonomuraea gerenzanensis ATCC 39727. Front. Microbiol. 2020, 11, 8. [Google Scholar] [CrossRef]
- Pelzer, S.; Süßmuth, R.; Heckmann, D.; Recktenwald, J.; Huber, P.; Jung, G.; Wohlleben, W. Identification and analysis of the balhimycin biosynthetic gene cluster and its use for manipulating glycopeptide biosynthesis in Amycolatopsis mediterranei DSM5908. Antimicrob. Agents Chemother. 1999, 43, 1565–1573. [Google Scholar] [CrossRef] [PubMed]
- Li, T.L.; Huang, F.; Haydock, S.F.; Mironenko, T.; Leadlay, P.F.; Spencer, J.B. Biosynthetic gene cluster of the glycopeptide antibiotic teicoplanin: Characterization of two glycosyltransferases and the key acyltransferase. Chem. Biol. 2004, 11, 107–119. [Google Scholar] [CrossRef] [PubMed]
- Sosio, M.; Kloosterman, H.; Bianchi, A.; de Vreugd, P.; Dijkhuizen, L.; Donadio, S. Organization of the teicoplanin gene cluster in Actinoplanes teichomyceticus. Microbiology 2004, 150, 95–102. [Google Scholar] [CrossRef] [PubMed]
- Sosio, M.; Stinchi, S.; Beltrametti, F.; Lazzarini, A.; Donadio, S. The gene cluster for the biosynthesis of the glycopeptide antibiotic A40926 by Nonomuraea species. Chem. Biol. 2003, 10, 541–549. [Google Scholar] [CrossRef]
- Gust, B.; Chandra, G.; Jakimowicz, D.; Yuqing, T.; Bruton, C.J.; Chater, K.F. λ Red-mediated genetic manipulation of antibiotic-producing Streptomyces. Adv. Appl. Microbiol. 2004, 54, 107–128. [Google Scholar] [CrossRef] [PubMed]
- Jovetic, S.; Feroggio, M.; Marinelli, F.; Lancini, G. Factors influencing cell fatty acid composition and A40926 antibiotic complex production in Nonomuraea sp. ATCC 39727. J. Ind. Microbiol. Biotechnol. 2008, 35, 1131–1138. [Google Scholar] [CrossRef] [PubMed]
- Horbal, L.; Kobylyanskyy, A.; Yushchuk, O.; Zaburannyi, N.; Luzhetskyy, A.; Ostash, B.; Marinelli, F.; Fedorenko, V. Evaluation of heterologous promoters for genetic analysis of Actinoplanes teichomyceticus—Producer of teicoplanin, drug of last defense. J. Biotechnol. 2013, 168, 367–372. [Google Scholar] [CrossRef] [PubMed]
- Sherwood, E.J.; Hesketh, A.R.; Bibb, M.J. Cloning and analysis of the planosporicin lantibiotic biosynthetic gene cluster of Planomonospora alba. J. Bacteriol. 2013, 195, 2309–2321. [Google Scholar] [CrossRef]
- Yushchuk, O.; Vior, N.M.; Andreo-Vidal, A.; Berini, F.; Rückert, C.; Busche, T.; Binda, E.; Kalinowski, J.; Truman, A.W.; Marinelli, F. Genomic-led discovery of a novel glycopeptide antibiotic by Nonomuraea coxensis DSM 45129. ACS Chem. Biol. 2021, 16, 915–928. [Google Scholar] [CrossRef] [PubMed]
- Kieser, T.; Bibb, M.J.; Buttner, M.J.; Chater, K.F.; Hopwood, D.A. Practical Streptomyces Genetics; John Innes Foundation: Norwich, UK, 2000. [Google Scholar]
- Bibb, M.J.; Janssen, G.R.; Ward, J.M. Cloning and analysis of the promoter region of the erythromycin resistance gene (ermE) of Streptomyces erythraeus. Gene 1985, 38, 215–226. [Google Scholar] [CrossRef]
- Mascher, T.; Zimmer, S.L.; Smith, T.-A.; Helmann, J.D. Antibiotic-inducible promoter regulated by the cell envelope stress-sensing two-component system LiaRS of Bacillus subtilis. Antimicrob. Agents Chemother. 2004, 48, 2888–2896. [Google Scholar] [CrossRef]
- Shirling, E.B.; Gottlieb, D. Methods for characterization of Streptomyces species. Int. J. Syst. Evol. Microbiol. 1966, 16, 313–340. [Google Scholar] [CrossRef]
- Gonsior, M.; Mühlenweg, A.; Tietzmann, M.; Rausch, S.; Poch, A.; Süssmuth, R.D. Biosynthesis of the peptide antibiotic feglymycin by a linear nonribosomal peptide synthetase mechanism. ChemBioChem 2015, 16, 2610–2614. [Google Scholar] [CrossRef] [PubMed]
- Nazari, B.; Forneris, C.C.; Gibson, M.I.; Moon, K.; Schramma, K.R.; Seyedsayamdost, M.R. Nonomuraea sp. ATCC 55076 harbours the largest actinomycete chromosome to date and the kistamicin biosynthetic gene cluster. Medchemcomm 2017, 8, 780–788. [Google Scholar] [CrossRef] [PubMed]
- Boeck, L.D.; Mertz, F.P. A47934, a novel glycopeptide-aglycone antibiotic produced by a strain of Streptomyces toyocaensis: Taxonomy and fermentation studies. J. Antibiot. 1986, 39, 1533–1540. [Google Scholar] [CrossRef]
- Pootoolal, J.; Thomas, M.G.; Marshall, C.G.; Neu, J.M.; Hubbard, B.K.; Walsh, C.T.; Wright, G.D. Assembling the glycopeptide antibiotic scaffold: The biosynthesis of A47934 from Streptomyces toyocaensis NRRL15009. Proc. Natl. Acad. Sci. USA 2002, 99, 8962–8967. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Wang, W.; Waglechner, N.; Culp, E.J.; Guitor, A.K.; Wright, G.D. GPAHex—A synthetic biology platform for type IV–V glycopeptide antibiotic production and discovery. Nat. Commun. 2020, 11, 5232. [Google Scholar] [CrossRef]
- Gust, B.; Kieser, T.; Chater, K.F. PCR targeting system in Streptomyces coelicolor A3(2). John Innes Cent. 2000, 3, 1–39. [Google Scholar]
- Marcone, G.L.; Beltrametti, F.; Binda, E.; Carrano, L.; Foulston, L.; Hesketh, A.; Bibb, M.; Marinelli, F. Novel mechanism of glycopeptide resistance in the A40926 producer Nonomuraea sp. ATCC 39727. Antimicrob. Agents Chemother. 2010, 54, 2465–2472. [Google Scholar] [CrossRef]
- Bierman, M.; Logan, R.; O’Brien, K.; Seno, E.T.; Nagaraja Rao, R.; Schoner, B.E.; Rao, R.N.; Schoner, B.E. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene 1992, 116, 43–49. [Google Scholar] [CrossRef]
- Paranthaman, S.; Dharmalingam, K. Intergeneric conjugation in Streptomyces peucetius and Streptomyces sp. strain C5: Chromosomal integration and expression of recombinant plasmids carrying the chiC gene. Appl. Environ. Microbiol. 2003, 69, 84–91. [Google Scholar] [CrossRef]
- Mascher, T.; Margulis, N.G.; Wang, T.; Ye, R.W.; Helmann, J.D. Cell wall stress responses in Bacillus subtilis: The regulatory network of the bacitracin stimulon. Mol. Microbiol. 2003, 50, 1591–1604. [Google Scholar] [CrossRef]
- Ha, H.S.; Hwang, Y.I.; Choi, S.U. Application of conjugation using φC31 att/int system for Actinoplanes teichomyceticus, a producer of teicoplanin. Biotechnol. Lett. 2008, 30, 1233–1238. [Google Scholar] [CrossRef] [PubMed]
| Name | Nucleotide Sequence (5′-3′) | Purpose |
|---|---|---|
| dbv3KO_R | TTTACTAGTCCCCGATGAGCCTCGGTCC | Amplification of dbv3 with 2 Kbp flanks |
| dbv3KO_F | TTTAAGCTTGCCGATCAGACCGGTGCCG | |
| dbv4KO_R | CCTCGCGAGCTCGTAGCCG | Amplification of dbv4 with 2 Kbp flanks |
| dbv4KO_F | CGTGGAAGCGCAGTGCCTC | |
| 3/4KOVER_R | GATATCCTGCCCGAGGCCG | Verification of dbv3, dbv4, and dbv3–4 knockouts |
| 3/4KOVER_F | CCAGATGCTGCAGGCGCGA | |
| dbv3_P1 | GCAAACCAAGTCGACGAACCGCTTGGGGGACGAGCAAGAATTCCGGGGATCCGTCGACC | Replacement of dbv3 with oriT-aadA within A40Y |
| dbv3_P2 | GCCCGAGGCCGGCGAATTCGGCTTGTCGAACTCTTCGCTTGTAGGCTGGAGCTGCTTC | |
| dbv4_P1 | CCCCGGCTCCGATATGACGCTAATCGAATCGGAGGCTAGATTCCGGGGATCCGTCGACC | Replacement of dbv4 with oriT-aadA within A40Y |
| dbv4_P2 | GTCGCTCTACATACGGCCGCCCGGCTCATCCACTCGTGCTGTAGGCTGGAGCTGCTTC | |
| dbv3_FWpIJ | AAAACATATGCTGTTCGGGCGAGATCGT | Cloning of dbv3 into pIJ12551 |
| dbv3_RVpIJ | AAAAGCGGCCGCCTACAGCCGCACTGCCTC | |
| dbv4_FWpIJ | AAAAAACATATGGACCCGACGGGAGTTGACATA | Cloning of dbv4 into pIJ12551 |
| dbv4_RVpIJ | TTTATTAGCGGCCGCTCATCCAGCGGCCAGATCGGTCG | |
| tei15_FWpIJ | GGGCATATGACACCTGACGAAGAG | Cloning of tei15* into pIJ12551 |
| tei15_RVpIJ | AAAAGCGGCCGCTCAGCTCGCCATC | |
| pSET_ver_F | GCATCGGCCGCGCTCCCGA | Verification of tei15* and tei16* cloned into pSET152A |
| tei15*_ver_R | CAGCTCAGCGCCGCTGAGCA | Verification of tei15* cloned into pSET152Atei15* |
| tei16*_ver_R | CTCGCACACGCCCGGGCC | Verification of tei16* cloned into pSET152Atei16* |
| hygR_F | GATACACCAAGGAAAGTCT | Amplification of hygR |
| hygR_R | TGTAGGCTGGAGCTGCTTC | |
| aac(3)IV_Fw | ACCGACTGGACCTTCCTTCT | Verification of apramycin resistance cassette |
| aac(3)IV_Rv | TCGGTCAGCTTCTCAACCTT |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Andreo-Vidal, A.; Yushchuk, O.; Marinelli, F.; Binda, E. Cross-Talking of Pathway-Specific Regulators in Glycopeptide Antibiotics (Teicoplanin and A40926) Production. Antibiotics 2023, 12, 641. https://doi.org/10.3390/antibiotics12040641
Andreo-Vidal A, Yushchuk O, Marinelli F, Binda E. Cross-Talking of Pathway-Specific Regulators in Glycopeptide Antibiotics (Teicoplanin and A40926) Production. Antibiotics. 2023; 12(4):641. https://doi.org/10.3390/antibiotics12040641
Chicago/Turabian StyleAndreo-Vidal, Andrés, Oleksandr Yushchuk, Flavia Marinelli, and Elisa Binda. 2023. "Cross-Talking of Pathway-Specific Regulators in Glycopeptide Antibiotics (Teicoplanin and A40926) Production" Antibiotics 12, no. 4: 641. https://doi.org/10.3390/antibiotics12040641
APA StyleAndreo-Vidal, A., Yushchuk, O., Marinelli, F., & Binda, E. (2023). Cross-Talking of Pathway-Specific Regulators in Glycopeptide Antibiotics (Teicoplanin and A40926) Production. Antibiotics, 12(4), 641. https://doi.org/10.3390/antibiotics12040641









