Identification of Vibrio metschnikovii and Vibrio injensis Isolated from Leachate Ponds: Characterization of Their Antibiotic Resistance and Virulence-Associated Genes
Abstract
:1. Introduction
2. Results
2.1. Collection of Leachate Samples
2.2. Identification of the Isolates of Vibrio spp.
2.3. Antibiotic Susceptibility Profile
2.4. Genome Sequencing of Isolate L7-12
3. Discussion
3.1. Identification of Isolates of Vibrio spp.
3.2. Antibiotic Susceptibility Profile of Vibrio spp. Isolates from Leachates
3.3. Virulence Factors in Isolate L7-12 of Vibrio injensis
4. Materials and Methods
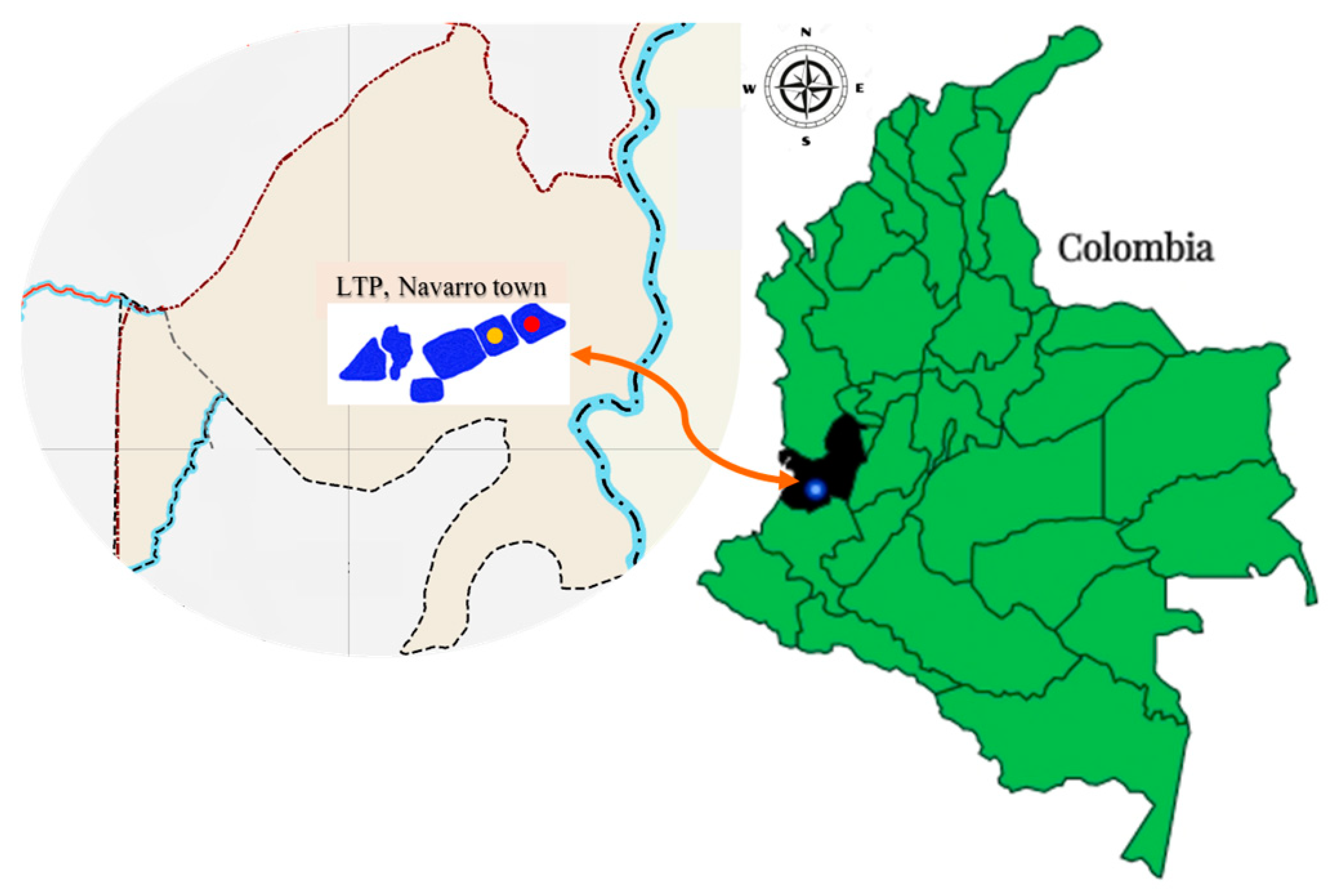
4.1. Sampling and Collection Sites of Leachate Samples
4.2. Biochemical Tests and Identification of Isolates of Vibrio spp.
4.3. Antibiotic Susceptibility Profile of the Isolates of Vibrio spp.
4.4. Genome Sequencing of Isolate L7-12
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Christensen, T.H.; Kjeldsen, P.; Hans-Jørgen, A.; Albrechtsen, A.; Heron, G.; Nielsen, P.H.; Bjerg, P.L.; Holm, P.E. Attenuation of Landfill Leachate Pollutants in Aquifers. Crit. Rev. Environ. Sci. Technol. 1994, 24, 119–202. [Google Scholar] [CrossRef]
- European Centre for Disease Prevention and Control & World Health Organization. Regional Office for Europe Antimicrobial Resistance Surveillance in Europe 2022–2020 Data; World Health Organization Regional Office European: Solna, Sweden, 2022; pp. xxi–136. [Google Scholar]
- Kjeldsen, P.; Barlaz, M.A.; Rooker, A.P.; Baun, A.; Ledin, A.; Christensen, T.H. Present and Long-Term Composition of MSW Landfill Leachate: A Review. Crit. Rev. Environ. Sci. Technol. 2002, 32, 297–336. [Google Scholar] [CrossRef]
- Martinez-Lopez, A.G.; Padrón-Hernández, W.; Rodríguez-Bernal, O.F.; Chiquito-Coyotl, O.; Escarola-Rosas, M.A.; Hernández-Lara, J.M.; Elvira-Hernández, E.A.; Méndez, G.A.; Tinoco-Magaña, J.C.; Martínez-Castillo, J. Alternativas Actuales Del Manejo de Lixiviados. Av. Quim. 2014, 9, 37–47. [Google Scholar]
- Torres, P.; Barba, L.E.; Pizarro, C. Mitigación de La Toxicidad Anaerobia de Lixiviados Mediante Mezclas Con Agua Residual Doméstica. Rev. Fac. Ing. 2010, 53, 64–74. [Google Scholar]
- Song, L.; Yang, S.; Gong, Z.; Wang, J.; Shi, X.; Wang, Y.; Zhang, R.; Wu, Y.; Wager, Y.Z. Antibiotics and Antibiotic-Resistant Genes in Municipal Solid Waste Landfills: Current Situation and Perspective. Curr. Opin. Environ. Sci. Health 2023, 31, 100421. [Google Scholar] [CrossRef]
- Wang, J.Y.; An, X.L.; Huang, F.Y.; Su, J.Q. Antibiotic Resistome in a Landfill Leachate Treatment Plant and Effluent-Receiving River. Chemosphere 2020, 242, 125207. [Google Scholar] [CrossRef]
- Almagro-Moreno, S.; Martinez-Urtaza, J.; Pukatzki, S. Vibrio Infections and the Twenty-First Century. Adv. Exp. Med. Biol. 2023, 1404, 1–16. [Google Scholar] [CrossRef]
- World Health Organization Cholera. Available online: https://www.who.int/news-room/fact-sheets/detail/cholera (accessed on 16 July 2023).
- Janda, J.M.; Newton, A.E.; Bopp, C.A. Vibriosis. Clin. Lab. Med. 2015, 35, 273–288. [Google Scholar] [CrossRef]
- Martinez-Urtaza, J.; Baker-Austin, C. Vibrio parahaemolyticus. Trends Microbiol. 2020, 28, 867–868. [Google Scholar] [CrossRef]
- Baker-Austin, C.; Oliver, J.D.; Alam, M.; Ali, A.; Waldor, M.K.; Qadri, F.; Martinez-Urtaza, J. Vibrio Spp. Infections. Nat. Rev. Dis. Prim. 2018, 4, 8. [Google Scholar] [CrossRef]
- Hecht, J.; Borowiak, M.; Fortmeier, B.; Dikou, S.; Gierer, W.; Klempien, I.; Nekat, J.; Schaefer, S.; Strauch, E. Case Report: Vibrio fluvialis Isolated from a Wound Infection after a Piercing Trauma in the Baltic Sea. Access Microbiol. 2022, 4, 000312. [Google Scholar] [CrossRef]
- Yang, A.; Yassin, M.; Phan, T. Vibrio Mimicus Wound Infection in a Burn Patient. Radiol. Case Rep. 2021, 16, 1348–1351. [Google Scholar] [CrossRef] [PubMed]
- Gusman, V.P.; Strajin, Z.R. Vibrio metschnikovii Isolated from Cosmetic Products as Potential Cause of Skin Infection. Future Microbiol. 2022, 17, 485–489. [Google Scholar] [CrossRef] [PubMed]
- Konechnyi, Y.; Khorkavyi, Y.; Ivanchuk, K.; Kobza, I.; Sękowska, A.; Korniychuk, O. Vibrio metschnikovii: Current State of Knowledge and Discussion of Recently Identified Clinical Case. Clin. Case Rep. 2021, 9, 2236–2244. [Google Scholar] [CrossRef]
- Mohebi, S.; Saboorian, R.; Shams, S. The First Report of Vibrio fluvialis Isolated from a Clinical Sample in Iran. Iran. J. Microbiol. 2022, 14, 677–682. [Google Scholar] [CrossRef]
- Dutta, D.; Kaushik, A.; Kumar, D.; Bag, S. Foodborne Pathogenic Vibrios: Antimicrobial Resistance. Front. Microbiol. 2021, 12, 638331. [Google Scholar] [CrossRef]
- Valáriková, J.; Korcová, J.; Ziburová, J.; Rosinský, J.; Čížová, A.; Bieliková, S.; Sojka, M.; Farkaš, P. Potential Pathogenicity and Antibiotic Resistance of Aquatic Vibrio Isolates from Freshwater in Slovakia. Folia Microbiol. 2020, 65, 545–555. [Google Scholar] [CrossRef]
- Páll, E.; Niculae, M.; Brudașcă, G.F.; Ravilov, R.K.; Șandru, C.D.; Cerbu, C.; Olah, D.; Zăblău, S.; Potârniche, A.V.; Spinu, M.; et al. Assessment and Antibiotic Resistance Profiling in Vibrio Species Isolated from Wild Birds Captured in Danube Delta Biosphere Reserve, Romania. Antibiotics 2021, 10, 333. [Google Scholar] [CrossRef]
- Vu, T.T.T.; Hoang, T.T.H.; Fleischmann, S.; Pham, H.N.; Lai, T.L.H.; Cam, T.T.H.; Truong, L.O.; Le, V.P.; Alter, T. Quantification and Antimicrobial Resistance of Vibrio parahaemolyticus in Retail Seafood in Hanoi, Vietnam. J. Food Prot. 2022, 85, 786–791. [Google Scholar] [CrossRef]
- Mohammed, Y.; Aboderin, A.O.; Okeke, I.N.; Olayinka, A.T. Antimicrobial Resistance of Vibrio cholerae from Sub-Saharan Africa: A Systematic Review. Afr. J. Lab. Med. 2018, 7, 7. [Google Scholar] [CrossRef]
- Das, B.; Verma, J.; Kumar, P.; Ghosh, A.; Ramamurthy, T. Antibiotic Resistance in Vibrio cholerae: Understanding the Ecology of Resistance Genes and Mechanisms. Vaccine 2020, 38, A83–A92. [Google Scholar] [CrossRef]
- Ambler, R.P. The Structure of Beta-Lactamases. Philos. Trans. R. Soc. B 1980, 289, 320–331. [Google Scholar] [CrossRef]
- Jang, K.S.; Kim, Y.H. Rapid and Robust MALDI-TOF MS Techniques for Microbial Identification: A Brief Overview of Their Diverse Applications. J. Microbiol. 2018, 56, 209–216. [Google Scholar] [CrossRef] [PubMed]
- Singhal, N.; Kumar, M.; Kanaujia, P.K.; Virdi, J.S. MALDI-TOF Mass Spectrometry: An Emerging Technology for Microbial Identification and Diagnosis. Front. Microbiol. 2015, 6, 791. [Google Scholar] [CrossRef] [PubMed]
- Santos, I.C.; Hildenbrand, Z.L.; Schug, K.A. Applications of MALDI-TOF MS in Environmental Microbiology. Analyst 2016, 141, 2827–2837. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Oh, H.S.; Park, S.C.; Chun, J. Towards a Taxonomic Coherence between Average Nucleotide Identity and 16S RRNA Gene Sequence Similarity for Species Demarcation of Prokaryotes. Int. J. Syst. Evol. Microbiol. 2014, 64, 346–351. [Google Scholar] [CrossRef]
- Jolley, K.A.; Bray, J.E.; Maiden, M.C.J. Open-Access Bacterial Population Genomics: BIGSdb Software, the PubMLST.Org Website and Their Applications. Wellcome Open Res. 2018, 3, 124. [Google Scholar] [CrossRef]
- Rahman, M.S.; Martino, M.E.; Cardazzo, B.; Facco, P.; Bordin, P.; Mioni, R.; Novelli, E.; Fasolato, L. Vibrio Trends in the Ecology of the Venice Lagoon. Appl. Environ. Microbiol. 2014, 80, 2372–2380. [Google Scholar] [CrossRef]
- Håkonsholm, F.; Lunestad, B.T.; Aguirre Sánchez, J.R.; Martinez-Urtaza, J.; Marathe, N.P.; Svanevik, C.S. Vibrios from the Norwegian Marine Environment: Characterization of Associated Antibiotic Resistance and Virulence Genes. Microbiologyopen 2020, 9, e1093. [Google Scholar] [CrossRef]
- Okoh, A.I.; Igbinosa, E.O. Antibiotic Susceptibility Profiles of Some Vibrio Strains Isolated from Wastewater Final Effluents in a Rural Community of the Eastern Cape Province of South Africa. BMC Microbiol. 2010, 10, 143. [Google Scholar] [CrossRef]
- Flores, B.; González, N.; Bravo, A.; Mora-Sánchez, B.; Torres, D.; Jirón, W.; Sheleby-Elías, J.; Balcázar, J.L. Identification of Pathogenic Bacteria in Fishes Caught in the Pacific off Nicaragua. Cienc. Mar. 2021, 47, 175–184. [Google Scholar] [CrossRef]
- Xiao, Z.; Li, X.; Xue, M.; Zhang, M.; Liu, W.; Fan, Y.; Chen, X.; Chu, Z.; Gong, F.; Zeng, L.; et al. Vibrio metschnikovii, a Potential Pathogen in Freshwater-Cultured Hybrid Sturgeon. Animals 2022, 12, 1101. [Google Scholar] [CrossRef] [PubMed]
- Dorman, M.J.; Kane, L.; Domman, D.; Turnbull, J.D.; Cormie, C.; Fazal, M.A.; Goulding, D.A.; Russell, J.E.; Alexander, S.; Thomson, N.R. The History, Genome and Biology of NCTC 30: A Non-Pandemic Vibrio cholerae Isolate from World War One. Proc. R. Soc. B Biol. Sci. 2019, 286, 20182025. [Google Scholar] [CrossRef]
- Chiou, J.; Li, R.; Chen, S. CARB-17 Family of β-Lactamases Mediates Intrinsic Resistance to Penicillins in Vibrio parahaemolyticus. Antimicrob. Agents Chemother. 2015, 59, 3593–3595. [Google Scholar] [CrossRef] [PubMed]
- Castle, S.S. Carbenicillin. In xPharm: The Comprehensive Pharmacology Reference; Elsevier: Amsterdam, The Netherlands, 2007; pp. 1–5. ISBN 9780080552323. [Google Scholar]
- Pariente Martín, M.; Escribano Garaizábal, E.; Liria Sánchez, P.J.; Crespo Sánchez, M.D. Vibrio metschnikovii from a Human Infected Leg Ulcer. Rev. Inst. Med. Trop. Sao Paulo 2008, 50, 311–312. [Google Scholar] [CrossRef] [PubMed]
- Wallet, F.; Tachon, M.; Nseir, S.; Courcol, R.J.; Roussel-Delvallez, M. Vibrio metschnikovii Pneumonia. Emerg. Infect. Dis. 2005, 11, 1641–1642. [Google Scholar] [CrossRef]
- Andersen, J.L.; He, G.X.; Kakarla, P.; Ranjana, K.C.; Kumar, S.; Lakra, W.S.; Mukherjee, M.M.; Ranaweera, I.; Shrestha, U.; Tran, T.; et al. Multidrug Efflux Pumps from Enterobacteriaceae, Vibrio cholerae and Staphylococcus Aureus Bacterial Food Pathogens. Int. J. Environ. Res. Public Health 2015, 12, 1487–1547. [Google Scholar] [CrossRef]
- Sylvie, G.-T.; Kristin, J.L. Mechanisms of Resistance to Aminoglycoside Antibiotics: Overview and Perspectives. Medchemcomm 2016, 7, 11–24. [Google Scholar] [CrossRef]
- Krause, K.M.; Serio, A.W.; Kane, T.R.; Connolly, L.E. Aminoglycosides: An Overview. Cold Spring Harb. Perspect. Med. 2016, 6, a027029. [Google Scholar] [CrossRef]
- Rizzo, L.; Manaia, C.; Merlin, C.; Schwartz, T.; Dagot, C.; Ploy, M.C.; Michael, I.; Fatta-Kassinos, D. Urban Wastewater Treatment Plants as Hotspots for Antibiotic Resistant Bacteria and Genes Spread into the Environment: A Review. Sci. Total Environ. 2013, 447, 345–360. [Google Scholar] [CrossRef]
- Watts, J.E.M.; Schreier, H.J.; Lanska, L.; Hale, M.S. The Rising Tide of Antimicrobial Resistance in Aquaculture: Sources, Sinks and Solutions. Mar. Drugs 2017, 15, 158. [Google Scholar] [CrossRef] [PubMed]
- Bier, N.; Schwartz, K.; Guerra, B.; Strauch, E. Survey on Antimicrobial Resistance Patterns in Vibrio vulnificus and Vibrio cholerae Non-O1/Non-O139 in Germany Reveals Carbapenemase-Producing Vibrio cholerae in Coastal Waters. Front. Microbiol. 2015, 6, 1179. [Google Scholar] [CrossRef]
- Le Roux, F.; Wegner, K.M.; Baker-Austin, C.; Vezzulli, L.; Osorio, C.R.; Amaro, C.; Ritchie, J.M.; Defoirdt, T.; Destoumieux-Garzón, D.; Blokesch, M.; et al. The Emergence of Vibrio Pathogens in Europe: Ecology, Evolution and Pathogenesis (Paris, 11–12th March 2015). Front. Microbiol. 2015, 6, 830. [Google Scholar] [CrossRef]
- Froelich, B.A.; Daines, D.A. In Hot Water: Effects of Climate Change on Vibrio–human Interactions. Environ. Microbiol. 2020, 22, 4101–4111. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Reytor, D.; Jaña, V.; Pavez, L.; Navarrete, P.; García, K. Accessory Toxins of Vibriopathogens and Their Role in Epithelial Disruption during Infection. Front. Microbiol. 2018, 9, 2248. [Google Scholar] [CrossRef]
- Jones, M.K.; Oliver, J.D. Vibrio vulnificus: Disease and Pathogenesis. Infect. Immun. 2009, 77, 1723–1733. [Google Scholar] [CrossRef]
- Duong-Nu, T.M.; Jeong, K.; Hong, S.H.; Puth, S.; Kim, S.Y.; Tan, W.; Lee, K.H.; Lee, S.E.; Rhee, J.H. A Stealth Adhesion Factor Contributes to Vibrio vulnificus Pathogenicity: Flp Pili Play Roles in Host Invasion, Survival in the Blood Stream and Resistance to Complement Activation. PLoS Pathog. 2019, 15, e1007767. [Google Scholar] [CrossRef]
- Li, L.; Meng, H.; Gu, D.; Li, Y.; Jia, M. Molecular Mechanisms of Vibrio parahaemolyticus Pathogenesis. Microbiol. Res. 2019, 222, 43–51. [Google Scholar] [CrossRef]
- You-Chul, J.; Mi-Ae, L.; Kyu-Ho, L. Role of Flagellin-Homologous Proteins in Biofilm Formation by Pathogenic Vibrio Species. mBio 2019, 10, e01793-19. [Google Scholar] [CrossRef]
- Ruenchit, P.; Reamtong, O.; Siripanichgon, K.; Chaicumpa, W.; Diraphat, P. New Facet of Non-O1/Non-O139 Vibrio cholerae Hemolysin a: A Competitive Factor in the Ecological Niche. FEMS Microbiol. Ecol. 2017, 93, 1–12. [Google Scholar] [CrossRef]
- Raghunath, P. Roles of Thermostable Direct Hemolysin (TDH) and TDH-Related Hemolysin (TRH) in Vibrio parahaemolyticus. Front. Microbiol. 2015, 5, 2010–2013. [Google Scholar] [CrossRef] [PubMed]
- Matsuda, S.; Kodama, T.; Okada, N.; Okayama, K.; Honda, T.; Iida, T. Association of Vibrio parahaemolyticus Thermostable Direct Hemolysin with Lipid Rafts Is Essential for Cytotoxicity but Not Hemolytic Activity. Infect. Immun. 2010, 78, 603–610. [Google Scholar] [CrossRef] [PubMed]
- Honda, T.; Ni, Y.; Miwatani, T.; Adachi, T.; Kim, J. The Thermostable Direct Hemolysin of Vibrio parahaemolyticus Is a Pore-Forming Toxin. Can. J. Microbiol. 1992, 38, 1175–1180. [Google Scholar] [CrossRef] [PubMed]
- Miyake, M.; Honda, T.; Miwatani, T. Effects of Divalent Cations and Polysaccharide on Vibrio metschnikovii Cytolysin-Induced Hemolysis of Rabbit Erythrocytes. Infect. Immun. 1989, 57, 158–163. [Google Scholar] [CrossRef] [PubMed]
- Paek, J.; Shin, J.H.; Shin, Y.; Park, I.S.; Kim, H.; Kook, J.K.; Kang, S.S.; Kim, D.S.; Park, K.H.; Chang, Y.H. Vibrio Injenensis Sp. Nov., Isolated from Human Clinical Specimens. Antonie Leeuwenhoek 2016, 110, 145–152. [Google Scholar] [CrossRef] [PubMed]
- Ghenem, L.; Elhadi, N.; Alzahrani, F.; Nishibuchi, M. Vibrio Parahaemolyticus: A Review on Distribution, Pathogenesis, Virulence Determinants and Epidemiology. Saudi J. Med. Med. Sci. 2017, 5, 93. [Google Scholar] [CrossRef]
- Kustusch, R.J.; Kuehl, C.J.; Crosa, J.H. Power Plays: Iron Transport and Energy Transduction in Pathogenic Vibrios. BioMetals 2011, 24, 559–566. [Google Scholar] [CrossRef]
- Broberg, C.A.; Calder, T.J.; Orth, K. Vibrio parahaemolyticus Cell Biology and Pathogenicity Determinants. Microbes Infect. 2011, 13, 992–1001. [Google Scholar] [CrossRef]
- Tanabe, T.; Funahashi, T.; Nakao, H.; Miyoshi, S.I.; Shinoda, S.; Yamamoto, S. Identification and Characterization of Genes Required for Biosynthesis and Transport of the Siderophore Vibrioferrin in Vibrio parahaemolyticus. J. Bacteriol. 2003, 185, 6938–6949. [Google Scholar] [CrossRef]
- León-Sicairos, N.; Angulo-Zamudio, U.A.; de la Garza, M.; Velázquez-Román, J.; Flores-Villaseñor, H.M.; Canizalez-Román, A. Strategies of Vibrio parahaemolyticus to Acquire Nutritional Iron during Host Colonization. Front. Microbiol. 2015, 6, 702. [Google Scholar] [CrossRef]
- Ramamurthy, T.; Nandy, R.K.; Mukhopadhyay, A.K.; Dutta, S.; Mutreja, A.; Okamoto, K.; Miyoshi, S.I.; Nair, G.B.; Ghosh, A. Virulence Regulation and Innate Host Response in the Pathogenicity of Vibrio cholerae. Front. Cell. Infect. Microbiol. 2020, 10, 1–22. [Google Scholar] [CrossRef]
- Pettis, G.S.; Mukerji, A.S. Structure, Function, and Regulation of the Essential Virulence Factor Capsular Polysaccharide of Vibrio vulnificus. Int. J. Mol. Sci. 2020, 21, 3259. [Google Scholar] [CrossRef] [PubMed]
- Khan, F.; Tabassum, N.; Anand, R.; Kim, Y.M. Motility of Vibrio Spp.: Regulation and Controlling Strategies. Appl. Microbiol. Biotechnol. 2020, 104, 8187–8208. [Google Scholar] [CrossRef]
- Tsou, A.; Frey, E.; Hsiao, A.; Liu, Z.; Zhu, J. Coordinated Regulation of Virulence by Quorum Sensing and Motility Pathways during the Initial Stages of Vibrio cholerae Infection. Commun. Integr. Biol. 2008, 1, 42–44. [Google Scholar] [CrossRef] [PubMed]
- Syed, K.A.; Beyhan, S.; Correa, N.; Queen, J.; Liu, J.; Peng, F.; Satchell, K.J.F.; Yildiz, F.; Klose, K.E. The Vibrio cholerae Flagellar Regulatory Hierarchy Controls Expression of Virulence Factors. J. Bacteriol. 2009, 191, 6555–6570. [Google Scholar] [CrossRef]
- Ligthart, K.; Belzer, C.; de Vos, W.M.; Tytgat, H.L.P. Bridging Bacteria and the Gut: Functional Aspects of Type IV Pili. Trends Microbiol. 2020, 28, 340–348. [Google Scholar] [CrossRef]
- Ayers, M.; Howell, P.L.; Burrows, L.L. Architecture of the Type II Secretion and Type IV Pilus Machineries. Future Microbiol. 2010, 5, 1203–1218. [Google Scholar] [CrossRef]
- Reichow, S.L.; Korotkov, K.V.; Gonen, M.; Sun, J.; Delarosa, J.R.; Hol, W.G.J.; Gonen, T. The Binding of Cholera Toxin to the Periplasmic Vestibule of the Type II Secretion Channel. Channels 2011, 5, 215–218. [Google Scholar] [CrossRef]
- Zheng, L.; Zhu, L.W.; Jing, J.; Guan, J.Y.; Lu, G.J.; Xie, L.H.; Ji, X.; Chu, D.; Sun, Y.; Chen, P.; et al. Pan-Genome Analysis of Vibrio cholerae and Vibrio metschnikovii Strains Isolated From Migratory Birds at Dali Nouer Lake in Chifeng, China. Front. Vet. Sci. 2021, 8, 638820. [Google Scholar] [CrossRef]
- Echazarreta, M.A.; Klose, K.E. Vibrio Flagellar Synthesis. Front. Cell. Infect. Microbiol. 2019, 9, 131. [Google Scholar] [CrossRef]
- Lutz, C.; Erken, M.; Noorian, P.; Sun, S.; McDougald, D. Environmental Reservoirs and Mechanisms of Persistence of Vibrio cholerae. Front. Microbiol. 2013, 4, 375. [Google Scholar] [CrossRef] [PubMed]
- Yildiz, F.H.; Visick, K.L. Vibrio Biofilms: So Much the Same yet so Different. Trends Microbiol. 2009, 17, 109–118. [Google Scholar] [CrossRef]
- Chiavelli, D.A.; Marsh, J.W.; Taylor, R.K. The Mannose-Sensitive Hemagglutinin of Vibrio cholerae Promotes Adherence to Zooplankton. Appl. Environ. Microbiol. 2001, 67, 3220–3225. [Google Scholar] [CrossRef] [PubMed]
- Mondragón-Quiguanas, A.; Villaquirán-Muriel, M.Á.; Rivera, S.P.; Rosero-García, D.; Aranaga, C.; Correa, A.; Falco, A. Beta-Lactam-Resistant Enterobacterales Isolated from Landfill Leachates. Pathogens 2022, 11, 1077. [Google Scholar] [CrossRef] [PubMed]
- Bartholomew, J.W.M.T. The Gram Stain. Bacteriol. Rev. 1952, 16, 1–29. [Google Scholar] [CrossRef]
- Lu, J.J.; Perng, C.L.; Lee, S.Y.; Wan, C.C. Use of PCR with Universal Primers and Restriction Endonuclease Digestions for Detection and Identification of Common Bacterial Pathogens in Cerebrospinal Fluid. J. Clin. Microbiol. 2000, 38, 2076–2080. [Google Scholar] [CrossRef]
- Clinical and Laboratory Standards Institute CLSI. Methods for Antimicrobial Dilution and Disk Susceptibility Testing of Infrequently Isolated or Fastidious Bacteria, 3rd ed.; CLSI Guideline M45; CLSI: Wayne, PE, USA, 2016; ISBN 1562389173. [Google Scholar]
- Clinical and Laboratory Standards Institute CLSI. M100 Performance Standards for Antimicrobial; CLSI: Wayne, PE, USA, 2020; Volume 40, ISBN 9781684400669. [Google Scholar]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An Ultra-Fast All-in-One FASTQ Preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Prjibelski, A.; Antipov, D.; Meleshko, D.; Lapidus, A.; Korobeynikov, A. Using SPAdes De Novo Assembler. Curr. Protoc. Bioinform. 2020, 70, e102. [Google Scholar] [CrossRef]
- Mikheenko, A.; Prjibelski, A.; Saveliev, V.; Antipov, D.; Gurevich, A. Versatile Genome Assembly Evaluation with QUAST-LG. Bioinformatics 2018, 34, i142–i150. [Google Scholar] [CrossRef]
- Florensa, A.F.; Kaas, R.S.; Clausen, P.T.L.C.; Aytan-Aktug, D.; Aarestrup, F.M. ResFinder—An Open Online Resource for Identification of Antimicrobial Resistance Genes in next-Generation Sequencing Data and Prediction of Phenotypes from Genotypes. Microb. Genom. 2022, 8, 000748. [Google Scholar] [CrossRef]
- Olson, R.D.; Assaf, R.; Brettin, T.; Conrad, N.; Cucinell, C.; Davis, J.J.; Dempsey, D.M.; Dickerman, A.; Dietrich, E.M.; Kenyon, R.W.; et al. Introducing the Bacterial and Viral Bioinformatics Resource Center (BV-BRC): A Resource Combining PATRIC, IRD and ViPR. Nucleic Acids Res. 2023, 51, D678–D689. [Google Scholar] [CrossRef] [PubMed]
- Arkin, A.P.; Cottingham, R.W.; Henry, C.S.; Harris, N.L.; Stevens, R.L.; Maslov, S.; Dehal, P.; Ware, D.; Perez, F.; Canon, S.; et al. KBase: The United States Department of Energy Systems Biology Knowledgebase. Nat. Biotechnol. 2018, 36, 566–569. [Google Scholar] [CrossRef] [PubMed]

| Sampling Site | Isolate Name | Oxidase Test | Antibiotic Susceptibility Profile (Kirby–Bauer Test) | |||||
|---|---|---|---|---|---|---|---|---|
| CTX | CAZ | IPM | CIP | AM | CN | |||
| Lagoon 3 | L3-2 | − | S | S | S | S | R | I |
| L3-5 | − | S | S | S | S | R | S | |
| L3-7 | − | S | S | S | S | I | I | |
| L3-18 | − | R | S | S | S | S | S | |
| L3-19 | − | S | S | S | S | S | S | |
| Lagoon 7 | L7-1 | − | S | S | S | S | R | I |
| L7-2 | − | S | S | S | S | R | S | |
| L7-3 | − | S | S | S | S | I | S | |
| L7-4 | − | S | R | S | S | R | I | |
| L7-5 | − | S | S | S | S | R | I | |
| L7-6 | − | S | S | S | S | I | S | |
| L7-7 | − | S | S | S | S | R | I | |
| L7-9 | − | S | S | S | S | R | S | |
| L7-10 | − | S | I | S | S | R | R | |
| L7-11 | − | S | S | S | S | I | S | |
| L7-12 | + | R | R | S | S | R | I | |
| L7-13 | − | S | I | S | S | R | S | |
| L7-14 | − | S | S | S | S | R | I | |
| L7-15 | − | S | R | S | S | S | S | |
| L7-16 | − | S | S | S | S | S | S | |
| L7-17 | − | I | I | S | S | R | R | |
| L7-18 | − | S | I | S | S | I | S | |
| L7-20 | − | I | S | S | S | S | S | |
| Genes | Functions |
|---|---|
| Sigma factors | |
| rpoS | Survival due to nutritional, oxidative, and osmotic stress |
| rpoE | |
| Antibiotic resistance | |
| blaCARB-9 | Carbenicillin (Beta-lactam) resistance |
| Efflux pumps | |
| norM | MATE |
| adeF, adeG, adeH | RND |
| Virulence factors | |
| tlh, tdh, hlyA, rxtC | Hemolysins |
| fur, vibB | Iron uptake |
| wecH | Capsular antiphagocytosis polysaccharides |
| glyA-1 | Intestinal colonization |
| flhA, fliEGI, fliR, fliA, flhB, fliMNQ, flaBD, fleN | Flagellar synthesis |
| cheY2, cheZ, cheAB, cheW1 | Chemotaxis |
| pilA, pilQ, pilT | Type IV pilus |
| mshA | Mannose-sensitive Hemagglutinin |
| gspk, gspF, hcp, vgrG-2 | Extracellular protein secretion |
| luxPQ, luxO, luxU | Quorum sensing |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Falco, A.; Villaquirán-Muriel, M.Á.; Gallo Pérez, J.D.; Mondragón-Quiguanas, A.; Aranaga, C.; Correa, A. Identification of Vibrio metschnikovii and Vibrio injensis Isolated from Leachate Ponds: Characterization of Their Antibiotic Resistance and Virulence-Associated Genes. Antibiotics 2023, 12, 1571. https://doi.org/10.3390/antibiotics12111571
Falco A, Villaquirán-Muriel MÁ, Gallo Pérez JD, Mondragón-Quiguanas A, Aranaga C, Correa A. Identification of Vibrio metschnikovii and Vibrio injensis Isolated from Leachate Ponds: Characterization of Their Antibiotic Resistance and Virulence-Associated Genes. Antibiotics. 2023; 12(11):1571. https://doi.org/10.3390/antibiotics12111571
Chicago/Turabian StyleFalco, Aura, Miguel Ángel Villaquirán-Muriel, José David Gallo Pérez, Alejandra Mondragón-Quiguanas, Carlos Aranaga, and Adriana Correa. 2023. "Identification of Vibrio metschnikovii and Vibrio injensis Isolated from Leachate Ponds: Characterization of Their Antibiotic Resistance and Virulence-Associated Genes" Antibiotics 12, no. 11: 1571. https://doi.org/10.3390/antibiotics12111571
APA StyleFalco, A., Villaquirán-Muriel, M. Á., Gallo Pérez, J. D., Mondragón-Quiguanas, A., Aranaga, C., & Correa, A. (2023). Identification of Vibrio metschnikovii and Vibrio injensis Isolated from Leachate Ponds: Characterization of Their Antibiotic Resistance and Virulence-Associated Genes. Antibiotics, 12(11), 1571. https://doi.org/10.3390/antibiotics12111571






