Phylotypic Profiling, Distribution of Pathogenicity Island Markers, and Antimicrobial Susceptibility of Escherichia coli Isolated from Retail Chicken Meat and Humans
Abstract
1. Introduction
2. Results
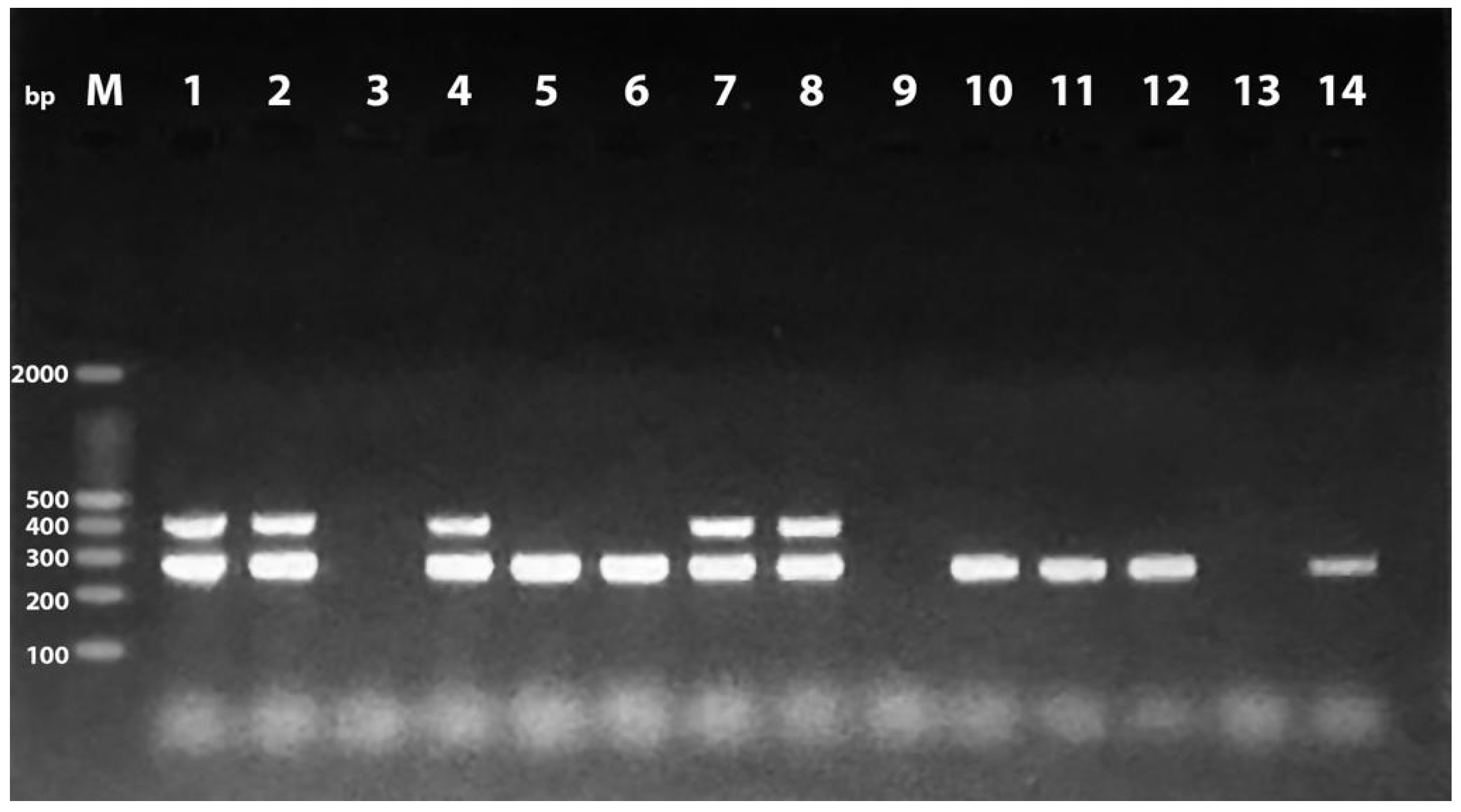
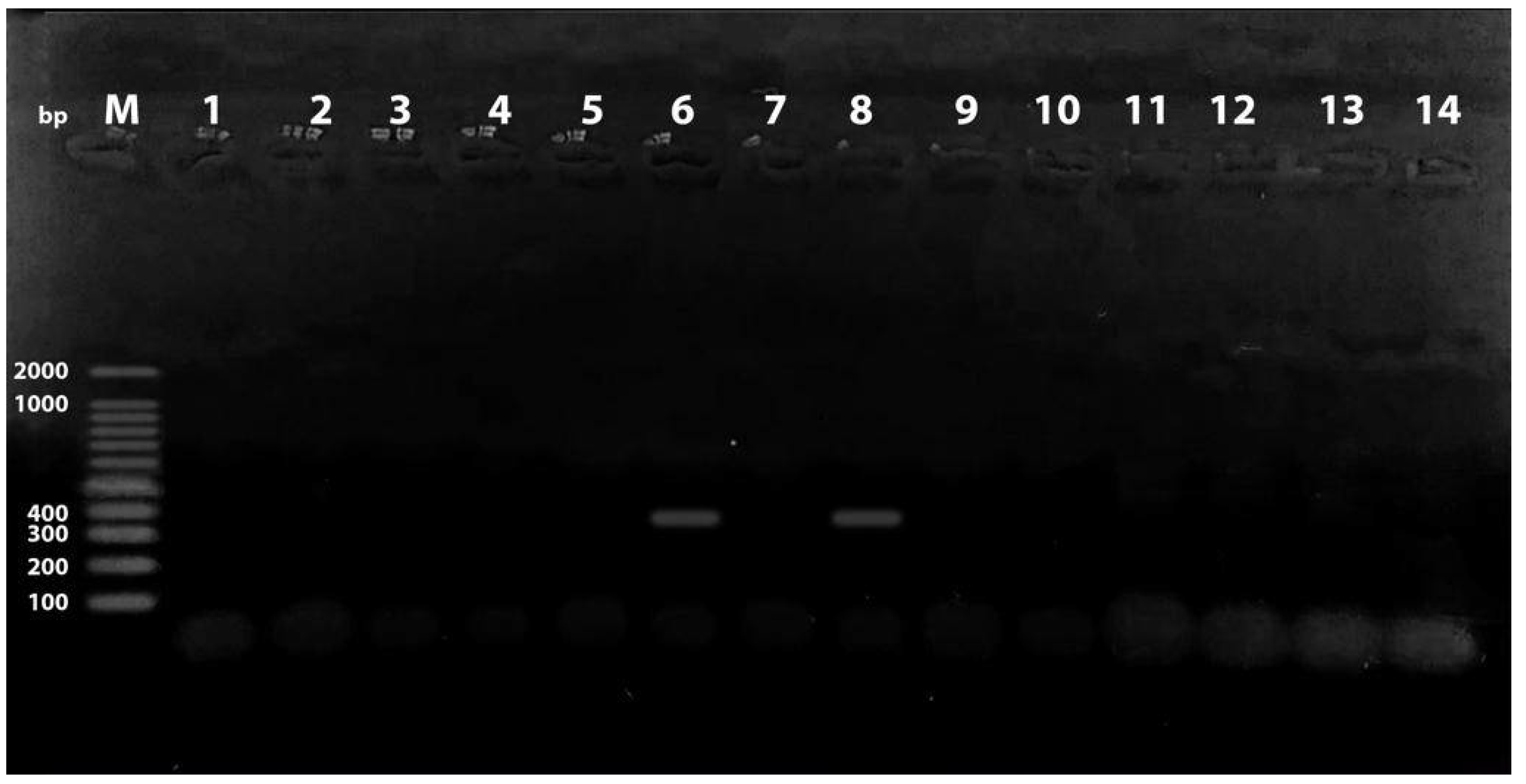
2.1. Prevalence of Phylogenetic Groups and PAIs among the Examined E. coli Isolates
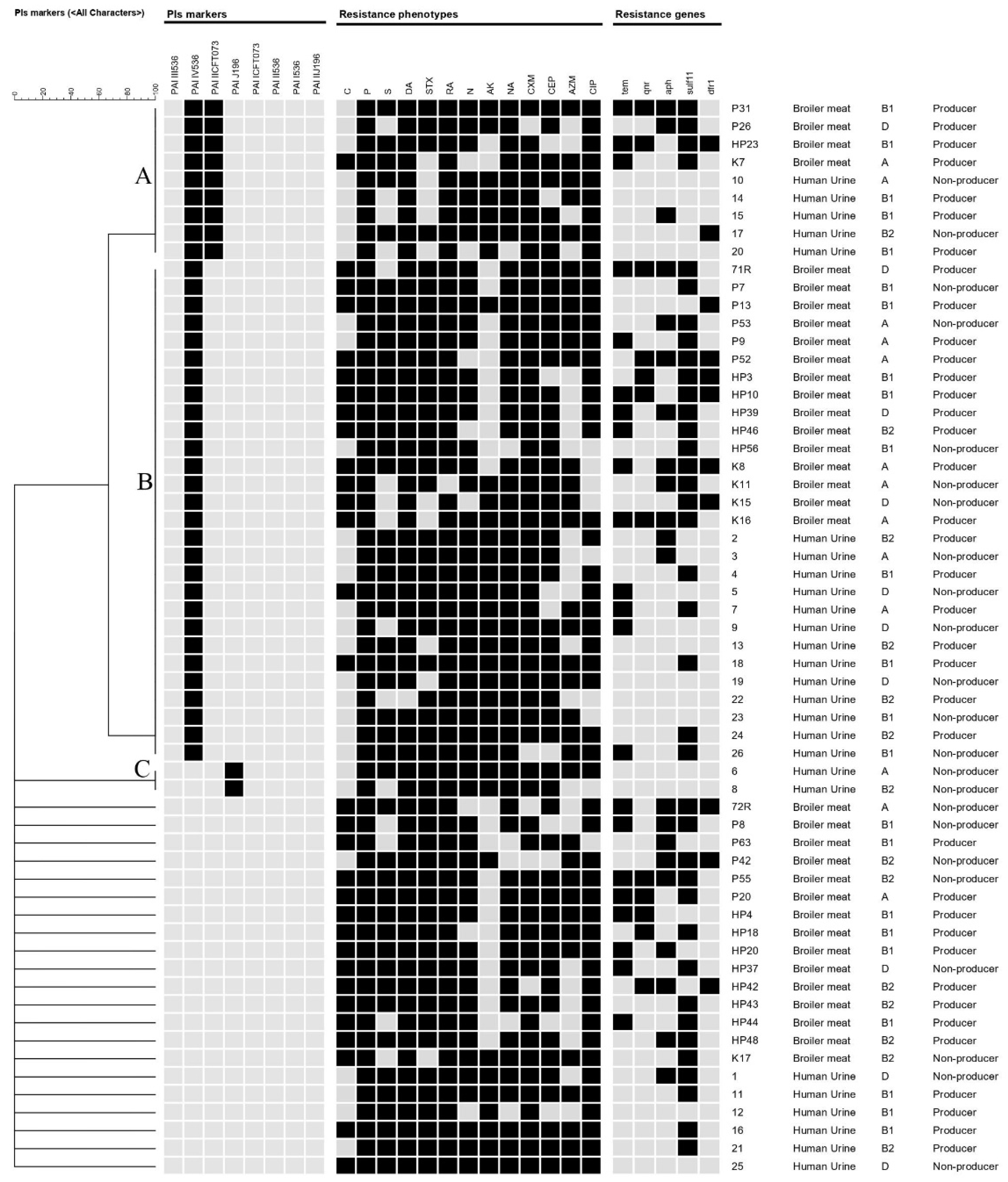
2.2. Distribution of PAIs among the Examined E. coli Isolates in Relation to Phylogenetic Group
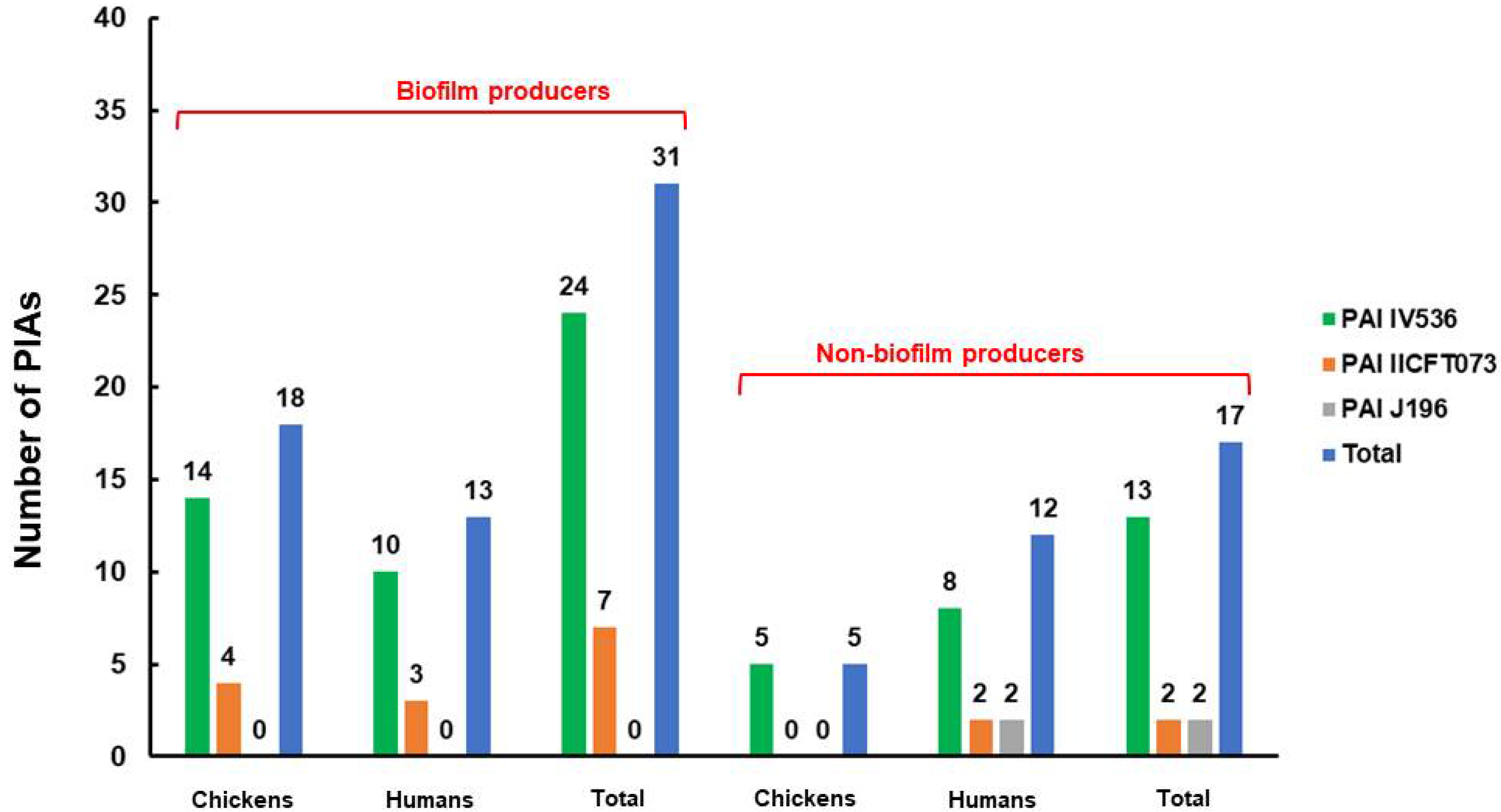
2.3. Antimicrobial Resistance Phenotypes, Genotypes, and in Vitro Biofilm Production of the Examined E. coli Isolates
3. Discussion
4. Materials and Methods
4.1. Sample Collection and Conventional Isolation of E. coli
4.2. Molecular Characterization of E. coli
4.3. PCR-based Phylotyping
4.4. Detection of Pathogenicity Islands Markers
4.5. Antimicrobial Susceptibility Testing
4.6. Molecular Characterization of Antimicrobial Resistance Genes
4.7. Biofilm-Formation Assay
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Correction Statement
References
- Leimbach, A.; Hacker, J.; Dobrindt, U. E. coli as an All-Rounder: The thin line between commensalism and pathogenicity. In Between Pathogenicity and Commensalism; Dobrindt, U., Hacker, J.H., Svanborg, C., Eds.; Springer: Berlin/Heidelberg, Germany, 2013; pp. 3–32. [Google Scholar]
- Croxen, M.A.; Finlay, B.B. Molecular mechanisms of Escherichia coli pathogenicity. Nat. Rev. Microbiol. 2010, 8, 26–38. [Google Scholar] [CrossRef] [PubMed]
- Köhler, C.D.; Dobrindt, U. What defines extraintestinal pathogenic Escherichia coli? Int. J. Med. Microbiol. 2011, 301, 642–647. [Google Scholar] [CrossRef] [PubMed]
- Ewers, C.; Li, G.; Wilking, H.; Kieβling, S.; Alt, K.; Antáo, E.-M.; Laturnus, C.; Diehl, I.; Glodde, S.; Homeier, T.; et al. Avian pathogenic, uropathogenic, and newborn meningitis-causing Escherichia coli: How closely related are they? Int. J. Med. Microbiol. 2007, 297, 163–176. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Siek, K.E.; Giddings, C.W.; Doetkott, C.; Johnson, T.J.; Fakhr, M.K.; Nolan, L.K. Comparison of Escherichia coli isolates implicated in human urinary tract infection and avian colibacillosis. Microbiology 2005, 151, 2097–2110. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Gao, S.; Huan, H.; Xu, X.; Zhu, X.; Yang, W.; Gao, Q.; Liu, X. Comparison of virulence factors and expression of specific genes between uropathogenic Escherichia coli and avian pathogenic E. coli in a murine urinary tract infection model and a chicken challenge model. Microbiol. Read. 2009, 155, 1634–1644. [Google Scholar] [CrossRef]
- Maluta, R.P.; Logue, C.M.; Casas, M.R.T.; Meng, T.; Guastalli, E.A.L.; Rojas, T.C.G.; Montelli, A.C.; Sadatsune, T.; de Carvalho, R.M.; Nolan, L.K.; et al. Overlapped sequence types (STs) and serogroups of avian pathogenic (APEC) and human extra-intestinal pathogenic (ExPEC) Escherichia coli isolated in Brazil. PLoS ONE 2014, 9, e105016. [Google Scholar] [CrossRef]
- Nandanwar, N.; Janssen, T.; Kühl, M.; Ahmed, N.; Ewers, C.; Wieler, L.H. Extraintestinal pathogenic Escherichia coli (ExPEC) of human and avian origin belonging to sequence type complex 95 (STC95) portray indistinguishable virulence features. Int. J. Med. Microbiol. 2014, 304, 835–842. [Google Scholar] [CrossRef] [PubMed]
- Bélanger, L.; Garenaux, A.; Harel, J.; Boulianne, M.; Nadeau, E.; Dozois, C.M. Escherichia coli from animal reservoirs as a potential source of human extraintestinal pathogenic E. Coli. FEMS Immunol. Med. Microbiol. 2011, 62, 1–10. [Google Scholar] [CrossRef]
- Clermont, O.; Bonacorsi, S.; Bingen, E. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl. Environ. Microbiol. 2000, 66, 4555–4558. [Google Scholar] [CrossRef]
- Johnson, J.R.; Delavari, P.; Kuskowski, M.; Stell, A.L. Phylogenetic distribution of extraintestinal virulence-associated traits in Escherichia coli. J. Infect. Dis. 2001, 183, 78–88. [Google Scholar] [CrossRef]
- Navidinia, M.; Najareerayeh, S.; Fallah, F.; Bakhshi, B.; Adabian, S.; Alimehr, S.; Gholinejad, Z. Distribution of the Pathogenicity Islands Markers (PAIs) in uropathogenic E. coli isolated from children in Mofid children hospital. Arch. Pediatr. Infect. Dis. 2013, 1, 75–79. [Google Scholar] [CrossRef]
- Navidinia, M.; Peerayeh, S.N.; Fallah, F.; Bakhshi, B.; Sajadinia, R.S. Phylogenetic grouping and pathotypic comparison of urine and fecal Escherichia coli isolates from children with urinary tract infection. Braz. J. Microbiol. 2014, 45, 509–514. [Google Scholar] [CrossRef] [PubMed]
- Neamati, F.; Firoozeh, F.; Saffari, M.; Zibaei, M. Virulence genes and antimicrobial resistance pattern in uropathogenic Escherichia coli isolated from hospitalized patients in Kashan, Iran. Jundishapur. J. Microbiol. 2015, 8, e17514. [Google Scholar] [CrossRef]
- Hacker, J.; Blum-Oehler, G.; Mühldorfer, I.; Tschäpe, H. Pathogenicity islands of virulent bacteria: Structure, function and impact on microbial evolution. Mol. Microbiol. 1997, 23, 1089–1097. [Google Scholar] [CrossRef]
- Oelschlaeger, T.A.; Dobrindt, U.; Hacker, J. Pathogenicity islands of uropathogenic E. coli and the evolution of virulence. Int. J. Antimicrob. Agents 2002, 19, 517–521. [Google Scholar] [CrossRef] [PubMed]
- Tangi, S.C.; Tajbakhsh, E.; Soleimani, N.A.; Shahraki, M.M. Prevalence of pathogenicity island markers genes in uropathogenic Escherichia coli isolated from patients with urinary tract infectious. Asian Pac. J. Trop. Dis. 2015, 5, 662–666. [Google Scholar] [CrossRef]
- Schmidt, H.; Hensel, M. Pathogenicity islands in bacterial pathogenesis. Clin. Microbiol. Rev. 2004, 17, 14–56. [Google Scholar] [CrossRef]
- Bingen-Bidois, M.; Clermont, O.; Bonacorsi, S.; Terki, M.; Brahimi, N.; Loukil, C.; Barraud, D.; Bingen, E. Phylogenetic analysis and prevalence of urosepsis strains of escherichia coli bearing pathogenicity island-like domains. Infect. Immun. 2002, 70, 3216–3226. [Google Scholar] [CrossRef]
- Chroma, M.; Kolar, M. Genetic methods for detection of antibiotic resistance: Focus on extended-spectrum β-lactamases. Biomed. Pap. 2010, 154, 289–296. [Google Scholar] [CrossRef]
- Russo, T.A.; Johnson, J.R. Medical and economic impact of extraintestinal infections due to Escherichia coli: Focus on an increasingly important endemic problem. Microbes. Infect. 2003, 5, 449–456. [Google Scholar] [CrossRef]
- Johnson, T.J.; Nolan, L.K. Pathogenomics of the virulence plasmids of Escherichia coli. Microbiol. Mol. Biol. Rev. 2009, 73, 750–754. [Google Scholar] [CrossRef] [PubMed]
- Hammerum, A.M.; Heuer, O.E. Human Health Hazards from Antimicrobial-Resistant Escherichia coli of Animal Origin. Clin. Infect. Dis. 2009, 48, 916–921. [Google Scholar] [CrossRef] [PubMed]
- Overdevest, I.; Willemsen, I.; Rijnsburger, M.; Eustace, A.; Xu, L.; Hawkey, P.; Heck, M.; Savelkoul, P.; Vandenbroucke-Grauls, C.; van der Zwaluw, K.; et al. Extended-spectrum β-lactamase genes of Escherichia coli in chicken meat and humans, The Netherlands. Emerg. Infect. Dis. J. 2011, 17, 1216–1222. [Google Scholar] [CrossRef] [PubMed]
- Dewanti, R.; Wong, A.C.L. Influence of culture conditions on biofilm formation by Escherichia coli O157:H7. Int. J. Food Microbiol. 1995, 26, 147–164. [Google Scholar] [CrossRef]
- Berlanga, M.; Guerrero, R. Living together in biofilms: The microbial cell factory and its biotechnological implications. Microb. Cell Factories 2016, 15, 165. [Google Scholar] [CrossRef] [PubMed]
- Johnson, J.R.; Kuskowski, M.A.; Smith, K.; O’Bryan, T.T.; Tatini, S. Antimicrobial-resistant and extraintestinal pathogenic Escherichia coli in retail foods. J. Infect. Dis. 2005, 191, 1040–1049. [Google Scholar] [CrossRef] [PubMed]
- Mellata, M.; Johnson, J.R.; Curtiss, R., III. Escherichia coli isolates from commercial chicken meat and eggs cause sepsis, meningitis and urinary tract infection in rodent models of human infections. Zoonoses Public Health 2018, 65, 103–113. [Google Scholar] [CrossRef]
- Johnson, J.R.; Porter, S.B.; Johnston, B.; Thuras, P.; Clock, S.; Crupain, M.; Rangan, U. Extraintestinal pathogenic and antimicrobial-resistant Escherichia coli, including sequence type 131 (st131), from retail chicken breasts in the United States in 2013. Appl. Environ. Microbiol. 2017, 83, e02956-16. [Google Scholar] [CrossRef]
- Lima-Filho, J.V.; Martins, L.V.; Nascimento, D.C.d.O.; Ventura, R.F.; Batista, J.E.C.; Silva, A.F.B.; Ralph, M.T.; Vaz, R.V.; Rabello, C.B.-V.; Silva, I.d.M.M.d.; et al. Zoonotic potential of multidrug-resistant extraintestinal pathogenic Escherichia coli obtained from healthy poultry carcasses in Salvador, Brazil. Braz. J. Infect. Dis. 2013, 17, 54–61. [Google Scholar] [CrossRef]
- Giufrè, M.; Graziani, C.; Accogli, M.; Luzzi, I.; Busani, L.; Cerquetti, M.; Escherichia coli Study Group. Escherichia coli of human and avian origin: Detection of clonal groups associated with fluoroquinolone and multidrug resistance in Italy. J. Antimicrob. Chemother. 2012, 67, 860–867. [Google Scholar] [CrossRef]
- Kluytmans, J.A.J.W.; Overdevest, I.T.M.A.; Willemsen, I.; Kluytmans-van den Bergh, M.F.Q.; van der Zwaluw, K.; Heck, M.; Rijnsburger, M.; Vandenbroucke-Grauls, C.M.J.E.; Savelkoul, P.H.M.; Johnston, B.D.; et al. Extended-spectrum β-Lactamase–producing Escherichia coli from retail chicken meat and humans: Comparison of strains, plasmids, resistance genes, and virulence factors. Clin. Infect. Dis. 2012, 56, 478–487. [Google Scholar] [CrossRef] [PubMed]
- Tóth, I.; Dobrindt, U.; Koscsó, B.; Kósa, A.; Herpay, M.; Nagy, B. Genetic and phylogenetic analysis of avian extraintestinal and intestinal Escherichia coli. Acta Microbiol. Immunol. Hung. 2012, 59, 393–409. [Google Scholar] [CrossRef]
- Literak, I.; Reitschmied, T.; Bujnakova, D.; Dolejska, M.; Cizek, A.; Bardon, J.; Pokludova, L.; Alexa, P.; Halova, D.; Jamborova, I. Broilers as a source of quinolone-resistant and extraintestinal pathogenic Escherichia coli in the Czech Republic. Microb. Drug Resist. 2012, 19, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Parham, N.J.; Pollard, S.J.; Chaudhuri, R.R.; Beatson, S.A.; Desvaux, M.; Russell, M.A.; Ruiz, J.; Fivian, A.; Vila, J.; Henderson, I.R. Prevalence of pathogenicity island IICFT073 genes among extraintestinal clinical isolates of Escherichia coli. J. Clin. Microbiol. 2005, 43, 2425–2434. [Google Scholar] [CrossRef]
- Ostblom, A.; Adlerberth, I.; Wold, A.E.; Nowrouzian, F.L. Pathogenicity island markers, virulence determinants malX and usp, and the capacity of Escherichia coli to persist in infants’ commensal microbiotas. Appl. Env. Microbiol. 2011, 77, 2303–2308. [Google Scholar] [CrossRef] [PubMed]
- Barbieri, N.L.; Oliveira, A.L.D.; Tejkowski, T.M.; Pavanelo, D.B.; Matter, L.B.; Pinheiro, S.R.S.; Vaz, T.M.I.; Nolan, L.K.; Logue, C.M.; Brito, B.G.D.; et al. Molecular Characterization and Clonal Relationships Among Escherichia coli Strains Isolated from Broiler Chickens with Colisepticemia. Foodborne Pathog. Dis. 2014, 12, 74–83. [Google Scholar] [CrossRef]
- Ewers, C.; Antão, E.-M.; Diehl, I.; Philipp, H.-C.; Wieler, L.H. Intestine and environment of the chicken as reservoirs for extraintestinal pathogenic Escherichia coli strains with zoonotic potential. Appl. Environ. Microbiol. 2009, 75, 184–192. [Google Scholar] [CrossRef]
- Cortés, P.; Blanc, V.; Mora, A.; Dahbi, G.; Blanco, J.E.; Blanco, M.; López, C.; Andreu, A.; Navarro, F.; Alonso, M.P.; et al. Isolation and characterization of potentially pathogenic antimicrobial-resistant Escherichia coli strains from chicken and pig farms in Spain. Appl. Environ. Microbiol. 2010, 76, 2799–2805. [Google Scholar] [CrossRef]
- Duriez, P.; Clermont, O.; Bonacorsi, S.; Bingen, E.; Chaventré, A.; Elion, J.; Picard, B.; Denamur, E. Commensal Escherichia coli isolates are phylogenetically distributed among geographically distinct human populations. Microbiology 2001, 147, 1671–1676. [Google Scholar] [CrossRef]
- Derakhshandeh, A.; Firouzi, R.; Moatamedifar, M.; Motamedi, A.; Bahadori, M.; Naziri, Z. Phylogenetic analysis of Escherichia coli strains isolated from human samples. Mol. Biol. Res. Commun. 2013, 2, 143–149. [Google Scholar]
- Bingen, E.; Picard, B.; Brahimi, N.; Mathy, S.; Desjardins, P.; Elion, J.; Denamur, E. Phylogenetic analysis of Escherichia coli strains causing neonatal meningitis suggests horizontal gene transfer from a predominant pool of highly virulent b2 group strains. J. Infect. Dis. 1998, 177, 642–650. [Google Scholar] [CrossRef] [PubMed]
- Johnson, J.R.; Manges, A.R.; O’Bryan, T.T.; Riley, L.W. A disseminated multidrug-resistant clonal group of uropathogenic Escherichia coli in pyelonephritis. Lancet 2002, 359, 2249–2251. [Google Scholar] [CrossRef]
- Sannes, M.R.; Kuskowski, M.A.; Owens, K.; Gajewski, A.; Johnson, J.R. Virulence factor profiles and phylogenetic background of Escherichia coli isolates from veterans with bacteremia and uninfected control subjects. J. Infect. Dis. 2004, 190, 2121–2128. [Google Scholar] [CrossRef]
- Zhang, L.; Foxman, B.; Marrs, C. Both urinary and rectal Escherichia coli isolates are dominated by strains of phylogenetic group B2. J. Clin. Microbiol. 2002, 40, 3951–3955. [Google Scholar] [CrossRef]
- Kuznetsova, M.V.; Maslennikova, I.L.; Pospelova, J.S.; Bertok D, Ž.; Erjavec, M.S. Differences in recipient ability of uropathogenic Escherichia coli strains in relation with their pathogenic potential. Infect. Genet. Evol. 2022, 97, 105160. [Google Scholar]
- Lee, J.C.; Lee, N.Y.; Lee, H.C.; Huang, W.H.; Tsui, K.C.; Chang, C.M.; Lee, C.C.; Chen, P.L.; Wu, C.J.; Hsueh, P.R.; et al. Clinical characteristics of urosepsis caused by extended-spectrum beta-lactamase-producing Escherichia coli or Klebsiella pneumonia and their emergence in the community. J. Microbiol. Immunol. Infect. 2012, 45, 127–133. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, A.M.; Shimamoto, T.; Shimamoto, T. Molecular characterization of multidrug-resistant avian pathogenic Escherichia coli isolated from septicemic broilers. Int. J. Med. Microbiol. 2013, 303, 475–483. [Google Scholar] [CrossRef] [PubMed]
- Che, D.; Hasan, M.S.; Chen, B. Identifying pathogenicity islands in bacterial pathogenomics using computational approaches. Pathogens 2014, 3, 36–56. [Google Scholar] [CrossRef]
- Naderi, G.; Haghi, F.; Zeighami, H.; Hemati, F.; Masoumian, N. Distribution of pathogenicity island (PAI) markers and phylogenetic groups in diarrheagenic and commensal Escherichia coli from young children. Gastroenterol. Hepatol. Bed Bench 2016, 9, 316–324. [Google Scholar]
- Middendorf, B.; Hochhut, B.; Leipold, K.; Dobrindt, U.; Blum-Oehler, G.; Hacker, J. Instability of pathogenicity islands in uropathogenic Escherichia coli 536. J. Bacteriol. 2004, 186, 3086–3096. [Google Scholar] [CrossRef]
- Johnson, J.R.; Stell, A.L. Extended virulence genotypes of Escherichia coli strains from patients with urosepsis in relation to phylogeny and host compromise. J. Infect. Dis. 2000, 181, 261–272. [Google Scholar] [CrossRef]
- Samei, A.; Haghi, F.; Zeighami, H. Distribution of pathogenicity island markers in commensal and uropathogenic Escherichia coli isolates. Folia Microbiol. 2016, 61, 261–268. [Google Scholar] [CrossRef] [PubMed]
- Firoozeh, F.; Saffari, M.; Neamati, F.; Zibaei, M. Detection of virulence genes in Escherichia coli isolated from patients with cystitis and pyelonephritis. Int. J. Infect. Dis. 2014, 29, 219–222. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Sun, J.-Y.; Han, L.-Z.; Huang, X.-H.; Fu, Q.; Ni, Y.-X. Phylogenetic groups and pathogenicity island markers in fecal Escherichia coli isolates from asymptomatic humans in China. Appl. Environ. Microbiol. 2010, 76, 6698–6700. [Google Scholar] [CrossRef]
- Sabaté, M.; Moreno, E.; Pérez, T.; Andreu, A.; Prats, G. Pathogenicity island markers in commensal and uropathogenic Escherichia coli isolates. Clin. Microbiol. Infect. 2006, 12(9), 880–886. [Google Scholar] [CrossRef]
- Gultekin, E.O.; Gultekin, E.O.; Ulger, S.T.; Delialioğlu, N. Distribution of Pathogenicity Island Markers and Virulence Factors Genes of Extraintestinal Pathogenic Escherichia coli Isolates. Jundishapur J. Microbiol. 2022, 15, e121044. [Google Scholar]
- El-Shaer, S.; Abdel-Rhman, S.H.; Barwa, R.; Hassan, R. Genetic characterization of extended-spectrum β-Lactamase-and carbapenemase-producing Escherichia coli isolated from Egyptian hospitals and environments. PloS One 2021, 16, e0255219. [Google Scholar] [CrossRef] [PubMed]
- El-Mahdy, R.; Mahmoud, R.; Shrief, R. Characterization of E. coli phylogroups causing catheter-associated urinary tract infection. Infect. Drug Resist. 2021, 14, 3183. [Google Scholar] [CrossRef] [PubMed]
- Gal-Mor, O.; Finlay, B.B. Pathogenicity islands: A molecular toolbox for bacterial virulence. Cell Microbiol. 2006, 8, 1707–1719. [Google Scholar] [CrossRef]
- Desvaux, M.; Dalmasso, G.; Beyrouthy, R.; Barnich, N.; Delmas, J.; Bonnet, R. Pathogenicity factors of genomic islands in intestinal and extraintestinal Escherichia coli. Front. Microbiol. 2020, 11, 2065. [Google Scholar] [CrossRef]
- Carattoli, A. Plasmids and the spread of resistance. Int. J. Med. Microbiol. 2013, 303, 298–304. [Google Scholar] [CrossRef]
- Hung, C.S.; Henderson, J.P. Emerging concepts of biofilms in infectious diseases. Mo. Med. 2009, 106, 292–296. [Google Scholar]
- Costerton, J.W.; Lewandowski, Z.; Caldwell, D.E.; Korber, D.R.; Lappin-Scott, H.M. Microbial biofilms. Annu. Rev. Microbiol. 1995, 49, 711–745. [Google Scholar] [CrossRef] [PubMed]
- Nandakumar, V.; Chittaranjan, S.; Kurian, V.M.; Doble, M. Characteristics of bacterial biofilm associated with implant material in clinical practice. Polym. J. 2013, 45, 137–152. [Google Scholar] [CrossRef]
- Davies, J.; Davies, D. Origins and evolution of antibiotic resistance. Microbiol. Mol. Biol. Rev. MMBR 2010, 74, 417–433. [Google Scholar] [CrossRef] [PubMed]
- Skyberg, J.A.; Siek, K.E.; Doetkott, C.; Nolan, L.K. Biofilm formation by avian Escherichia coli in relation to media, source and phylogeny. J. Appl. Microbiol. 2007, 102, 548–554. [Google Scholar] [CrossRef]
- Katongole, P.; Nalubega, F.; Florence, N.C.; Asiimwe, B.; Andia, I. Biofilm formation, antimicrobial susceptibility and virulence genes of Uropathogenic Escherichia coli isolated from clinical isolates in Uganda. BMC Infect. Dis. 2020, 20, 453. [Google Scholar] [CrossRef]
- Quinn, P.; Markey, B.; Carter, M.; Donnelly, W.; Leonard, F. Veterinary Microbiology and Microbial Disease; Blackwell Science Company: Hoboken, NJ, USA; State University Press: Detroit, MI, USA, 2002. [Google Scholar]
- Alexopoulou, K.; Foka, A.; Petinaki, E.; Jelastopulu, E.; Dimitracopoulos, G.; Spiliopoulou, I. Comparison of two commercial methods with PCR restriction fragment length polymorphism of the tuf gene in the identification of coagulase-negative staphylococci. Lett. Appl. Microbiol. 2006, 43, 450–454. [Google Scholar] [CrossRef]
- Teichmann, A.; Agra, H.N.; de Souza Nunes, L.; da Rocha, M.P.; Renner, J.D.; Possuelo, L.G.; Carneiro, M.; Rieger, A.; Benitez, L.B.; Valim, A.R. Antibiotic resistance and detection of the sul2 gene in urinary isolates of Escherichia coli in patients from Brazil. J. Infect. Dev. Ctries. 2014, 8, 39–43. [Google Scholar] [CrossRef]
- Higgins, J.; Hohn, C.; Hornor, S.; Frana, M.; Denver, M.; Joerger, R. Genotyping of Escherichia coli from environmental and animal samples. J. Microbiol. Methods 2007, 70, 227–235. [Google Scholar] [CrossRef]
- Belaaouaj, A.; Lapoumeroulie, C.; Caniça, M.M.; Vedel, G.; Névot, P.; Krishnamoorthy, R.; Paul, G. Nucleotide sequences of the genes coding for the TEM-like β-lactamases IRT-1 and IRT-2 (formerly called TRI-1 and TRI-2). FEMS Microbiol. Lett. 1994, 120, 75–80. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Maynard, C.; Fairbrother, J.M.; Bekal, S.; Sanschagrin, F.; Levesque, R.C.; Brousseau, R.; Masson, L.; Larivière, S.; Harel, J. Antimicrobial Resistance Genes in Enterotoxigenic Escherichia coli O149:K91 Isolates Obtained over a 23-Year Period from Pigs. Antimicrob. Agents Chemother. 2003, 47, 3214–3221. [Google Scholar] [CrossRef] [PubMed]
- Toro, C.S.; FarfÁN, M.; Contreras, I.; Flores, O.; Navarro, N.; Mora, G.C.; Prado, V. Genetic analysis of antibiotic-resistance determinants in multidrug-resistant Shigella strains isolated from Chilean children. Epidemiol. Infect. 2005, 133, 81–86. [Google Scholar] [CrossRef] [PubMed]
- Robicsek, A.; Strahilevitz, J.; Sahm, D.F.; Jacoby, G.A.; Hooper, D.C. qnr prevalence in ceftazidime-resistant Enterobacteriaceae isolates from the United States. Antimicrob. Agents Chemother. 2006, 50, 2872–2874. [Google Scholar] [CrossRef] [PubMed]
- Bonacorsi, S.P.P.; Clermont, O.; Tinsley, C.; Le Gall, I.; Beaudoin, J.-C.; Elion, J.; Nassif, X.; Bingen, E. Identification of regions of the Escherichia coli chromosome specific for neonatal meningitis-associated strains. Infect. Immun. 2000, 68, 2096–2101. [Google Scholar] [CrossRef] [PubMed]
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing: Twenty-Seventh Informational Supplement M100-S27; CLSI: Wayne, PA, USA, 2017. [Google Scholar]
- Titilawo, Y.; Sibanda, T.; Obi, L.; Okoh, A. Multiple antibiotic resistance indexing of Escherichia coli to identify high-risk sources of faecal contamination of water. Environ. Sci. Pollut. Res. 2015, 22, 10969–10980. [Google Scholar] [CrossRef]
- Krumperman, P.H. Multiple antibiotic resistance indexing of Escherichia coli to identify high-risk sources of fecal contamination of foods. Appl. Environ. Microbiol. 1983, 46, 165–170. [Google Scholar] [CrossRef]
- Kadam, S.R.; den Besten, H.M.W.; van der Veen, S.; Zwietering, M.H.; Moezelaar, R.; Abee, T. Diversity assessment of Listeria monocytogenes biofilm formation: Impact of growth condition, serotype and strain origin. Int. J. Food Microbiol. 2013, 165, 259–264. [Google Scholar] [CrossRef]
- Wolfe, A.J.; Millikan, D.S.; Campbell, J.M.; Visick, K.L. Vibrio fischeri sigma54 controls motility, biofilm formation, luminescence, and colonization. Appl. Env. Microbiol. 2004, 70, 2520–2524. [Google Scholar] [CrossRef]
- Sadat, A.; El-Sherbiny, H.; Zakaria, A.; Ramadan, H.; Awad, A. Prevalence, antibiogram and virulence characterization of Vibrio isolates from fish and shellfish in Egypt: A possible zoonotic hazard to humans. J. Appl. Microbiol. 2020, 131, 485–498. [Google Scholar]
- Sadat, A.; Shata, R.R.; Farag, A.M.M.; Ramadan, H.; Alkhedaide, A.; Soliman, M.M.; Elbadawy, M.; Abugomaa, A.; Awad, A. Prevalence and Characterization of PVL-Positive Staphylococcus aureus Isolated from Raw Cow’s Milk. Toxins 2022, 14, 97. [Google Scholar] [CrossRef]

| Source of Isolates | Phylogenetic Group | |||||
|---|---|---|---|---|---|---|
| Pathogenic Phylogroups | Commensal Phylogroups | |||||
| Group B2 | Group D | Total | Group A | Group B1 | Total | |
| Chicken (n = 34) | 7 (20.6%) | 5 (14.7%) | 12 (35.3%) | 9 (26.5%) | 13 (38.3%) | 22 (64.7%) |
| Human (n = 26) | 7 (26.9%) | 5 (19.2%) | 12 (46.2) | 4 (15.4%) | 10 (38.5%) | 14 (53.8%) |
| Source of Isolates | PAI III536 | PAI IV536 | PAI IICFT073 | PAI J196 | PAI ICFT073 | PAI II536 | PAI I536 | PAI IIJ196 |
|---|---|---|---|---|---|---|---|---|
| Chicken | - | 19 (55.9%) | 4 (11.8%) | - | - | - | - | - |
| Human | - | 18 (69.2%) | 5 (19.2%) | 2(7.7%) | - | - | - | - |
| Isolates | PAIs/Phylogenetic Group | PAI III536 | PAI IV536 | PAI IICFT073 | PAI J196 | PAI ICFT073 | PAI II536 | PAI I536 | PAI IIJ196 | Total |
|---|---|---|---|---|---|---|---|---|---|---|
| Chicken (n = 34) | B2 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 (4.34%) |
| D | 0 | 4 | 1 | 0 | 0 | 0 | 0 | 0 | 5 (21.7%) | |
| A | 0 | 7 | 1 | 0 | 0 | 0 | 0 | 0 | 8 (34.8%) | |
| B1 | 0 | 7 | 2 | 0 | 0 | 0 | 0 | 0 | 9 (39.1%) | |
| Total | 0 | 19 | 4 | 0 | 0 | 0 | 0 | 0 | 23 (100%) | |
| Human (n = 26) | B2 | 0 | 5 | 1 | 1 | 0 | 0 | 0 | 0 | 7 (28%) |
| D | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 3 (12%) | |
| A | 0 | 3 | 1 | 1 | 0 | 0 | 0 | 0 | 5 (20%) | |
| B1 | 0 | 7 | 3 | 0 | 0 | 0 | 0 | 0 | 10 (40%) | |
| Total | 0 | 18 | 5 | 2 | 0 | 0 | 0 | 0 | 25 (100%) |
| Antibiotics | Family | Disc Code | CPD | Chicken (34) | Human (26) | ||
|---|---|---|---|---|---|---|---|
| Resistant | Sensitive | Resistant | Sensitive | ||||
| Penicillin | β-lactam | P | 10 | 34 (100%) | 0 (0.00%) | 26 (100%) | 0 (0.00%) |
| Cefuroxime | Cephalosporin | CXM | 30 | 30 (88.2%) | 4 (11.8%) | 25 (96.2%) | 1 (3.9%) |
| Cefoperazone | CEP | 30 | 29 (85.3%) | 5 (14.7%) | 21 (80.8%) | 5 (19.2%) | |
| Amikacin | Aminoglycoside | AK | 30 | 8 (23.5%) | 26 (76.5%) | 26 (100%) | 0 (0.00%) |
| Streptomycin | S | 15 | 25 (73.5%) | 9 (26.5%) | 20 (76.9%) | 6 (23.1%) | |
| Neomycin | N | 5 | 28 (82.4%) | 6 (17.6%) | 24 ( 92.3%) | 2 ((7.7%) | |
| Azithromycin | Macrolide | AZM | 15 | 20 (58.8%) | 14 (41.2%) | 15 (57.7%) | 11 (42.3%) |
| Nalidixic acid | Quinolone | NAL | 30 | 30 (88.2%) | 4 (11.8%) | 24 (92.3%) | 2 (7.7%) |
| Trimethoprim–sulfamethoxazole | Sulfonamide | SXT | 25 | 30 (88.2%) | 4 (11.8%) | 20 (76.9%) | 6 (23.1%) |
| Clindamycin | Lincosamide | DA | 2 | 34 (100%) | 0 (0.00%) | 25 (96.2%) | 1 (3.8%) |
| Ciprofloxacin | Fluoroquinolone | CIP | 5 | 29 (85.3%) | 5 (14.7%) | 22 (84.6%) | 4 (15.4%) |
| Chloramphenicol | Phenicols | C | 30 | 27 (79.4%) | 7 (20.6%) | 4 (15.4%) | 22 (84.6%) |
| Rifampin | Rifamycin | Rifampin | 5 | 33 (97.1%) | 1 (2.9%) | 26 (100%) | 0 (0.00%) |
| Antibiotypes | Resistance Pattern | Isolates No. (%) | MAR | MAR Index | |
|---|---|---|---|---|---|
| Chicken | I | C, P, DA, STX, RA, N, CXM, CIP | 1 (2.9%) | 8 | 0.62 |
| II | P, S, DA, STX, RA, N, CXM, CEP | 1 (2.9%) | 6 | 0.62 | |
| III | C, P, S, DA, SXT, RA, NA, CEP, CIP | 1 (2.9%) | 9 | 0.7 | |
| IV | C, P, DA, STX, RA, N, NA, CXM, CIP | 1 (2.9%) | 9 | 0.7 | |
| V | C, P, DA, STX, RA, N, CXM, CEP, AZM | 1 (2.9%) | 8 | 0.7 | |
| VI | P, S, DA, STX, RA, N, AK, AZM, CIP | 1 (2.9%) | 8 | 0.7 | |
| VII | P, DA, STX, RA, N, AK, NA, CEP, CIP | 1 (2.9%) | 8 | 0.7 | |
| VIII | P, S, DA, STX, RA, N, NA, CXM, CIP | 1 (2.9%) | 8 | 0.7 | |
| IX | C, P, DA, RA, AK, NA, CXM, CEP, AZM | 1 (2.9%) | 8 | 0.7 | |
| X | C, P, S, DA, STX, RA, N, NA, CXM, CIP | 1 (2.9%) | 9 | 0.77 | |
| XI | C, P, S, DA, STX, RA, N, NA, CEP, CIP | 1 (2.9%) | 9 | 0.77 | |
| XII | C, P, S, DA, STX, RA, NA, CXM, CEP, CIP | 1 (2.9%) | 9 | 0.77 | |
| XIII | C, P, S, DA, RA, NA, CXM, CEP, AZM, CIP | 1 (2.9%) | 9 | 0.77 | |
| XIV | C, P, DA, STX, N, AK, NA, CXM, CEP, AZM | 1 (2.9%) | 8 | 0.77 | |
| XV | C, P, DA, STX, RA, N, NA, CXM, CEP, AZM, CIP | 1 (2.9%) | 10 | 0.84 | |
| XVI | P, S, DA, STX, RA, N, NA, CXM, CEP, AZM, CIP | 2 (5.8%) | 9 | 0.84 | |
| XVII | C, P, S, DA, STX, RA, NA, CXM, CEP, AZM. CIP | 2 (5.8%) | 10 | 0.84 | |
| XVIII | C, P, S, DA, STX, RA, N, NA, CXM, CEP, CIP | 5 (14.7%) | 9 | 0.84 | |
| XIX | C, P, S, DA, STX, RA, N, NA, CXM, CEP, AZM | 1 (2.9%) | 9 | 0.84 | |
| XX | C, P, DA, RA, N, AK, NA, CXM, CEP, AZM, CIP | 2 (5.8%) | 9 | 0.84 | |
| XXI | C, P, S, DA, STX, RA, N, NA, CXM, CEP, AZM, CIP | 5 (17.7%) | 10 | 0.9 | |
| XXII | P, S, DA, STX, RA, N, AK, NA, CXM, CEP, AZM, CIP | 1 (2.9%) | 9 | 0.9 | |
| XXIII | C, P, S, DA, STX, RA, N, AK, NA, CXM, CEP, AZM, CIP | 1(2.9%) | 10 | 1 | |
| Human | I | P, DA, RA, AK, CXM, CEP, CIP | 1 (3.8%) | 6 | 0.54 |
| II | P, S, DA, STX, RA, AK, CXM, CIP | 1 (3.8%) | 7 | 0.62 | |
| III | P, STX, RA, N, AK, NA, CXM, CEP | 1 (3.8%) | 7 | 0.62 | |
| IV | P, DA, SXT, RA, N, AK, NA, CXM, CEP | 1 (3.8%) | 8 | 0.7 | |
| V | P, DA, RA, N, AK, NA, CXM, AZM, CIP | 1 (3.8%) | 8 | 0.7 | |
| VI | P, DA, RA, N, AK, NA, CXM, CEP, CIP | 1 (3.8%) | 7 | 0.7 | |
| VII | P, S, DA, STX, RA, N, AK, NA, CXM, CEP | 1 (3.8%) | 8 | 0.77 | |
| VIII | P, S, DA, RA, N, AK, NA, CXM, CEP, CIP | 1 (3.8%) | 7 | 0.77 | |
| IX | P, S, DA, STX, RA, N, AK, NA, AZM, CIP | 1 (3.8%) | 8 | 0.77 | |
| X | P, S, DA, SXT, RA, N, AK, NA, CXM, CEP, CIP | 3 (11.5%) | 8 | 0.85 | |
| XI | C, P, S, DA, SXT, RA, N, AK, NA, CXM, CIP | 1 (3.8%) | 9 | 0.85 | |
| XII | P, S, DA, STX, RA, N, AK, NA, CXM, AZM, CIP | 1 (3.8%) | 10 | 0.85 | |
| XIII | P, DA, STX, RA, N, AK, NA, CXM, CEP, AZM, CIP | 1 (3.8%) | 9 | 0.85 | |
| XIV | P, S, DA, RA, N, AK, NA, CXM, CEP, AZM, CIP | 2 (7.7%) | 8 | 0.85 | |
| XV | P, S, DA, STX, RA, N, AK, NA, CXM, CEP, AZM | 1 (3.8%) | 8 | 0.85 | |
| XVI | P, S, DA, STX, RA, N, AK, NA, CXM, CEP, AZM, CIP | 5 (19.2%) | 9 | 0.92 | |
| XVII | C, P, S, DA, STX, RA, N, AK, NA, CXM, CEP, AZM, CIP | 3 (11.5%) | 10 | 1 |
| Point of Differences | Items Tested | Chicken Isolates (34) | Human Isolates (26) | Total(60) |
|---|---|---|---|---|
| Resistant genes | blaTEM | 18 (52.9%) | 4 (15.4%) | 22 (36.7%) |
| qnrA | 12 (35.3%) | 0 (0.00%) | 12 (20%) | |
| aphA1 | 17 (50%) | 4 (15.4%) | 21 (35%) | |
| sul2 | 29 (85.3%) | 9 (34.6%) | 38 (63.3%) | |
| dfrA1 | 10 (29.4%) | 1 (3.8%) | 11 (18.3%) | |
| Biofilm production | Negative | 11 (32.4%) | 12 (46.2%) | 23 (38.3%) |
| Weak positive | 4 (11.8%) | 3 (11.5%) | 7 (11.7%) | |
| Moderate | 11 (32.4%) | 4 (15.4%) | 15 (25%) | |
| Strong positive | 8 (23.5%) | 7 (26.9%) | 15 (25%) | |
| Overall positive | 23 (67.6%) | 14 (53.8%) | 37 (61.7%) |
| Gene | Primer Name | Primer Sequences (5’ to 3’) | (bp) | Reference |
|---|---|---|---|---|
| 16S rRNA | 16S-F | F-GCGGACGGGTGAGTAATGT | 200 | [71] |
| 16S-R | R-TCATCCTCTCAGACCAGCTA | |||
| Pathogenicity Island (PAI) Genes | ||||
| PAI I536 | I.9 | F-TAATGCCGGAGATTCATTGTC | 1800 | [56] |
| 1.10 | R-AGGATTTGTCTCAGGGCTTT | |||
| PAI II536 | orf1 up | F-CCATGTCCAAAGCTCGAGC | 1000 | |
| orf1 down | R-CTACGTCAGGCTGGCTTTG | |||
| PAI III536 | sfaAI.1 | F-CGGGCATGCATCAATTATCTTTG | 200 | [3] |
| sfaAI.2 | R-TGTGTAGATGCAGTCACTCCG | |||
| PAI IV536 | IRP2 FP | F-AAGGATTCGCTGTTACCGGAC | 300 | [3] |
| IRP2 RP | R-TCGTCGGGCAGCGTTTCTTCT | |||
| PAI IICFT073 | cft073.2Ent1 | F-ATGGATGTTGTATCGCGCP | 400 | [56] |
| cft073.2Ent2 | R-ACGAGCATGTGGATCTGC | |||
| PAI J196 | papGIf | F-TCGTGCTCAGGTCCGGAATTT | 400 | [13] |
| papGIR | R-TGGCATCCCACATTATCG | |||
| PAI IIJ196 | Hlyd | F-GGATCCATGAAAACATGGTTAATGGG | 2300 | |
| Cnf | R-GATATTTTTGTTGCCATTGGTTACC | |||
| PAI ICFT073 | RPAi | F-GGACATCCTGTTACAGCACGCA | 930 | |
| RPAf | R-TCGCCACCAATCACAGCCGAAC | |||
| Phylogenetic Island Genes | ||||
| ChuA | ChuA-F | F-GACGAACCAACGGTCAGGAT | 279 | [72] |
| ChuA-R | R-TGCCGCCAGTACCAAAGACA | |||
| YjaA | YjaA-F | F-TGAAGTGTCAGGAGACGCTG | 211 | |
| YjaA-R | R-ATGGAGAATGCGTTCCTCAAC | |||
| TspE4C2 | TspE4C2-F | F-GAGTAATGTCGGGGCATTCA | 152 | |
| TspE4C2-R | R-CGCGCCAACAAAGTATTACG | |||
| Antimicrobial Resistance Genes | ||||
| blaTEM | Tem-F | ATTCTTGAAGACGAAAGG | 1150 | [73] |
| Tem-R | ACGCTCAGTGGAACGAAAAC | |||
| sul2 | Sul2-F | CGGCATCGTCAACATAACC | 722 | [74] |
| Sul2-R | GTGTGCGGATGAAGTCAG | |||
| dfrA1 | dfrA1-F | GGAGTGCCAAAGGTGAACAGC | 367 | [75] |
| dfrA1-R | GAGGCGAAGTCTTGGGTAAAAAC | |||
| qnrA | QnrA-F | ATTTCTCACGCCAGGATTTG | 514 | [76] |
| QnrA-R | GATCGGCAAAGGTTAGGTCA | |||
| aphA1 | aphA1-F | ATGGGCTCGCGATAATGTC | 600 | [74] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sadat, A.; Ramadan, H.; Elkady, M.A.; Hammad, A.M.; Soliman, M.M.; Aboelenin, S.M.; Al-Harthi, H.F.; Abugomaa, A.; Elbadawy, M.; Awad, A. Phylotypic Profiling, Distribution of Pathogenicity Island Markers, and Antimicrobial Susceptibility of Escherichia coli Isolated from Retail Chicken Meat and Humans. Antibiotics 2022, 11, 1197. https://doi.org/10.3390/antibiotics11091197
Sadat A, Ramadan H, Elkady MA, Hammad AM, Soliman MM, Aboelenin SM, Al-Harthi HF, Abugomaa A, Elbadawy M, Awad A. Phylotypic Profiling, Distribution of Pathogenicity Island Markers, and Antimicrobial Susceptibility of Escherichia coli Isolated from Retail Chicken Meat and Humans. Antibiotics. 2022; 11(9):1197. https://doi.org/10.3390/antibiotics11091197
Chicago/Turabian StyleSadat, Asmaa, Hazem Ramadan, Mohamed A. Elkady, Amal Mahmoud Hammad, Mohamed M. Soliman, Salama M. Aboelenin, Helal F. Al-Harthi, Amira Abugomaa, Mohamed Elbadawy, and Amal Awad. 2022. "Phylotypic Profiling, Distribution of Pathogenicity Island Markers, and Antimicrobial Susceptibility of Escherichia coli Isolated from Retail Chicken Meat and Humans" Antibiotics 11, no. 9: 1197. https://doi.org/10.3390/antibiotics11091197
APA StyleSadat, A., Ramadan, H., Elkady, M. A., Hammad, A. M., Soliman, M. M., Aboelenin, S. M., Al-Harthi, H. F., Abugomaa, A., Elbadawy, M., & Awad, A. (2022). Phylotypic Profiling, Distribution of Pathogenicity Island Markers, and Antimicrobial Susceptibility of Escherichia coli Isolated from Retail Chicken Meat and Humans. Antibiotics, 11(9), 1197. https://doi.org/10.3390/antibiotics11091197











