Biochemical Characterizations of the Putative Endolysin Ecd09610 Catalytic Domain from Clostridioides difficile
Abstract
1. Introduction
2. Results
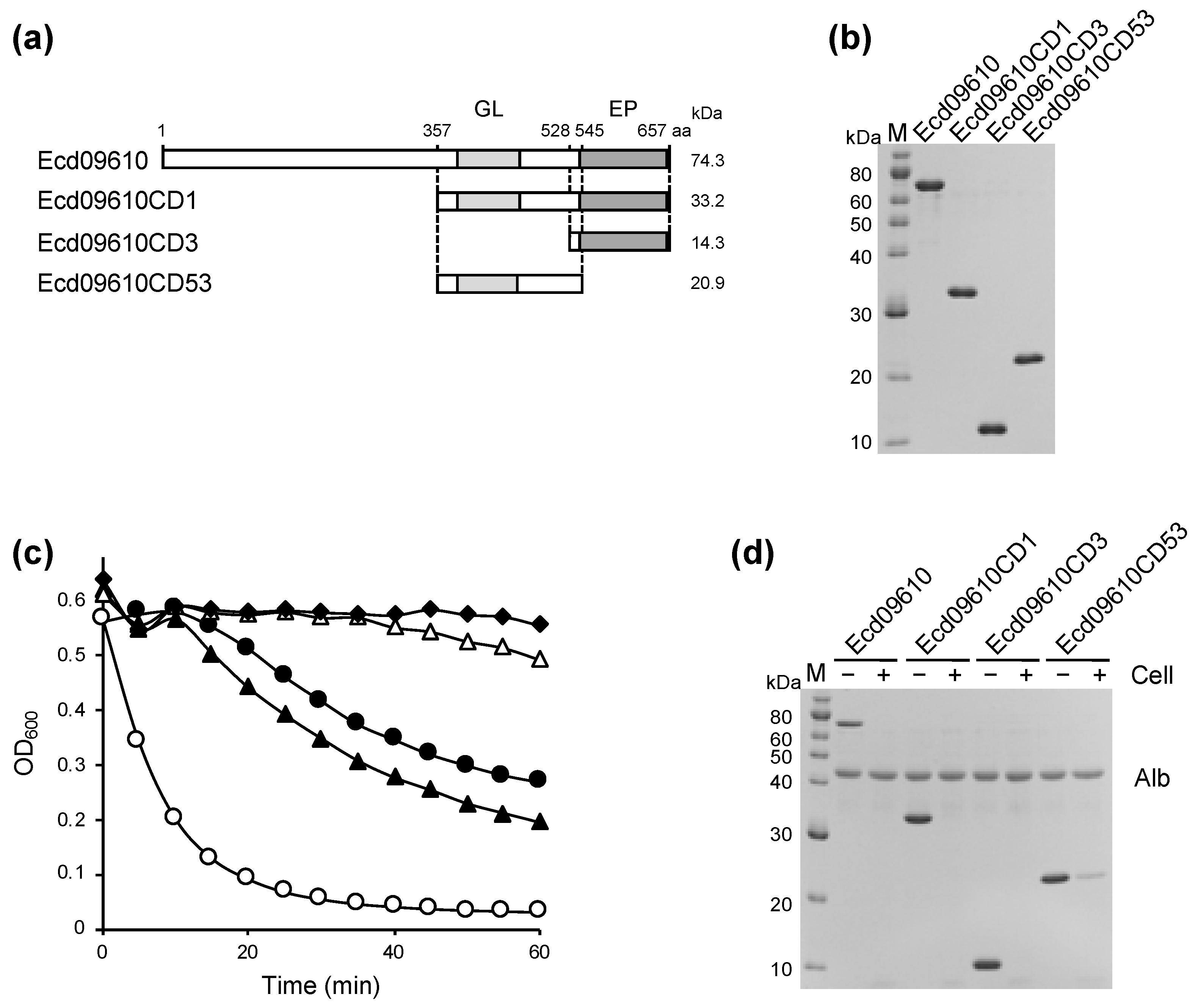
2.1. Identification, Cloning, Expression, and Purification of Ecd09610 and Its Derivatives
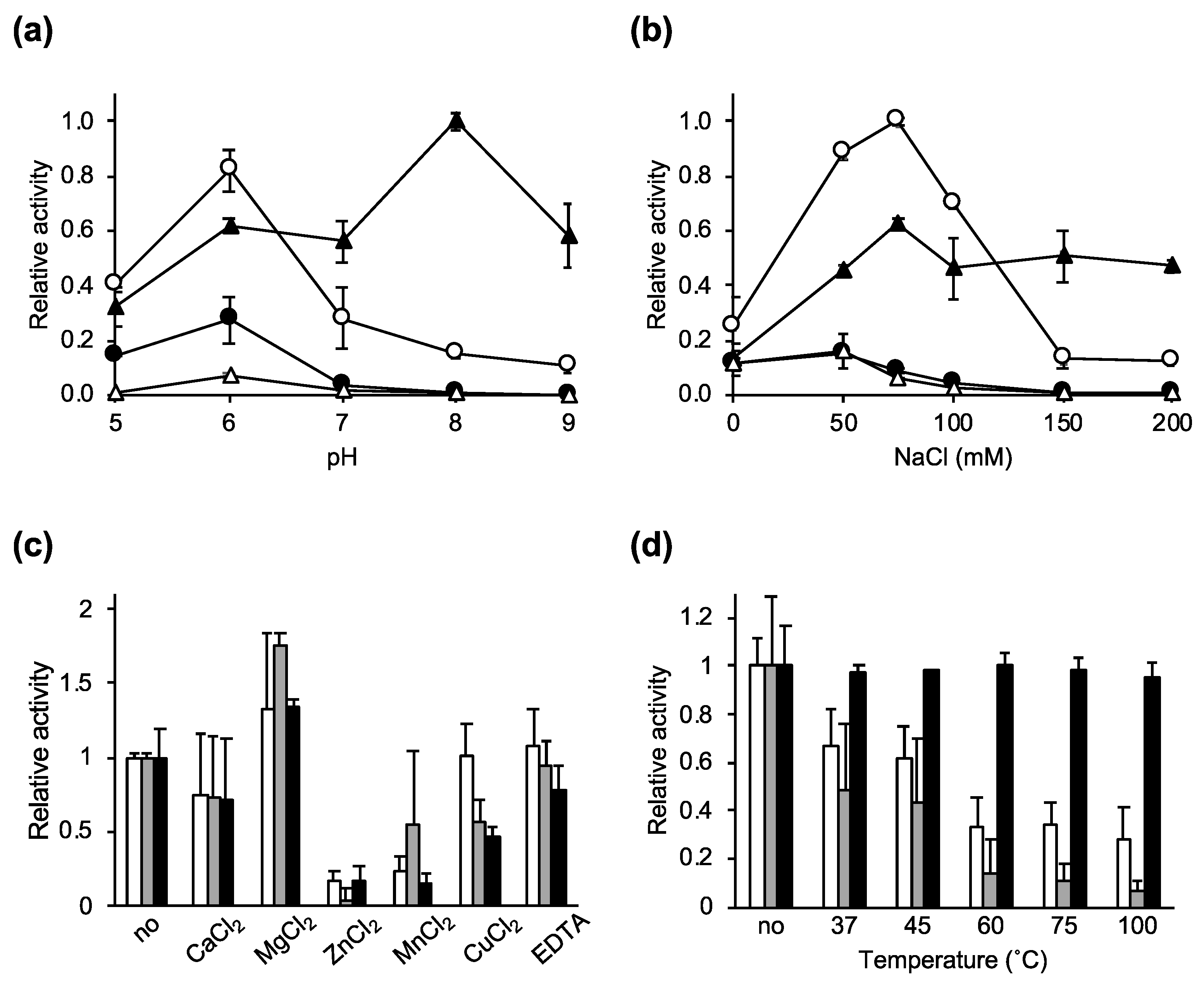
2.2. Characterization of the Lytic Activity of Ecd09610 and Its Derivatives
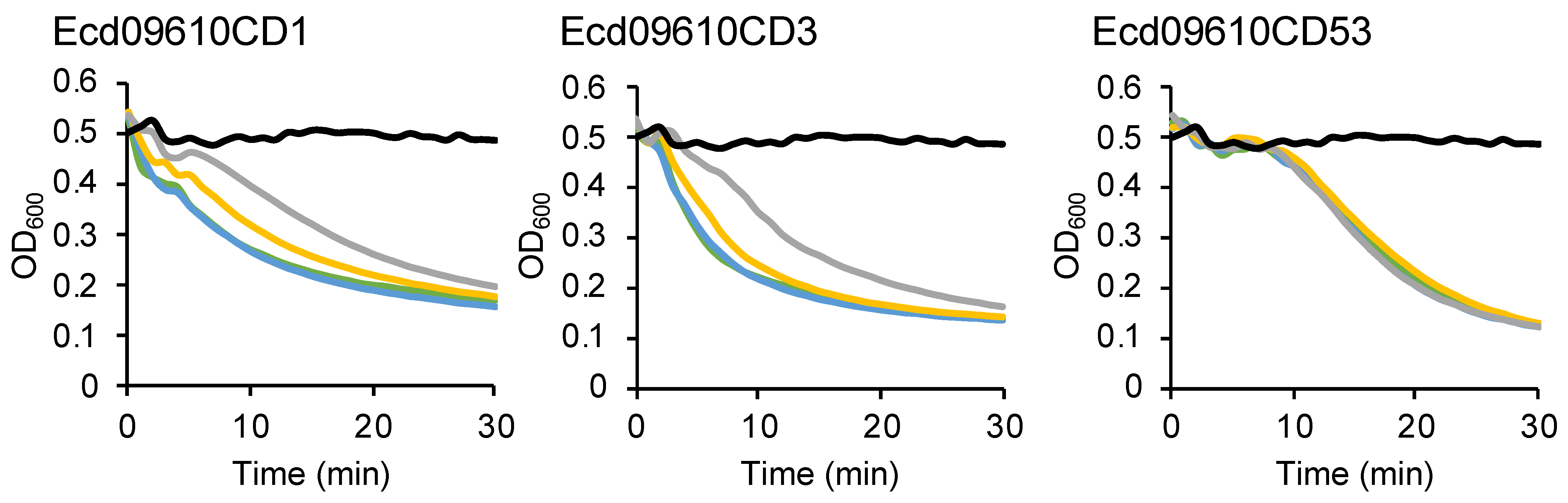
2.3. Bacterial Specificity of Ecd09610 Derivatives
2.4. Long-Term Stability of Ecd09610 Derivatives
3. Discussion
4. Materials and Methods
4.1. Construction of Plasmids
4.2. Preparation of Proteins
4.3. Lytic Activity Assay
4.4. Cell Binding Assay
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Burke, K.E.; Lamont, J.T. Clostridium difficile infection: A worldwide disease. Gut Liver 2014, 8, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Monaghan, T.; Boswell, T.; Mahida, Y.R. Recent advances in Clostridium difficile-associated disease. Postgrad. Med. J. 2009, 85, 152–162. [Google Scholar] [CrossRef] [PubMed]
- Venugopal, A.A.; Johnson, S. Fidaxomicin: A novel macrocyclic antibiotic approved for treatment of Clostridium difficile infection. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2012, 54, 568–574. [Google Scholar] [CrossRef]
- Bartsch, S.M.; Umscheid, C.A.; Fishman, N.; Lee, B.Y. Is fidaxomicin worth the cost? An economic analysis. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2013, 57, 555–561. [Google Scholar] [CrossRef] [PubMed]
- Rohlke, F.; Stollman, N. Fecal microbiota transplantation in relapsing Clostridium difficile infection. Ther. Adv. Gastroenterol. 2012, 5, 403–420. [Google Scholar] [CrossRef]
- Kalakuntla, A.S.; Nalakonda, G.; Nalakonda, K.; Pidikiti, C.V.; Aasim, S.A. Probiotics and Clostridium difficile: A Review of Dysbiosis and the Rehabilitation of Gut Microbiota. Cureus 2019, 11, e5063. [Google Scholar] [CrossRef] [PubMed]
- O’Horo, J.C.; Jindai, K.; Kunzer, B.; Safdar, N. Treatment of recurrent Clostridium difficile infection: A systematic review. Infection 2014, 42, 43–59. [Google Scholar] [CrossRef]
- Broendum, S.S.; Buckle, A.M.; McGowan, S. Catalytic diversity and cell wall binding repeats in the phage-encoded endolysins. Mol. Microbiol. 2018, 110, 879–896. [Google Scholar] [CrossRef]
- Smith, T.J.; Blackman, S.A.; Foster, S.J. Autolysins of Bacillus subtilis: Multiple enzymes with multiple functions. Microbiol. Read. Engl. 2000, 146 Pt 2, 249–262. [Google Scholar] [CrossRef]
- Vollmer, W.; Joris, B.; Charlier, P.; Foster, S. Bacterial peptidoglycan (murein) hydrolases. FEMS Microbiol. Rev. 2008, 32, 259–286. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhong, X.; Lu, P.; Zhu, Y.; Dong, W.; Roy, S.; Hejair, H.M.A.; Pan, Z.; Ma, J.; Yao, H. A novel autolysin AtlASS mediates bacterial cell separation during cell division and contributes to full virulence in Streptococcus suis. Vet. Microbiol. 2019, 234, 92–100. [Google Scholar] [CrossRef] [PubMed]
- Young, R. Bacteriophage lysis: Mechanism and regulation. Microbiol. Rev. 1992, 56, 430–481. [Google Scholar] [CrossRef]
- Fischetti, V.A. Bacteriophage lysins as effective antibacterials. Curr. Opin. Microbiol. 2008, 11, 393–400. [Google Scholar] [CrossRef] [PubMed]
- Nariya, H.; Miyata, S.; Tamai, E.; Sekiya, H.; Maki, J.; Okabe, A. Identification and characterization of a putative endolysin encoded by episomal phage phiSM101 of Clostridium perfringens. Appl. Microbiol. Biotechnol. 2011, 90, 1973–1979. [Google Scholar] [CrossRef] [PubMed]
- Tamai, E.; Yoshida, H.; Sekiya, H.; Nariya, H.; Miyata, S.; Okabe, A.; Kuwahara, T.; Maki, J.; Kamitori, S. X-ray structure of a novel endolysin encoded by episomal phage phiSM101 of Clostridium perfringens. Mol. Microbiol. 2014, 92, 326–337. [Google Scholar] [CrossRef] [PubMed]
- Sekiya, H.; Tamai, E.; Kawasaki, J.; Murakami, K.; Kamitori, S. Structural and biochemical characterizations of the novel autolysin Acd24020 from Clostridioides difficile and its full-function catalytic domain as a lytic enzyme. Mol. Microbiol. 2021, 115, 684–698. [Google Scholar] [CrossRef] [PubMed]
- Kirk, J.A.; Banerji, O.; Fagan, R.P. Characteristics of the Clostridium difficile cell envelope and its importance in therapeutics. Microb. Biotechnol. 2017, 10, 76–90. [Google Scholar] [CrossRef] [PubMed]
- Roach, D.R.; Donovan, D.M. Antimicrobial bacteriophage-derived proteins and therapeutic applications. Bacteriophage 2015, 5, e1062590. [Google Scholar] [CrossRef]
- Nelson, D.C.; Schmelcher, M.; Rodriguez-Rubio, L.; Klumpp, J.; Pritchard, D.G.; Dong, S.; Donovan, D.M. Endolysins as antimicrobials. Adv. Virus Res. 2012, 83, 299–365. [Google Scholar]
- Schmelcher, M.; Donovan, D.M.; Loessner, M.J. Bacteriophage endolysins as novel antimicrobials. Future Microbiol. 2012, 7, 1147–1171. [Google Scholar] [CrossRef]
- Nelson, D.; Loomis, L.; Fischetti, V.A. Prevention and elimination of upper respiratory colonization of mice by group A streptococci by using a bacteriophage lytic enzyme. Proc. Natl. Acad. Sci. USA 2001, 98, 4107–4112. [Google Scholar] [CrossRef] [PubMed]
- Mondal, S.I.; Akter, A.; Draper, L.A.; Ross, R.P.; Hill, C. Characterization of an Endolysin Targeting Clostridioides difficile That Affects Spore Outgrowth. Int. J. Mol. Sci. 2021, 22, 5690. [Google Scholar] [CrossRef] [PubMed]
- Swift, S.M.; Reid, K.P.; Donovan, D.M.; Ramsay, T.G. Thermophile Lytic Enzyme Fusion Proteins that Target Clostridium perfringens. Antibiotics 2019, 8, 214. [Google Scholar] [CrossRef] [PubMed]
- Valero-Rello, A. Diversity, specificity and molecular evolution of the lytic arsenal of Pseudomonas phages: In silico perspective. Environ. Microbiol. 2019, 21, 4136–4150. [Google Scholar] [CrossRef] [PubMed]
- Qin, X.; Singh, K.V.; Xu, Y.; Weinstock, G.M.; Murray, B.E. Effect of disruption of a gene encoding an autolysin of Enterococcus faecalis OG1RF. Antimicrob. Agents Chemother. 1998, 42, 2883–2888. [Google Scholar] [CrossRef]
- García, P.; González, M.P.; García, E.; López, R.; García, J.L. LytB, a novel pneumococcal murein hydrolase essential for cell separation. Mol. Microbiol. 1999, 31, 1275–1281. [Google Scholar] [CrossRef]
- Camiade, E.; Peltier, J.; Bourgeois, I.; Couture-Tosi, E.; Courtin, P.; Antunes, A.; Chapot-Chartier, M.P.; Dupuy, B.; Pons, J.L. Characterization of Acp, a peptidoglycan hydrolase of Clostridium perfringens with N-acetylglucosaminidase activity that is implicated in cell separation and stress-induced autolysis. J. Bacteriol. 2010, 192, 2373–2384. [Google Scholar] [CrossRef] [PubMed]
- Shirakawa, D.; Wakinaka, T.; Watanabe, J. Identification of the putative N-acetylglucosaminidase CseA associated with daughter cell separation in Tetragenococcus halophilus. Biosci. Biotechnol. Biochem. 2020, 84, 1724–1735. [Google Scholar] [CrossRef] [PubMed]
- Bublitz, M.; Polle, L.; Holland, C.; Heinz, D.W.; Nimtz, M.; Schubert, W.D. Structural basis for autoinhibition and activation of Auto, a virulence-associated peptidoglycan hydrolase of Listeria monocytogenes. Mol. Microbiol. 2009, 71, 1509–1522. [Google Scholar] [CrossRef]
- Hashimoto, W.; Ochiai, A.; Momma, K.; Itoh, T.; Mikami, B.; Maruyama, Y.; Murata, K. Crystal structure of the glycosidase family 73 peptidoglycan hydrolase FlgJ. Biochem. Biophys. Res. Commun. 2009, 381, 16–21. [Google Scholar] [CrossRef]
- Bai, X.H.; Chen, H.J.; Jiang, Y.L.; Wen, Z.; Huang, Y.; Cheng, W.; Li, Q.; Qi, L.; Zhang, J.R.; Chen, Y.; et al. Structure of pneumococcal peptidoglycan hydrolase LytB reveals insights into the bacterial cell wall remodeling and pathogenesis. J. Biol. Chem. 2014, 289, 23403–23416. [Google Scholar] [CrossRef] [PubMed]
- Lipski, A.; Hervé, M.; Lombard, V.; Nurizzo, D.; Mengin-Lecreulx, D.; Bourne, Y.; Vincent, F. Structural and biochemical characterization of the β-N-acetylglucosaminidase from Thermotoga maritima: Toward rationalization of mechanistic knowledge in the GH73 family. Glycobiology 2015, 25, 319–330. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zaloba, P.; Bailey-Elkin, B.A.; Derksen, M.; Mark, B.L. Structural and Biochemical Insights into the Peptidoglycan Hydrolase Domain of FlgJ from Salmonella typhimurium. PLoS ONE 2016, 11, e0149204. [Google Scholar] [CrossRef] [PubMed]
- Tamai, E.; Sekiya, H.; Goda, E.; Makihata, N.; Maki, J.; Yoshida, H.; Kamitori, S. Structural and biochemical characterization of the Clostridium perfringens autolysin catalytic domain. FEBS Lett. 2017, 591, 231–239. [Google Scholar] [CrossRef]
- Bourgogne, T.; Vacheron, M.J.; Guinand, M.; Michel, G. Purification and partial characterization of the gamma-D-glutamyl-L-di-amino acid endopeptidase II from Bacillus sphaericus. Int. J. Biochem. 1992, 24, 471–476. [Google Scholar] [CrossRef]
- Yamaguchi, H.; Furuhata, K.; Fukushima, T.; Yamamoto, H.; Sekiguchi, J. Characterization of a new Bacillus subtilis peptidoglycan hydrolase gene, yvcE (named cwlO), and the enzymatic properties of its encoded protein. J. Biosci. Bioeng. 2004, 98, 174–181. [Google Scholar] [CrossRef]
- Ohnishi, R.; Ishikawa, S.; Sekiguchi, J. Peptidoglycan hydrolase LytF plays a role in cell separation with CwlF during vegetative growth of Bacillus subtilis. J. Bacteriol. 1999, 181, 3178–3184. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Sudek, S.; McMullan, D.; Miller, M.D.; Geierstanger, B.; Jones, D.H.; Krishna, S.S.; Spraggon, G.; Bursalay, B.; Abdubek, P.; et al. Structural basis of murein peptide specificity of a gamma-D-glutamyl-l-diamino acid endopeptidase. Structure 2009, 17, 303–313. [Google Scholar] [CrossRef] [PubMed]
- Squeglia, F.; Ruggiero, A.; Romano, M.; Vitagliano, L.; Berisio, R. Mutational and structural study of RipA, a key enzyme in Mycobacterium tuberculosis cell division: Evidence for the L-to-D inversion of configuration of the catalytic cysteine. Acta Crystallogr. Sect. D Biol. Crystallogr. 2014, 70 Pt 9, 2295–2300. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Abdubek, P.; Astakhova, T.; Axelrod, H.L.; Bakolitsa, C.; Cai, X.; Carlton, D.; Chen, C.; Chiu, H.J.; Chiu, M.; et al. Structure of the γ-D-glutamyl-L-diamino acid endopeptidase YkfC from Bacillus cereus in complex with L-Ala-γ-D-Glu: Insights into substrate recognition by NlpC/P60 cysteine peptidases. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2010, 66 Pt 10, 1354–1364. [Google Scholar] [CrossRef]
- Loeffler, J.M.; Fischetti, V.A. Synergistic lethal effect of a combination of phage lytic enzymes with different activities on penicillin-sensitive and -resistant Streptococcus pneumoniae strains. Antimicrob. Agents Chemother. 2003, 47, 375–377. [Google Scholar] [CrossRef] [PubMed]
- Sekiya, H.; Okada, M.; Tamai, E.; Shimamoto, T.; Shimamoto, T.; Nariya, H. A Putative Amidase Endolysin Encoded by Clostridium perfringens St13 Exhibits Specific Lytic Activity and Synergizes with the Muramidase Endolysin Psm. Antibiotics 2021, 10, 245. [Google Scholar] [CrossRef] [PubMed]
- Ho, S.N.; Hunt, H.D.; Horton, R.M.; Pullen, J.K.; Pease, L.R. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 1989, 77, 51–59. [Google Scholar] [CrossRef]
- Gerova, M.; Halgasova, N.; Ugorcakova, J.; Bukovska, G. Endolysin of bacteriophage BFK20: Evidence of a catalytic and a cell wall binding domain. FEMS Microbiol. Lett. 2011, 321, 83–91. [Google Scholar] [CrossRef] [PubMed]
| Bacteria | Relative Activity (%) | ||
|---|---|---|---|
| Ecd09610CD1 | Ecd09610CD3 | Ecd09610CD53 | |
| C. difficile 630 | 100.0 ± 14.1 | 100.0 ± 20.8 | 100.0 ± 10.00 |
| C. difficile ATCC43255 | 104.9 ± 9.00 | 97.6 ± 15.6 | 15.6 ± 5.20 |
| C. difficile ATCC9689 | 101.7 ± 21.8 | 119.3 ± 21.0 | 96.3 ± 21.0 |
| C. acetobutylicum ATCC824 | −10.2 ± 0.90 | −6.8 ± 4.10 | −16.9 ± 4.90 |
| C. coccoides ATCC29236 | −1.3 ± 0.70 | −0.8 ± 0.90 | −2.3 ± 7.40 |
| C. histolyticum JCM1403 | 12.6 ± 4.30 | 8.1 ± 2.10 | 5.8 ± 1.50 |
| C. lituseburense ATCC25759 | 3.0 ± 5.10 | 4.5 ± 3.80 | 3.3 ± 1.50 |
| C. novyi ATCC17861 | 22.4 ± 11.0 | −2.3 ± 6.10 | 10.9 ± 17.40 |
| C. perfringens strain13 | 5.6 ± 0.60 | 1.3 ± 3.50 | 3.7 ± 5.20 |
| C. ramosum ATCC25582 | 65.3 ± 1.70 | 17.3 ± 2.90 | 7.5 ± 2.30 |
| C. tetani KZ1113 | 34.8 ± 3.00 | 38.9 ± 1.00 | 11.0 ± 1.80 |
| A. fossor ATCC43386 | 15.0 ± 2.20 | 11.0 ± 2.50 | 5.0 ± 2.00 |
| B. adolescentis ATCC15703 | 0.3 ± 2.10 | −2.4 ± 3.50 | −4.9 ± 0.80 |
| E. cylindroides ATCC27805 | 1.4 ± 5.00 | −2.6 ± 2.20 | −2.3 ± 2.60 |
| B. subtilis ATCC6633 | 16.8 ± 3.20 | 8.9 ± 10.0 | 8.5 ± 3.90 |
| S. aureus FDA209P | 1.8 ± 1.90 | 2.6 ± 2.00 | −1.1 ± 4.90 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sekiya, H.; Yamaji, H.; Yoshida, A.; Matsunami, R.; Kamitori, S.; Tamai, E. Biochemical Characterizations of the Putative Endolysin Ecd09610 Catalytic Domain from Clostridioides difficile. Antibiotics 2022, 11, 1131. https://doi.org/10.3390/antibiotics11081131
Sekiya H, Yamaji H, Yoshida A, Matsunami R, Kamitori S, Tamai E. Biochemical Characterizations of the Putative Endolysin Ecd09610 Catalytic Domain from Clostridioides difficile. Antibiotics. 2022; 11(8):1131. https://doi.org/10.3390/antibiotics11081131
Chicago/Turabian StyleSekiya, Hiroshi, Hina Yamaji, Ayumi Yoshida, Risa Matsunami, Shigehiro Kamitori, and Eiji Tamai. 2022. "Biochemical Characterizations of the Putative Endolysin Ecd09610 Catalytic Domain from Clostridioides difficile" Antibiotics 11, no. 8: 1131. https://doi.org/10.3390/antibiotics11081131
APA StyleSekiya, H., Yamaji, H., Yoshida, A., Matsunami, R., Kamitori, S., & Tamai, E. (2022). Biochemical Characterizations of the Putative Endolysin Ecd09610 Catalytic Domain from Clostridioides difficile. Antibiotics, 11(8), 1131. https://doi.org/10.3390/antibiotics11081131






