Abstract
Little information is available on the local epidemiology of mobile genetic elements such as plasmids harboring acquired beta-lactamase genes in Western African Ghana. In the present study, we screened for plasmids in three Escherichia coli and four Klebsiella pneumoniae isolates expressing extended spectrum beta-lactamases (ESBL) mediated by the blaCTX-M-15 gene from chronically infected wounds of Ghanaian patients. Bacterial isolates were subjected to combined short-read and long-read sequencing to obtain the sequences of their respective plasmids. In the blaCTX-M-15-gene-carrying plasmids of the four ESBL-positive K. pneumoniae isolates, IncFIB/IncFII (n = 3) and FIA (n = 1) sequences were detected, while in the blaCTX-M-15-gene-carrying plasmids of the three ESBL-positive E. coli isolates, IncFIA/IncFIB (n = 2) and IncFIB (n = 1) sequences were found. The three IncFIB/IncFII sequence-containing plasmids were almost identical to a K. pneumoniae plasmid reported from France. They belonged to the clonal lineages ST17, ST36 and ST39 of K. pneumoniae, suggesting transversal spread of this obviously evolutionary successful plasmid in Ghana. Other resistance gene-encoding plasmids observed in the assessed Enterobacterales harbored IncFIA/IncR and IncFII sequences. International spread was confirmed by the high genetic similarity to resistance-mediating plasmids published from Asia, Australia, Europe and Northern America, including a blaCTX-M-15-gene-carrying plasmid isolated from a wild bird in Germany. In conclusion, the study contributed to the scarcely available information on the epidemiology of third-generation cephalosporine resistance-mediating plasmids in Ghana. Furthermore, the global spread of resistance-mediating plasmids provided hints on the evolutionary success of individual resistance-harboring plasmids by transversal spread among K. pneumoniae lineages in Ghana.
1. Introduction
In recent years, multidrug resistance has become a major concern in sub-Saharan Africa. It has made bacterial infections increasingly difficult to treat, especially those associated with Gram-negative pathogens [1,2,3,4,5,6]. Acquired antimicrobial drug resistance in Gram-negative bacteria is typically mediated by mobile genetic elements such as plasmids, whose horizontal spread is driven by conjugation-based transmission [7]. Their persistence in bacterial clones is influenced by both fitness costs for the bacterial hosts [8] as well as by compensatory mutations [7,9]. The latter comprise, e.g., mutations in intergenic regions and the selection of genes involved in anaerobic metabolism [10].
In Enterobacterales such as Escherichia coli and Klebsiella pneumoniae, multidrug resistance is frequently mediated by epidemic resistance plasmids of incompatibility (Inc) groups such as IncFII, IncA/C, IncL/M, IncN and IncI1, which carry genes for extended-spectrum beta-lactamases (ESBLs), AmpC beta-lactamases and carbapenemases [11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38]. Next-generation sequencing-based approaches have been introduced early for the identification of plasmid sequences [39,40,41].
Epidemiological information on the spread and distribution of resistance-mediating plasmids in bacteria in Ghana is scarce besides individual approaches such as a study on the diversity of plasmids in Ghanaian gonococci from the beginning of the 1980s [42]. Regional spread of a trimethoprim resistance gene cassette via a successful transposable element was reported for Escherichia coli strains isolated in Ghana between 2006 and 2008 [43]. The conjugation-based transfer of blaTEM-gene-mediated ESBL expression could be shown for two-thirds of blaTEM gene-positive Enterobacterales isolated at a Ghanaian tertiary hospital [44]. In Ghanaian salmonellae comprising ESBL-positive strains mediated by the beta-lactamase genes blaTEM52-B and blaCTX-M15, IncN-type, IncFII(S)/IncFIB(S)/IncQ1-type, IncX1-type and TrfA/IncHI2/IncHI2A-type plasmids have been reported [45]. In an Escherichia coli isolate of the ST410 sequence type, the IncHI-type transferrable plasmid p2189-NDM was described, carrying the resistance genes blaNDM-1, blaCTX-M-15, aadA1, aac(6’)-Ib, sul3, dfrA12 and cmlA1 [46]. In Klebsiella pneumoniae isolates from a teaching hospital, IncFIB(K)-type and ColRNAI-type plasmids harbored resistance genes such as blaCTX-M-15, blaSHV-11, blaTEM-1B, blaOXA-1, ac(3)-IIa, strB, strA, aadA16, qnrB66, oqxA and oqxB, [47]. Various other epidemiological studies conducted in Ghana provide information on locally abundant resistance mechanisms without further addressing transposable genetic elements [48,49,50,51,52,53].
Recently, a predominance of Gram-negative rod-shaped bacteria was identified in chronic wounds in rural Ghana [54] with only low to moderate resistance rates compared to other reports from Ghanaian hospitals [55]. Among the Enterobacterales, a minority of three E. coli and four K. pneumoniae expressing the blaCTX-M-15 gene with a resulting ESBL-phenotype [56] were identified.
In the present study, the mobile genetic elements within those ESBL-producing Enterobacterales from chronic wounds of Ghanaian patients were assessed. By doing so, the so far scarce knowledge on the local epidemiology of plasmids mediating acquired antimicrobial resistance in Enterobacterales from Ghana was investigated.
2. Results
From seven Enterobacterales isolates from chronic wounds in Ghana as characterized in the methods chapter, 28 plasmid sequences were detected in four assessed ESBL-positive K. pneumoniae strains and in three E. coli strains. The sizes of the recorded plasmid contigs ranged from 1538 to 224,675 base pairs (Table 1). GenBank accession numbers and typing results applying the software Plasmidfinder 2.0 and mob-typer are shown in Table 1. The most frequently detected PlasmidFinder 2.0 and mob-typer matches for Inc sequences comprised IncFII (n = 5), IncFIA (n = 4), IncFIB (n = 4), IncFIC (n = 1), IncP1 (n = 1) and IncR (n = 1) sequences in 9 out of 16 plasmid sequences from ESBL-positive K. pneumoniae isolates. The ESBL-encoding blaCTX-M-15 genes were carried on plasmids of the IncFIB/IncFII type in three out of four K. pneumoniae strains and in another instance on an IncFIA type plasmid. In the plasmids from the three ESBL-positive E. coli, IncFIB (n = 4), IncFIA (n = 2), IncFII (n = 1), IncFIC (n = 1) and IncY (n = 1) sequences were detected in 7 out of 12 plasmid sequences. The ESBL-encoding blaCTX-M-15 genes were located on plasmids of the IncFIA/IncFIB/IncFIC-like type, the IncFIA/IncFIB/IncFII type and the IncFIB-type in E. coli strains. The blaCTX-M-15-carrying plasmids of both K. pneumoniae and E. coli strains are visualized in Figure 1. As suggested by the mob-typer software, predicted mobilities of the plasmids comprised the following categories: conjugative (n = 8, including 4 blaCTX-M-15 gene harboring plasmids), mobilizable (n = 11, including 2 blaCTX-M-15 gene harboring plasmids) and non-mobilizable (n = 9, including 1 blaCTX-M-15 gene harboring plasmid) (Table 1).

Table 1.
Identified plasmids with information on size, typing results based on PlasmidFinder-2.0 and mob-typer, predicted mobility based on mob-typer and encoded resistance genes. Resistance genes occurring with more than one copy are marked with (*).
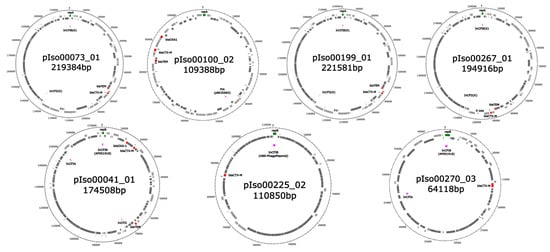
Figure 1.
Visualization of the blaCTX-M-gene-carrying plasmids. Upper row: Plasmids detected in K. pneumoniae. Lower row: Plasmids detected in E. coli. Genes located on the forward and reverse strand are colored in light and dark grey, respectively. Bla genes are shown in red. Genes coding for replication-associated proteins and marking the start gene of the sequence are shown in green. PlasmidFinder matches are shown in purple.
Resistance genes were located on 8 out of 16 plasmids of the four ESBL-positive K. pneumoniae isolates, as well as on 4 out of 12 plasmids of the three ESBL-positive E. coli isolates, as detailed in Table 1. In three out of four instances in K. pneumoniae, the blaCTX-M-15 gene was associated with the aminoglycoside-mediating gene aac(3)-IIa and the narrow-spectrum beta-lactamase gene blaTEM-1B on the same plasmid. One blaCTX-M-15 gene harboring plasmid in K. pneumoniae was associated with multiple resistance genes comprising macrolide resistance-mediating mph(A); aminoglycoside resistance-mediating aph(3′)-Ia aph(3″)-Ib, aph(6)-Id, aac(3)-IIa and aadA16; sulfonamide resistance-mediating sul1 and sul2; disinfectant resistance-mediating qacE; quinolone resistance-mediating qnrB6; trimethoprim-resistance-mediating dfrA27; rifampicin resistance-mediating ARR-3; fluoroquinolone and aminoglycoside-resistance-mediating aac(6′)-Ib-cr; the narrow-spectrum beta-lactamases blaTEM-1B and blaOXA-1; and phenicol resistance-mediating catB3. In the three ESBL-positive E. coli strains, the blaCTX-M-15 gene was the only resistance gene located on the plasmid in one instance. In another instance, it was associated with phenicol resistance-mediating catA1 and tetracycline resistance-mediating tet(B). In the third strain, an association with phenicol resistance-mediating catA1 and catB3, the narrow-spectrum beta-lactamases blaOXA-1 and blaTEM-1B, fluoroquinolone and aminoglycoside resistance-mediating aac(6′)-Ib-cr, disinfectant resistance-mediating sitABCD and qacE, tetracycline resistance-mediating tet(B), macrolide resistance-mediating mph(A), sulfonamide resistance-mediating sul1, aminoglycoside resistance-mediating aadA5 and aac(3)-IId, as well as trimethoprim resistance-mediating dfrA17, was recorded.
Of note, three out of four blaCTX-M-15-gene-carrying plasmids from K. pneumoniae isolates of different clonal lineages showed a very high genetic similarity to a plasmid isolated from a K. pneumoniae isolate from France (Appendix A, Table A1 and Table A2). This similarity was confirmed by a BlastN comparison of all blaCTX-M-15 gene harboring plasmids from the present study, confirming the high genetic similarity of the plasmids pIso00073_01, pIso00199_01 and pIso00267_01 from the K. pneumoniae clonal complex isolates ST39, ST17 and ST36, respectively (Table 2). In the mob-typer analysis, these plasmids were characterized as conjugative (Table 1).

Table 2.
Sequence homology as assessed by pairwise blastN analysis of blaCTX-M-gene containing plasmids/query coverages.
As indicated by another BlastN search (Appendix A), similar resistance-carrying plasmids to the ones from the present study have been globally isolated and sequenced in Europe, Asia, Australia and North America. The international isolations were performed not only from human samples but also from a sample taken from a wild bird, as reported from Germany (Appendix A).
3. Discussion
The study was conducted to add epidemiological information on the local epidemiology of third-generation cephalosporine resistance-mediating plasmids in Enterobacterales isolated from chronic wounds in Ghana. To do so, four K. pneumoniae and three E. coli strains carrying the ESBL-mediating blaCTX-M-15 genes were chosen from a previous study [54,56]. Wound isolates were chosen due to their likely etiological relevance for human infections, although etiologically irrelevant colonization cannot be completely ruled out at primarily non-sterile sampling sites such as superficial wounds in contact with the environment. Phenotypical strain characteristics did not affect the choice, which included all isolated blaCTX-M-15-gene-carrying Enterobacterales from the previous assessment. The strains were subjected to combined long-read and short-read sequencing to identify resistance-encoding plasmids and to compare the results with previous assessments.
In concordance with our results, IncFIB-type plasmids found in K. pneumoniae isolates from Ghana associated with the blaCTX-M-15 gene had been previously described in 2019 by Agyepong and colleagues [47]. In addition to the previously published results, we detected three plasmids with IncFIB/IncFII sequences that had previously been reported from France (GenBank accession number LR991402.1). Very similar, although not completely identical, plasmids were found in Ghanaian K. pneumoniae strains of the clonal lineages ST17, ST36 and ST39, suggesting the horizontal transmission of this plasmid within K. pneumoniae strains in Ghana, as also confirmed by mobility prediction with the mob-typer software. Of note, ESBL-positive ST39 K. pneumoniae strains have previously been reported to be highly prevalent in pigs and abattoir workers in Cameroon [57]. A blaCTX-M-15-gene-carrying plasmid of the FIA type found in another K. pneumoniae strain was previously described by Canadian scientists (GenBank accession number CP023950.1), confirming its international spread. Plasmids of the incompatibility groups IncFIA, IncFIB and IncFII carrying blaCTXM-15 genes have also been described from Eastern African Tanzania [58]. In Tanzanian K. pneumoniae strains, in particular, a blaCTXM-15-gene-harboring plasmid of the incompatibility group IncFIIK5/IncR has been associated with highly efficient horizontal transfer [59].
The description of plasmids carrying IncFIA/IncFIB/IncFIC, IncFIA/IncFIB/IncFII and IncFIB sequences associated with blaCTX-M-15 genes in E. coli strains is new and adds to the available information on the epidemiology of plasmids encoding resistance against third-generation cephalosporines in Gram-negative pathogens in Ghana [46,47]. Interestingly, genetically highly similar plasmid sequences have been reported from Australia (GenBank accession number LR890289.1), the United Kingdom (UK) [60] and Germany [61] before, comprising a human E. coli isolate from the UK and an E. coli strain isolated from a wild bird in Germany. It has recently been suggested [62] that not only international travel but also bacterial spread by migrating birds might contribute to the distribution of resistant bacterial isolates and their resistance-encoding plasmids. A single isolation from a bird cannot be considered as definitive proof because contamination from human sources remains an option but is nevertheless in line with this hypothesis. Interestingly, plasmids of the incompatibility group IncY have been linked to blaCTXM-15 gene-carriage in Tanzanian E. coli strains [63], while an IncY plasmid from one of the assessed Ghanaian E. coli strains did not harbor any resistance-associated genes.
Resistance against several antibiotic drugs other than beta-lactams, which had been phenotypically observed for the assessed Ghanaian Enterobacterales, was shown to be caused by co-occurring plasmids. In K. pneumoniae, such plasmids comprised the types IncFIA, IncFIA/IncR, IncFIA/IncFIC/IncFII and IncFIB/IncFII, while in E. coli, an IncFIA/IncFIB/IncFII-type plasmid encoded multiple resistance genes. Resistance-gene-carrying IncFIC plasmids in Africa have also been described in multidrug-resistant Salmonella enterica isolated in Kenya [64]. In contrast, ColRNAI-type plasmids, which were associated with antimicrobial resistance in Ghanaian K. pneumoniae strains in a previous study [47], did not encode resistance genes in our assessment.
As reported previously [39], linking of different contigs on the same plasmid can be challenging due to technical limitations of the sequencing technology. With focus on this technical issue, double sequencing with short-read (Illumina) and long-read (Nanopore) technologies was performed, followed by hybrid assembly of both data sets. Furthermore, evidence of plasmid replicon sequences was secured using three different methods (PlasmidFinder, MOB typing and BlastN versus the SRST2 database [65]) for all contigs. Furthermore, the plasmid nature of these shorter contigs is supported by normalized depths from 1.3 to 12.1 with an average of 4.2, as calculated by the Unicyler assembler (chromosome set to 1.0). Admittedly, these procedures do not provide 100% proof but were considered as sufficient justification for reporting the contigs as plasmids.
The study has a few limitations. First and most important, the number of ESBL-positive isolates available from the previous study [56] for inclusion in the plasmid assessment was considerably low. Accordingly, a regional representativeness cannot be ensured, although partial matching with previously published results from Ghana could be demonstrated [47]. Second, although the inclusion of etiologically relevant strains was aspired to by including strains from a wound infection study [54,56] instead of screening isolates, etiological relevance of the included strains is not definitely assured because the discrimination of causative infectious agents and colonizing bacterial flora is challenging at primarily non-sterile sites such as chronic wounds. Third, the postulated horizontal spread of plasmid-mediated third-generation cephalosporine resistance in Ghana was not confirmed by conjugation experiments. Such approaches would have been beyond both the scope and the financial options of this investigator-initiated, solely epidemiological study.
4. Materials and Methods
4.1. Sample Collection, Bacterial Culture, Antibiotic Susceptibility Testing and Whole Genome Sequencing
A total of seven ESBL-positive Enterobacterales carrying the blaCTX-M-15 gene were selected from a previous study on bacterial isolates from chronic wounds of patients from rural Ghana [54,56]. In summary, the strains were isolated from patients ≥ 15 years with infected chronic wounds at the Outpatient Department (OPD) of the Agogo Presbyterian Hospital in the Asante Akim North District of rural Ghana. Antibiotic resistance, as indicated in the Appendix A below, was assessed by the disk diffusion method and interpreted following the European Committee on Antimicrobial Susceptibility Testing (EUCAST) guidelines v.6.0 (http://www.eucast.org (accessed on 31 January 2016)). Both species identity and antibiotic susceptibility had been confirmed using the VITEK2 system (bioMérieux, Nürtingen, Germany), as described elsewhere [54,56].
Following nucleic acid extraction applying the MasterPure Complete DNA and RNA Purification Kit (LGC standards GmbH, Wesel, Germany), DNA was sent for whole genome sequencing (WGS) to BGI Europe, Denmark, Copenhagen. There, a BGISEQ-500 device was used for sequencing, generating 2 × 150 bp paired-end reads with an aimed coverage of 100×. Short-read archive (SRA) accession numbers of the obtained sequences are indicated in Table 1, linking to the original raw data as uploaded for public use to the short-read archive (SRA, NCBI) under the accession number PRJNA699140 [56]. Details on the chosen isolates are provided in Appendix A.
In addition, 1 µg DNA was sent to BGI Genomics C, Ltd., for long-read sequencing. In detail, sequencing analysis was performed on a PormethION (Nanopore) device using the flow-cell version R.9.4.1. Base-calling was performed with the Guppy software (https://nanoporetech.com, last accessed on 19 May 2022) applying the high-accuracy (HAC) model. After sequencing, about 3 mb of data was obtained from each sample.
4.2. Assessment of the Whole Genome Sequencing Data for Plasmids
Assembly of long-read sequences by Flye, v2.9 (https://github.com/fenderglass/Flye, last accessed on 30 March 2022) using (i) unfiltered fastq reads, (ii) fastq reads > 3 kb and (iii) fastq reads > 3 kb with subsequent polishing by Medaka v1.5.0 (https://github.com/nanoporetech/medaka, last accessed on 30 March 2022) did not reveal significant differences with respect to the assembled contigs. However, as expected, hybrid assemblies of long- and short-read sequences were of remarkably higher quality due to the resolution of sequencing errors in homo-nucleotide stretches (data not shown), a shortcoming of the nanopore technology [66]. Therefore, hybrid assemblies were performed by Unicycler [67] using unfiltered Oxford nanopore long-reads and BGI paired-end short-reads (after fastqc, trimmomatic). This resulted in assemblies containing one chromosome contig (K. pneumoniae 5.3–5.4 Mb; E. coli 4.7–5.0 Mb) and up to six plasmids per genome. Furthermore, the assembled plasmid contigs were analyzed by blastN (https://blast.ncbi.nlm.nih.gov/Blast.cgi, last accessed on 30 March 2022) vs. “Bacteria (taxid:2)” [68]. Results were filtered for the highest query coverage (as shown in Appendix A). Annotation was conducted by RAST (http://rast.nmpdr.org, last accessed on 30 March 2022). Analysis of the assembled genomes for resistance genes was conducted by ResFinder (http://cge.cbs.dtu.dk/services/ResFinder/—version 4.1, last accessed on 30 March 2022), with a nucleotide identity threshold of 90% and a minimum match length of 60%. Detection of plasmids was performed by PlasmidFinder, version 2.0.1 (https://cge.cbs.dtu.dk/services/PlasmidFinder-2.0/, last accessed on 30 March 2022) [39,69] and by the mob-typer software (https://github.com/phac-nml/mob-suite, last accessed on 14 April 2022) to identify matching incompatibility types [70] and to predict the mobility of the plasmids. As a third procedure, BlastN versus the SRST2 database [65] was conducted to ensure evidence of plasmid replicon sequences for all contigs, which were named IsoXXXXX_pXX. The sizes of the plasmids were based on the Unicycler results (short- and long-read hybrid assembly), which represents the most reliable approach currently available. Regarding the position of the blaCTX-M-15 genes within the plasmid sequences, the ResFinder results were independently confirmed by PROKKA annotation and Abricate [71] analysis, using the NCBI AMR Finder Plus Database, of the contigs of the above-mentioned hybrid assemblies (data not shown [72]). The ResFinder output provided the coordinates of the detected genes within the analyzed nucleotide sequences. For visualization purposes, the plasmid nucleotide sequences were annotated via RASTk using default settings [73]. The generated merged GenBank files were visualized using the tool DNAplotter [74].
4.3. Ethical Clearance
Ethical clearance was provided by the Committee on Human Research, Publications and Ethics, School of Medical Science, Kwame Nkrumah University of Science and Technology in Kumasi, Ghana (approval number CHRPE/AP/078/16).
5. Conclusions
Despite the above-mentioned limitations, which narrow the interpretability of the results, the study adds to the scarcely available data on the epidemiology of plasmids encoding third-generation cephalosporine resistance in ESBL-positive Enterobacterales in Western African Ghana. At least for K. pneumoniae, an individual conjugative blaCTX-M-15-gene-carrying plasmid was found in three out of four assessed strains of different clonal lineages, suggesting successful vertical transmission in Ghana. Furthermore, the assessment exemplarily demonstrated the international spread of such resistance-mediating plasmids in times of globalization, affected, e.g., by human travelling and the migration of wild birds. Future assessments should comprise more sampling sites, clinical conditions and geographic locations in Ghana to provide more robust and conclusive epidemiological data compared to the present hypothesis-forming study.
Author Contributions
U.L., F.P., S.T. and H.F. designed and coordinated this study. F.P. and S.T. performed the bioinformatic analysis. H.F. and U.L. wrote the first draft of this manuscript. F.P., S.T., D.D., T.T., K.O., C.W.A., M.L., A.J., M.K., S.S., K.T., H.F., J.M. and U.L., jointly supported the interpretation of the results, writing and editing the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This study was funded by institutional funds of the Bernhard Nocht Institute for Tropical Medicine (BNITM): reference number not applicable. We acknowledge support by the Open Access Publication Funds of Göttingen University.
Institutional Review Board Statement
The study was conducted according to guidelines of the Declaration of Helsinki. The Committee on Human Research, Publications and Ethics, School of Medical Science, Kwame Nkrumah University of Science and Technology in Kumasi, Ghana, approved this study (approval number CHRPE/AP/078/16).
Informed Consent Statement
Informed consent was obtained from all study participants.
Data Availability Statement
All relevant data have been provided in the paper. Raw data are available via the links indicated in the paper and can also be provided by the authors on reasonable request.
Acknowledgments
We thank all patients that participated in this study and the staff at the Agogo Presbyterian Hospital. Without their support, this research study would not have been possible.
Conflicts of Interest
The authors declare no conflict of interest.
Appendix A

Table A1.
Plasmids carrying antimicrobial resistance genes and disinfectant resistance genes and genetic similarity according to BlastN matching with previously published plasmids in the NCBI database, as well as the previous sites of detection.
Table A1.
Plasmids carrying antimicrobial resistance genes and disinfectant resistance genes and genetic similarity according to BlastN matching with previously published plasmids in the NCBI database, as well as the previous sites of detection.
| Strain/Sequence Type (ST)/GenBank Accession Number of Genomic DNA | Plasmid Number; GenBank Accession Number | Best Hit with Respect to the Query Coverage | Query Coverage (%) | Nucleotide Identity (%) | Geographic Site of NCBI Sequence Submission (Bacterial Species and Source) [Reference] |
|---|---|---|---|---|---|
| K. pneumoniae Iso00073/ST39/CP095150 | pIso00073_01; CP095151 | LR991402.1 | 97% | 99.99% | France (K. pneumoniae, source: unknown) [none] |
| pIso00073_02; CP095152 | CP063009.1 | 94% | 99.99% | Russian Federation (K. pneumoniae, source: human) [75] | |
| pIso00073_03; CP095153 | CP054171.1 | 95% | 99.86% | India (K. pneumoniae, source: human) [none] | |
| K. pneumoniae Iso00100/ST152/CP095145 | pIso00100_01; CP095146 | CP065826.1 | 79% | 99.95% | United States of America, (K. pneumoniae, source: human) [none] |
| pIso00100_02; CP095147 | CP023950.1 | 80% | 99.48% | Canada (K. pneumoniae, source: human) [none] | |
| K. pneumoniae Iso00199/ST17/CP095140 | pIso00199_01; CP095141 | LR991402.1 | 96% | 99.99% | France (K. pneumoniae, source: unknown) [none] |
| K. pneumoniae Iso00267/ST36/CP095132 | pIso00267_01; CP095133 | LR991402.1 | 94% | 99.99% | France (K. pneumoniae, source: unknown) [none] |
| pIso00267_02; CP095134 | CP016810.1 | 72% | 99.97% | United States of America, (K. pneumoniae, source: human) [none] | |
| E. coli Iso00041/ST 2/CP095155 | pIso00041_01; CP095156 | LR890289.1 | 99% | 99.98% | Australia (E. coli, source: unknown) [none] |
| E. coli Iso00225/ST506/CP095137 | pIso00225_01; CP095138 | CP088462.1 | 100% | 100% | South Korea (E. coli, source: human) [none] |
| pIso00225_02; CP095139 | MW590712.1 | 100% | 100% | United Kingdom (E. coli, source: human) [60] | |
| E. coli Iso00270/ST2/CP095125 | pIso00270_03; CP095128 | CP023816.1 | 96% | 99.11% | Germany (E. coli, source: wild bird) [61] |

Table A2.
Strain-specific details of the 7 Enterobacterales included in the screening for mobile genetic elements mediating antimicrobial resistance. Further details are provided elsewhere [56].
Table A2.
Strain-specific details of the 7 Enterobacterales included in the screening for mobile genetic elements mediating antimicrobial resistance. Further details are provided elsewhere [56].
| Species and Strain Number (73) | Sequence Type | Acquired Antimicrobial Resistance Genes | Recorded Phenotypic Resistance Against Apart From Penicillins and Cephalosporines * | Short-Read Archive (SRA) Accession Number |
|---|---|---|---|---|
| Klebsiella pneumoniae (73) | ST39 | blaTEM-1B, blaCTX-M-15,sul1, fosA, dfrA27, erm(B), mph(A), tet(D), oqxB, oqxA, aac(6′)-Ib-cr, qnrB2, catA2-like, aadA16, aac(3)-IIa, aph(3″)-Ib, aph(6)-Id | gentamicin, ciprofloxacin, moxifloxacin, trimethoprim/sulfamethoxazole | SRR13617236 |
| Klebsiella pneumoniae (100) | ST152 | blaCTX-M-15, blaOXA-1, blaTEM-1B,sul2, sul1, dfrA1, dfrA27, mph(A), aac(6′)-Ib-cr, oqxB, qnrB6, oqxA, catB3, catA1, ARR-3, aac(3)-IIa, aph(6)-Id, aph(3″)-Ib, aadA1, aadA16, aph(3′)-Ia | gentamicin, ciprofloxacin, moxifloxacin, trimethoprim/sulfamethoxazole | SRR13617311 |
| Klebsiella pneumoniae (199) | ST17 | blaCTX-M-15, blaTEM-1B,sul2, sul1, fosA-like, dfrA16, oqxA, oqxB, aadA2b, aac(3)-IIa | gentamicin, trimethoprim/sulfamethoxazole | SRR13617280 |
| Klebsiella pneumoniae (267) | ST36 | blaCTX-M-15, blaTEM-1B,sul2, sul1, fosA, dfrA27, tet(D), aac(6′)-Ib-cr, oqxA, oqxB, catA2-like, ARR-3, aph(6)-Id, aph(3″)-Ib, aadA16, aac(3)-IIa | gentamicin, trimethoprim/sulfamethoxazole | SRR13617257 |
| Escherichia coli (41) | ST2 | blaOXA-1, blaTEM-1B, blaCTX-M-15,sul1, dfrA17, mph(A), tet(B), aac(6′)-Ib-cr, catB3, catA1, aac(3)-IId, aadA5, mdf(A) | gentamicin, ciprofloxacin, moxifloxacin, trimethoprim/sulfamethoxazole | SRR13617294 |
| Escherichia coli (225) | ST506 | blaTEM-1D, blaCTX-M-15,sul1, sul2, dfrA17, mph(A), tet(A), catA1, aadA5, aph(6)-Id, aph(3″)-Ib, mdf(A)-like | moxifloxacin, trimethoprim/sulfamethoxazole | SRR13617270 |
| Escherichia coli (270) | ST2 | blaCTX-M-15, tet(B), catA1, mdf(A) | ciprofloxacin, moxifloxacin | SRR13617256 |
* Only resistance according to EUCAST is recorded, while intermediate susceptibility has been attributed to the susceptibility group. All strains were phenotypically resistant against ampicillin, ampicillin/sulbactam, piperacillin/tazobactam, cefuroxime, cefuroxime axetil, cefpodoxime, cefotaxime and ceftazidime. Other tested antimicrobial drugs comprised ertapenem, imipenem, meropenem, gentamicin, ciprofloxacin, moxifloxacin, tigecycline and trimethoprim/sulfamethoxazole.
References
- Mama, M.; Abdissa, A.; Sewunet, T. Antimicrobial susceptibility pattern of bacterial isolates from wound infection and their sensitivity to alternative topical agents at Jimma University Specialized Hospital, South-West Ethiopia. Ann. Clin. Microbiol. Antimicrob. 2014, 13, 14. [Google Scholar] [CrossRef] [PubMed]
- Moremi, N.; Mushi, M.F.; Fidelis, M.; Chalya, P.; Mirambo, M.; Mshana, S.E. Predominance of multi-resistant gram-negative bacteria colonizing chronic lower limb ulcers (CLLUs) at Bugando Medical Center. BMC Res. Notes 2014, 7, 211. [Google Scholar] [CrossRef] [PubMed]
- Kassam, N.A.; Damian, D.J.; Kajeguka, D.; Nyombi, B.; Kibiki, G.S. Spectrum and antibiogram of bacteria isolated from patients presenting with infected wounds in a Tertiary Hospital, northern Tanzania. BMC Res. Notes 2017, 10, 757. [Google Scholar] [CrossRef] [PubMed]
- Janssen, H.; Janssen, I.; Cooper, P.; Kainyah, C.; Pellio, T.; Quintel, M.; Monnheimer, M.; Groß, U.; Schulze, M.H. Antimicrobial-Resistant Bacteria in Infected Wounds, Ghana, 2014. Emerg. Infect. Dis. 2018, 24, 916–919. [Google Scholar] [CrossRef]
- Kazimoto, T.; Abdulla, S.; Bategereza, L.; Juma, O.; Mhimbira, F.; Weisser, M.; Utzinger, J.; von Müller, L.; Becker, S.L. Causative agents and antimicrobial resistance patterns of human skin and soft tissue infections in Bagamoyo, Tanzania. Acta Trop. 2018, 186, 102–106. [Google Scholar] [CrossRef] [PubMed]
- Lai, P.S.; Bebell, L.M.; Meney, C.; Valeri, L.; White, M.C. Epidemiology of antibiotic-resistant wound infections from six countries in Africa. BMJ Glob. Health 2018, 2 (Suppl. S4), e000475. [Google Scholar] [CrossRef]
- San Millan, A. Evolution of Plasmid-Mediated Antibiotic Resistance in the Clinical Context. Trends Microbiol. 2018, 26, 978–985. [Google Scholar] [CrossRef]
- San Millan, A.; MacLean, R.C. Fitness Costs of Plasmids: A Limit to Plasmid Transmission. Microbiol. Spectr. 2017, 5, 5. [Google Scholar] [CrossRef]
- Loftie-Eaton, W.; Bashford, K.; Quinn, H.; Dong, K.; Millstein, J.; Hunter, S.; Thomason, M.K.; Merrikh, H.; Ponciano, J.M.; Top, E.M. Compensatory mutations improve general permissiveness to antibiotic resistance plasmids. Nat. Ecol. Evol. 2017, 1, 1354–1363. [Google Scholar] [CrossRef]
- Dunn, S.J.; Connor, C.; McNally, A. The evolution and transmission of multi-drug resistant Escherichia coli and Klebsiella pneumoniae: The complexity of clones and plasmids. Curr. Opin. Microbiol. 2019, 51, 51–56. [Google Scholar] [CrossRef]
- Carattoli, A. Plasmids in Gram negatives: Molecular typing of resistance plasmids. Int. J. Med. Microbiol. 2011, 301, 654–658. [Google Scholar] [CrossRef] [PubMed]
- Shin, J.; Choi, M.J.; Ko, K.S. Replicon sequence typing of IncF plasmids and the genetic environments of blaCTX-M-15 indicate multiple acquisitions of blaCTX-M-15 in Escherichia coli and Klebsiella pneumoniae isolates from South Korea. J. Antimicrob. Chemother. 2012, 67, 1853–1857. [Google Scholar] [CrossRef] [PubMed]
- Machuca, J.; López-Cerero, L.; Fernández-Cuenca, F.; Mora-Navas, L.; Mediavilla-Gradolph, C.; López-Rodríguez, I.; Pascual, Á. OXA-48-Like-Producing Klebsiella pneumoniae in Southern Spain in 2014-2015. Antimicrob. Agents Chemother. 2018, 63, e01396-18. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Tang, Y.; Fu, P.; Tian, D.; Yu, L.; Huang, Y.; Li, G.; Li, M.; Wang, Y.; Yang, Z.; et al. The type I-E CRISPR-Cas system influences the acquisition of bla(KPC)-IncF plasmid in Klebsiella pneumoniae. Emerg. Microbes Infect. 2020, 9, 1011–1022. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Fu, P.; Zhou, Y.; Xie, Y.; Jin, J.; Wang, B.; Yu, L.; Huang, Y.; Li, G.; Li, M.; et al. Absence of the type I-E CRISPR-Cas system in Klebsiella pneumoniae clonal complex 258 is associated with dissemination of IncF epidemic resistance plasmids in this clonal complex. J. Antimicrob. Chemother. 2020, 75, 890–895. [Google Scholar] [CrossRef]
- Yu, X.; Zhang, W.; Zhao, Z.; Ye, C.; Zhou, S.; Wu, S.; Han, L.; Han, Z.; Ye, H. Molecular characterization of carbapenem-resistant Klebsiella pneumoniae isolates with focus on antimicrobial resistance. BMC Genom. 2019, 20, 822. [Google Scholar] [CrossRef]
- Feng, Y.; Liu, L.; McNally, A.; Zong, Z. Coexistence of three bla(KPC-2) genes on an IncF/IncR plasmid in ST11 Klebsiella pneumoniae. J. Glob. Antimicrob. Resist. 2019, 17, 90–93. [Google Scholar] [CrossRef]
- Peirano, G.; Bradford, P.A.; Kazmierczak, K.M.; Chen, L.; Kreiswirth, B.N.; Pitout, J.D. Importance of Clonal Complex 258 and IncF(K2-like) Plasmids among a Global Collection of Klebsiella pneumoniae with bla(KPC). Antimicrob. Agents Chemother. 2017, 61, e02610-16. [Google Scholar] [CrossRef]
- Pérez-Vázquez, M.; Sola Campoy, P.J.; Ortega, A.; Bautista, V.; Monzón, S.; Ruiz-Carrascoso, G.; Mingorance, J.; González-Barberá, E.M.; Gimeno, C.; Aracil, B.; et al. Emergence of NDM-producing Klebsiella pneumoniae and Escherichia coli in Spain: Phylogeny, resistome, virulence and plasmids encoding blaNDM-like genes as determined by WGS. J. Antimicrob. Chemother. 2019, 74, 3489–3496. [Google Scholar] [CrossRef]
- Villa, L.; García-Fernández, A.; Fortini, D.; Carattoli, A. Replicon sequence typing of IncF plasmids carrying virulence and resistance determinants. J. Antimicrob. Chemother. 2010, 65, 2518–2529. [Google Scholar] [CrossRef]
- Poirel, L.; Potron, A.; Nordmann, P. OXA-48-like carbapenemases: The phantom menace. J. Antimicrob. Chemother. 2012, 67, 1597–1606. [Google Scholar] [CrossRef] [PubMed]
- Gaibani, P.; Scaltriti, E.; Benni, C.; Pongolini, S.; Ambretti, S.; Landini, M.P.; Viale, P.; Giannella, M.; Re, M.C. Characterization of an IncL/M plasmid carrying blaOXA-48 in a Klebsiella pneumoniae strain from Italy. New Microbiol. 2017, 40, 284–285. [Google Scholar] [PubMed]
- Carattoli, A.; Seiffert, S.N.; Schwendener, S.; Perreten, V.; Endimiani, A. Differentiation of IncL and IncM Plasmids Associated with the Spread of Clinically Relevant Antimicrobial Resistance. PLoS ONE 2015, 10, e0123063. [Google Scholar] [CrossRef] [PubMed]
- Adamczuk, M.; Zaleski, P.; Dziewit, L.; Wolinowska, R.; Nieckarz, M.; Wawrzyniak, P.; Kieryl, P.; Plucienniczak, A.; Bartosik, D. Diversity and Global Distribution of IncL/M Plasmids Enabling Horizontal Dissemination of β-Lactam Resistance Genes among the Enterobacteriaceae. Biomed. Res. Int. 2015, 2015, 414681. [Google Scholar] [CrossRef]
- Oliveira, É.M.; Beltrão, E.M.B.; Scavuzzi, A.M.L.; Barros, J.F.; Lopes, A.C.S. High plasmid variability, and the presence of IncFIB, IncQ, IncA/C, IncHI1B, and IncL/M in clinical isolates of Klebsiella pneumoniae with blaKPC and blaNDM from patients at a public hospital in Brazil. Rev. Soc. Bras. Med. Trop. 2020, 53, e20200397. [Google Scholar] [CrossRef]
- Rada, A.M.; De La Cadena, E.; Agudelo, C.; Capataz, C.; Orozco, N.; Pallares, C.; Dinh, A.Q.; Panesso, D.; Ríos, R.; Diaz, L.; et al. Dynamics of bla(KPC-2) Dissemination from Non-CG258 Klebsiella pneumoniae to Other Enterobacterales via IncN Plasmids in an Area of High Endemicity. Antimicrob. Agents Chemother. 2020, 64, e01743-20. [Google Scholar] [CrossRef]
- Aires-de-Sousa, M.; Ortiz de la Rosa, J.M.; Gonçalves, M.L.; Pereira, A.L.; Nordmann, P.; Poirel, L. Epidemiology of Carbapenemase-Producing Klebsiella pneumoniae in a Hospital, Portugal. Emerg. Infect. Dis. 2019, 25, 1632–1638. [Google Scholar] [CrossRef]
- Yang, Y.; Higgins, C.H.; Rehman, I.; Galvao, K.N.; Brito, I.L.; Bicalho, M.L.; Song, J.; Wang, H.; Bicalho, R.C. Genomic Diversity, Virulence, and Antimicrobial Resistance of Klebsiella pneumoniae Strains from Cows and Humans. Appl. Environ. Microbiol. 2019, 85, e02654-18. [Google Scholar] [CrossRef]
- Hao, M.; He, Y.; Zhang, H.; Liao, X.P.; Liu, Y.H.; Sun, J.; Du, H.; Kreiswirth, B.N.; Chen, L. CRISPR-Cas9-Mediated Carbapenemase Gene and Plasmid Curing in Carbapenem-Resistant Enterobacteriaceae. Antimicrob. Agents Chemother. 2020, 64, e00843-20. [Google Scholar] [CrossRef]
- García-Fernández, A.; Villa, L.; Moodley, A.; Hasman, H.; Miriagou, V.; Guardabassi, L.; Carattoli, A. Multilocus sequence typing of IncN plasmids. J. Antimicrob. Chemother. 2011, 66, 1987–1991. [Google Scholar] [CrossRef]
- Jing, Y.; Jiang, X.; Yin, Z.; Hu, L.; Zhang, Y.; Yang, W.; Yang, H.; Gao, B.; Zhao, Y.; Zhou, D.; et al. Genomic diversification of IncR plasmids from China. J. Glob. Antimicrob. Resist. 2019, 19, 358–364. [Google Scholar] [CrossRef] [PubMed]
- Dong, N.; Liu, L.; Zhang, R.; Chen, K.; Xie, M.; Chan, E.W.C.; Chen, S. An IncR Plasmid Harbored by a Hypervirulent Carbapenem-Resistant Klebsiella pneumoniae Strain Possesses Five Tandem Repeats of the bla (KPC-2)::NTE(KPC)-Id Fragment. Antimicrob. Agents Chemother. 2019, 63, e01775-18. [Google Scholar] [CrossRef] [PubMed]
- Kocsis, E.; Gužvinec, M.; Butić, I.; Krešić, S.; Crnek, S.Š.; Tambić, A.; Cornaglia, G.; Mazzariol, A. blaNDM-1 Carriage on IncR Plasmid in Enterobacteriaceae Strains. Microb. Drug Resist. 2016, 22, 123–128. [Google Scholar] [CrossRef] [PubMed]
- Gamal, D.; Fernández-Martínez, M.; Salem, D.; El-Defrawy, I.; Montes, L.Á.; Ocampo-Sosa, A.A.; Martínez-Martínez, L. Carbapenem-resistant Klebsiella pneumoniae isolates from Egypt containing blaNDM-1 on IncR plasmids and its association with rmtF. Int. J. Infect. Dis. 2016, 43, 17–20. [Google Scholar] [CrossRef] [PubMed]
- Qian, C.; Zhu, X.; Lu, J.; Shen, K.; Chen, Q.; Zhou, W.; Liu, H.; Lu, W.; Zhou, D.; Sun, Z.; et al. Characterization of an IncR Plasmid with Two Copies of ISCR-Linked qnrB6 from ST968 Klebsiella pneumoniae. Int. J. Genom. 2020, 2020, 3484328. [Google Scholar] [CrossRef]
- Guo, Q.; Spychala, C.N.; McElheny, C.L.; Doi, Y. Comparative analysis of an IncR plasmid carrying armA, blaDHA-1 and qnrB4 from Klebsiella pneumoniae ST37 isolates. J. Antimicrob. Chemother. 2016, 71, 882–886. [Google Scholar] [CrossRef]
- Compain, F.; Frangeul, L.; Drieux, L.; Verdet, C.; Brisse, S.; Arlet, G.; Decré, D. Complete nucleotide sequence of two multidrug-resistant IncR plasmids from Klebsiella pneumoniae. Antimicrob. Agents Chemother. 2014, 58, 4207–4210. [Google Scholar] [CrossRef]
- Qu, D.; Shen, Y.; Hu, L.; Jiang, X.; Yin, Z.; Gao, B.; Zhao, Y.; Yang, W.; Yang, H.; Han, J.; et al. Comparative analysis of KPC-2-encoding chimera plasmids with multi-replicon IncR:Inc(pA1763-KPC):IncN1 or IncFII(pHN7A8):Inc(pA1763-KPC):IncN1. Infect. Drug Resist. 2019, 12, 285–296. [Google Scholar] [CrossRef]
- Carattoli, A.; Zankari, E.; García-Fernández, A.; Voldby Larsen, M.; Lund, O.; Villa, L.; Møller Aarestrup, F.; Hasman, H. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 2014, 58, 3895–3903. [Google Scholar] [CrossRef]
- De Maio, N.; Shaw, L.P.; Hubbard, A.; George, S.; Sanderson, N.D.; Swann, J.; Wick, R.; AbuOun, M.; Stubberfield, E.; Hoosdally, S.J.; et al. Comparison of long-read sequencing technologies in the hybrid assembly of complex bacterial genomes. Microb. Genom. 2019, 5, e000294. [Google Scholar] [CrossRef]
- Suzuki, Y.; Nishijima, S.; Furuta, Y.; Yoshimura, J.; Suda, W.; Oshima, K.; Hattori, M.; Morishita, S. Long-read metagenomic exploration of extrachromosomal mobile genetic elements in the human gut. Microbiome 2019, 7, 119. [Google Scholar] [CrossRef] [PubMed]
- Perine, P.L.; Totten, P.A.; Knapp, J.S.; Holmes, K.K.; Bentsi, C.; Klufio, C.A. Diversity of gonococcal plasmids, auxotypes, and serogroups in Ghana. Lancet 1983, 1, 1051–1052. [Google Scholar] [CrossRef]
- Labar, A.S.; Millman, J.S.; Ruebush, E.; Opintan, J.A.; Bishar, R.A.; Aboderin, A.O.; Newman, M.J.; Lamikanra, A.; Okeke, I.N. Regional dissemination of a trimethoprim-resistance gene cassette via a successful transposable element. PLoS ONE 2012, 7, e38142. [Google Scholar] [CrossRef] [PubMed]
- Oduro-Mensah, D.; Obeng-Nkrumah, N.; Bonney, E.Y.; Oduro-Mensah, E.; Twum-Danso, K.; Osei, Y.D.; Sackey, S.T. Genetic characterization of TEM-type ESBL-associated antibacterial resistance in Enterobacteriaceae in a tertiary hospital in Ghana. Ann. Clin. Microbiol Antimicrob. 2016, 15, 29. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kudirkiene, E.; Andoh, L.A.; Ahmed, S.; Herrero-Fresno, A.; Dalsgaard, A.; Obiri-Danso, K.; Olsen, J.E. The Use of a Combined Bioinformatics Approach to Locate Antibiotic Resistance Genes on Plasmids from Whole Genome Sequences of Salmonella enterica Serovars from Humans in Ghana. Front. Microbiol. 2018, 9, 1010. [Google Scholar] [CrossRef]
- Ayibieke, A.; Sato, W.; Mahazu, S.; Prah, I.; Addow-Thompson, J.; Ohashi, M.; Suzuki, T.; Iwanaga, S.; Ablordey, A.; Saito, R. Molecular characterisation of the NDM-1-encoding plasmid p2189-NDM in an Escherichia coli ST410 clinical isolate from Ghana. PLoS ONE 2018, 13, e0209623. [Google Scholar] [CrossRef]
- Agyepong, N.; Govinden, U.; Owusu-Ofori, A.; Amoako, D.G.; Allam, M.; Janice, J.; Pedersen, T.; Sundsfjord, A.; Essack, S. Genomic characterization of multidrug-resistant ESBL-producing Klebsiella pneumoniae isolated from a Ghanaian teaching hospital. Int. J. Infect. Dis. 2019, 85, 117–123. [Google Scholar] [CrossRef]
- Moirongo, R.M.; Lorenz, E.; Ntinginya, N.E.; Dekker, D.; Fernandes, J.; Held, J.; Lamshöft, M.; Schaumburg, F.; Mangu, C.; Sudi, L.; et al. Regional Variation of Extended-Spectrum Beta-Lactamase (ESBL)-Producing Enterobacterales, Fluoroquinolone-Resistant Salmonella enterica and Methicillin-Resistant Staphylococcus aureus Among Febrile Patients in Sub-Saharan Africa. Front. Microbiol. 2020, 11, 567235. [Google Scholar] [CrossRef]
- Acheampong, G.; Owusu, M.; Owusu-Ofori, A.; Osei, I.; Sarpong, N.; Sylverken, A.; Kung, H.J.; Cho, S.T.; Kuo, C.H.; Park, S.E.; et al. Chromosomal and plasmid-mediated fluoroquinolone resistance in human Salmonella enterica infection in Ghana. BMC Infect. Dis. 2019, 19, 898. [Google Scholar] [CrossRef]
- Eibach, D.; Nagel, M.; Lorenzen, S.; Hogan, B.; Belmar Campos, C.; Aepfelbacher, M.; Sarpong, N.; May, J. Extended-spectrum β-lactamase-producing Enterobacteriaceae among geckos (Hemidactylus brookii) in a Ghanaian hospital. Clin. Microbiol. Infect. 2019, 25, 1048–1050. [Google Scholar] [CrossRef]
- Adzitey, F.; Assoah-Peprah, P.; Teye, G.A. Whole-genome sequencing of Escherichia coli isolated from contaminated meat samples collected from the Northern Region of Ghana reveals the presence of multiple antimicrobial resistance genes. J. Glob. Antimicrob. Resist. 2019, 18, 179–182. [Google Scholar] [CrossRef] [PubMed]
- Falgenhauer, L.; Imirzalioglu, C.; Oppong, K.; Akenten, C.W.; Hogan, B.; Krumkamp, R.; Poppert, S.; Levermann, V.; Schwengers, O.; Sarpong, N.; et al. Detection and Characterization of ESBL-Producing Escherichia coli from Humans and Poultry in Ghana. Front. Microbiol. 2019, 9, 3358. [Google Scholar] [CrossRef] [PubMed]
- Ayibieke, A.; Kobayashi, A.; Suzuki, M.; Sato, W.; Mahazu, S.; Prah, I.; Mizoguchi, M.; Moriya, K.; Hayashi, T.; Suzuki, T.; et al. Prevalence and Characterization of Carbapenem-Hydrolyzing Class D β-Lactamase-Producing Acinetobacter Isolates from Ghana. Front. Microbiol. 2020, 11, 587398. [Google Scholar] [CrossRef] [PubMed]
- Krumkamp, R.; Oppong, K.; Hogan, B.; Strauss, R.; Frickmann, H.; Wiafe-Akenten, C.; Boahen, K.G.; Rickerts, V.; McCormick Smith, I.; Groß, U.; et al. Spectrum of antibiotic resistant bacteria and fungi isolated from chronically infected wounds in a rural district hospital in Ghana. PLoS ONE 2020, 15, e0237263. [Google Scholar] [CrossRef] [PubMed]
- Codjoe, F.S.; Donkor, E.S.; Smith, T.J.; Miller, K. Phenotypic and Genotypic Characterization of Carbapenem-Resistant Gram-Negative Bacilli Pathogens from Hospitals in Ghana. Microb. Drug Resist. 2019, 25, 1449–1457. [Google Scholar] [CrossRef]
- Dekker, D.; Pankok, F.; Thye, T.; Taudien, S.; Oppong, K.; Wiafe Akenten, C.; Lamshöft, M.; Jaeger, A.; Kaase, M.; Scheithauer, S.; et al. Clonal Clusters, Molecular Resistance Mechanisms and Virulence Factors of Gram-Negative Bacteria Isolated from Chronic Wounds in Ghana. Antibiotics 2021, 10, 339. [Google Scholar] [CrossRef]
- Founou, L.L.; Founou, R.C.; Allam, M.; Ismail, A.; Djoko, C.F.; Essack, S.Y. Genome Sequencing of Extended-Spectrum β-Lactamase (ESBL)-Producing Klebsiella pneumoniae Isolated from Pigs and Abattoir Workers in Cameroon. Front. Microbiol. 2018, 9, 188. [Google Scholar] [CrossRef]
- Minja, C.A.; Shirima, G.; Mshana, S.E. Conjugative Plasmids Disseminating CTX-M-15 among Human, Animals and the Environment in Mwanza Tanzania: A Need to Intensify One Health Approach. Antibiotics 2021, 10, 836. [Google Scholar] [CrossRef]
- Pedersen, T.; Tellevik, M.G.; Kommedal, Ø.; Lindemann, P.C.; Moyo, S.J.; Janice, J.; Blomberg, B.; Samuelsen, Ø.; Langeland, N. Horizontal Plasmid Transfer among Klebsiella pneumoniae Isolates Is the Key Factor for Dissemination of Extended-Spectrum β-Lactamases among Children in Tanzania. mSphere 2020, 5, e00428-20. [Google Scholar] [CrossRef]
- Bevan, E.R.; Powell, M.J.; Toleman, M.A.; Thomas, C.M.; Piddock, L.J.V.; Hawkey, P.M. Molecular characterization of plasmids encoding blaCTX-M from faecal Escherichia coli in travellers returning to the UK from South Asia. J. Hosp. Infect. 2021, 114, 134–143. [Google Scholar] [CrossRef]
- Schaufler, K.; Semmler, T.; Wieler, L.H.; Trott, D.J.; Pitout, J.; Peirano, G.; Bonnedahl, J.; Dolejska, M.; Literak, I.; Fuchs, S.; et al. Genomic and Functional Analysis of Emerging Virulent and Multidrug-Resistant Escherichia coli Lineage Sequence Type 648. Antimicrob. Agents Chemother. 2019, 63, e00243-19. [Google Scholar] [CrossRef] [PubMed]
- Bonnedahl, J.; Järhult, J.D. Antibiotic resistance in wild birds. Upsala J. Med. Sci. 2014, 119, 113–116. [Google Scholar] [CrossRef] [PubMed]
- Alonso, C.A.; Zarazaga, M.; Ben Sallem, R.; Jouini, A.; Ben Slama, K.; Torres, C. Antibiotic resistance in Escherichia coli in husbandry animals: The African perspective. Lett. Appl. Microbiol. 2017, 64, 318–334. [Google Scholar] [CrossRef] [PubMed]
- Mutai, W.C.; Waiyaki, P.G.; Kariuki, S.; Muigai, A.W.T. Plasmid profiling and incompatibility grouping of multidrug resistant Salmonella enterica serovar Typhi isolates in Nairobi, Kenya. BMC Res. Notes 2019, 12, 422. [Google Scholar] [CrossRef]
- Inouye, M.; Dashnow, H.; Raven, L.A.; Schultz, M.B.; Pope, B.J.; Tomita, T.; Zobel, J.; Holt, K.E. SRST2: Rapid genomic surveillance for public health and hospital microbiology labs. Genome Med. 2014, 6, 90. [Google Scholar] [CrossRef]
- Delahaye, C.; Nicolas, J. Sequencing DNA with nanopores: Troubles and biases. PLoS ONE 2021, 16, e0257521. [Google Scholar] [CrossRef]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef]
- Carattoli, A.; Hasman, H. PlasmidFinder and In Silico pMLST: Identification and Typing of Plasmid Replicons in Whole-Genome Sequencing (WGS). Methods Mol. Biol. 2020, 2075, 285–294. [Google Scholar]
- Johnson, T.J.; Nolan, L.K. Plasmid replicon typing. Methods Mol. Biol. 2009, 551, 27–35. [Google Scholar]
- Seemann, T. Abricate. Available online: https://github.com/tseemann/abricate (accessed on 11 May 2022).
- Feldgarden, M.; Brover, V.; Haft, D.H.; Prasad, A.B.; Slotta, D.J.; Tolstoy, I.; Tyson, G.H.; Zhao, S.; Hsu, C.H.; McDermott, P.F.; et al. Validating the AMRFinder Tool and Resistance Gene Database by Using Antimicrobial Resistance Genotype-Phenotype Correlations in a Collection of Isolates. Antimicrob. Agents Chemother. 2019, 63, e00483-19. [Google Scholar] [CrossRef] [PubMed]
- Brettin, T.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Olsen, G.J.; Olson, R.; Overbeek, R.; Parrello, B.; Pusch, G.D.; et al. RASTtk: A modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci. Rep. 2015, 5, 8365. [Google Scholar] [CrossRef] [PubMed]
- Carver, T.; Thomson, N.; Bleasby, A.; Berriman, M.; Parkhill, J. DNAPlotter: Circular and linear interactive genome visualization. Bioinformatics 2009, 25, 119–120. [Google Scholar] [CrossRef] [PubMed]
- Shelenkov, A.; Mikhaylova, Y.; Yanushevich, Y.; Samoilov, A.; Petrova, L.; Fomina, V.; Gusarov, V.; Zamyatin, M.; Shagin, D.; Akimkin, V. Molecular Typing, Characterization of Antimicrobial Resistance, Virulence Profiling and Analysis of Whole-Genome Sequence of Clinical Klebsiella pneumoniae Isolates. Antibiotics 2020, 9, 261. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).