Multidrug-Resistant Enterobacterales in Community-Acquired Urinary Tract Infections in Djibouti, Republic of Djibouti
Abstract
1. Introduction
2. Results
2.1. Bacterial Isolates and Patient Characteristics
2.2. Multidrug-Resistant Enterobacterales (MDR-E) Isolates
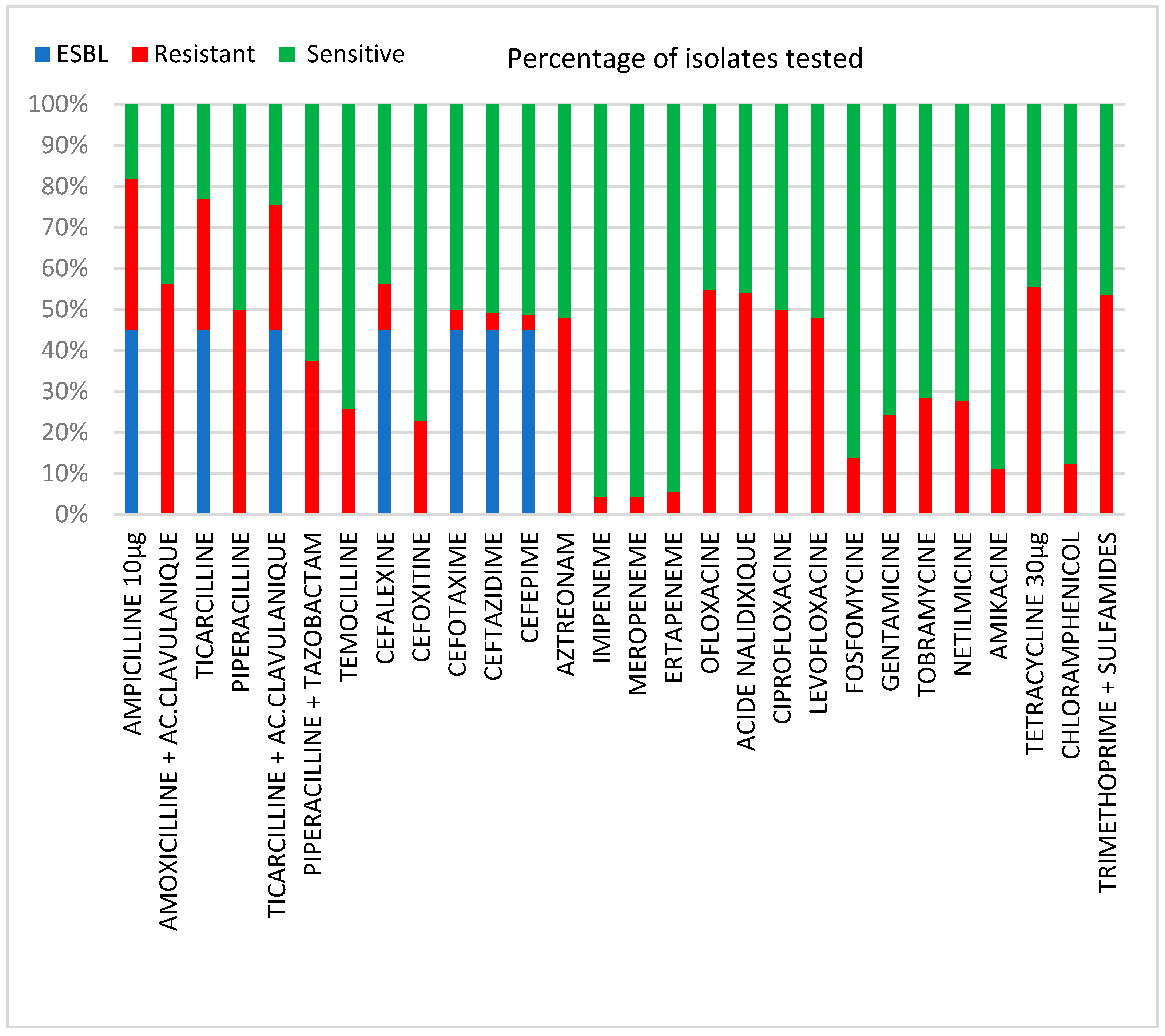
2.3. Antibiotic Resistance Patterns
2.4. Molecular Characterization of Beta-Lactamases and Encoding Genes
2.5. Molecular Epidemiology Typing
3. Discussion
4. Materials and Methods
4.1. Study Setting
4.2. Specimen Collection, Identification, and Antimicrobial Susceptibility Testing
4.3. Molecular Identification of ESBLs and Carbapenemases
4.4. Molecular Epidemiology Typing
4.5. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Larabi, K.; Masmoudi, A.; Fendri, C. Étude bactériologique et phénotypes de résistance des germes responsables d’infections urinaires dans un CHU de Tunis: À propos de 1930 cas. Méd. Mal. Infect. 2003, 33, 348–352. [Google Scholar] [CrossRef]
- Arpin, C.; Dubois, V.; Coulange, L.; André, C.; Fischer, I.; Noury, P.; Grobost, F.; Brochet, J.-P.; Jullin, J.; Dutilh, B.; et al. Extended-Spectrum β-Lactamase-Producing Enterobacteriaceae in Community and Private Health Care Centers. Antimicrob. Agents Chemother. 2003, 47, 3506–3514. [Google Scholar] [CrossRef]
- Pitout, J.D.; Laupland, K.B. Extended-Spectrum β-Lactamase-Producing Enterobacteriaceae: An Emerging Public-Health Concern. Lancet Infect. Dis. 2008, 8, 159–166. [Google Scholar] [CrossRef]
- Cantón, R.; Novais, A.; Valverde, A.; Machado, E.; Peixe, L.; Baquero, F.; Coque, T.M. Prevalence and Spread of Extended-Spectrum β-Lactamase-Producing Enterobacteriaceae in Europe. Clin. Microbiol. Infect. 2008, 14, 144–153. [Google Scholar] [CrossRef] [PubMed]
- Winokur, P.L.; Canton, R. Variations in the Prevalence of Strains Expressing an Extended-Spectrum b-Lactamase Phenotype and Characterization of Isolates from Europe, the Americas, and the Western Pacific Region. Clin. Infect. Dis. 2001, 32 (Suppl. 2), S94–S103. [Google Scholar] [CrossRef] [PubMed]
- Bauernfeind, A.; Schweighart, S.; Grimm, H. A New Plasmidic Cefotaximase in a Clinical Isolate of Escherichia coli. Infection 1990, 18, 294–298. [Google Scholar] [CrossRef]
- Bauernfeind, A.; Holley, M.; Jungwirth, R.; Mangold, P.; Röhnisch, T.; Schweighart, S.; Wilhelm, R.; Casellas, J.M.; Goldberg, M. A New Plasmidic Cefotaximase from Patients Infected with Salmonella typhimurium. Infection 1992, 20, 158–163. [Google Scholar] [CrossRef] [PubMed]
- Pitout, J.D.D.; Hanson, N.D.; Church, D.L.; Laupland, K.B. Population-Based Laboratory Surveillance for Escherichia coli–Producing Extended-Spectrum b-Lactamases: Importance of Community Isolates with BlaCTX-M Genes. Clin. Infect. Dis. 2004, 38, 1736–1741. [Google Scholar] [CrossRef][Green Version]
- Cantón, R.; Coque, T.M. The CTX-M β-Lactamase Pandemic. Curr. Opin. Microbiol. 2006, 9, 466–475. [Google Scholar] [CrossRef]
- Queenan, A.M.; Bush, K. Carbapenemases: The Versatile β-Lactamases. Clin. Microbiol. Rev. 2007, 20, 440–458. [Google Scholar] [CrossRef]
- Poirel, L.; Héritier, C.; Tolün, V.; Nordmann, P. Emergence of Oxacillinase-Mediated Resistance to Imipenem in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 2004, 48, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Nordmann, P.; Poirel, L. The Difficult-to-Control Spread of Carbapenemase Producers among Enterobacteriaceae Worldwide. Clin. Microbiol. Infect. 2014, 20, 821–830. [Google Scholar] [CrossRef] [PubMed]
- Carrër, A.; Poirel, L.; Yilmaz, M.; Akan, Ö.A.; Feriha, C.; Cuzon, G.; Matar, G.; Honderlick, P.; Nordmann, P. Spread of OXA-48-Encoding Plasmid in Turkey and Beyond. Antimicrob. Agents Chemother. 2010, 54, 1369–1373. [Google Scholar] [CrossRef] [PubMed]
- Wangkheimayum, J.; Paul, D.; Dhar, D.; Nepram, R.; Chetri, S.; Bhowmik, D.; Chakravarty, A.; Bhattacharjee, A. Occurrence of Acquired 16S RRNA Methyltransferase-Mediated Aminoglycoside Resistance in Clinical Isolates of Enterobacteriaceae within a Tertiary Referral Hospital of Northeast India. Antimicrob. Agents Chemother. 2017, 61, e01037-16. [Google Scholar] [CrossRef] [PubMed]
- Shamsrizi, P.; Gladstone, B.P.; Carrara, E.; Luise, D.; Cona, A.; Bovo, C.; Tacconelli, E. Variation of Effect Estimates in the Analysis of Mortality and Length of Hospital Stay in Patients with Infections Caused by Bacteria-Producing Extended-Spectrum Beta-Lactamases: A Systematic Review and Meta-Analysis. BMJ Open 2020, 10, e030266. [Google Scholar] [CrossRef] [PubMed]
- Paterson, D.L.; Bonomo, R.A. Extended-Spectrum β-Lactamases: A Clinical Update. Clin. Microbiol. Rev. 2005, 18, 657–686. [Google Scholar] [CrossRef]
- Lee, D.S.; Lee, S.-J.; Choe, H.-S. Community-Acquired Urinary Tract Infection by Escherichia coli in the Era of Antibiotic Resistance. BioMed Res. Int. 2018, 2018, 7656752. [Google Scholar] [CrossRef]
- Ali, G.H.; Yakout, M.A. Comparative Study of ESBL Production Among Uropathogenic Escherichia coli Clinical Isolates from Pre- and Post-Menopausal Women in Egypt. Curr. Microbiol. 2021, 78, 3516–3525. [Google Scholar] [CrossRef]
- Ramírez-Castillo, F.Y.; Moreno-Flores, A.C.; Avelar-González, F.J.; Márquez-Díaz, F.; Harel, J.; Guerrero-Barrera, A.L. An Evaluation of Multidrug-Resistant Escherichia coli Isolates in Urinary Tract Infections from Aguascalientes, Mexico: Cross-Sectional Study. Ann. Clin. Microbiol. Antimicrob. 2018, 17, 34. [Google Scholar] [CrossRef]
- Ángel Díaz, M.; Ramón Hernández, J.; Martínez-Martínez, L.; Rodríguez-Baño, J.; Pascual, Á. Escherichia coli y Klebsiella pneumoniae productoras de betalactamasas de espectro extendido en hospitales españoles: Segundo estudio multicéntrico (proyecto GEIH-BLEE 2006). Enferm. Infecc. Microbiol. Clín. 2009, 27, 503–510. [Google Scholar] [CrossRef]
- Sarkis, P.; Assaf, J.; Sarkis, J.; Zanaty, M.; Rehban, R. Profil de résistance aux antibiotiques dans les infections urinaires communautaires au Liban. Prog. Urol. 2017, 27, 727. [Google Scholar] [CrossRef]
- Fam, N.; Leflon-Guibout, V.; Fouad, S.; Aboul-Fadl, L.; Marcon, E.; Desouky, D.; El-Defrawy, I.; Abou-Aitta, A.; Klena, J.; Nicolas-Chanoine, M.-H. CTX-M-15-Producing Escherichia coli Clinical Isolates in Cairo (Egypt), Including Isolates of Clonal Complex ST10 and Clones ST131, ST73, and ST405 in Both Community and Hospital Settings. Microb. Drug Resist. 2011, 17, 67–73. [Google Scholar] [CrossRef] [PubMed]
- Abujnah, A.A.; Zorgani, A.; Sabri, M.A.M.; El-Mohammady, H.; Khalek, R.A.; Ghenghesh, K.S. Multidrug Resistance and Extended-Spectrum β-Lactamases Genes among Escherichia coli from Patients with Urinary Tract Infections in Northwestern Libya. Libyan J. Med. 2015, 10, 26412. [Google Scholar] [CrossRef]
- Valverde, A.; Coque, T.M.; Sanchez-Moreno, M.P.; Rollan, A.; Baquero, F.; Canton, R. Dramatic Increase in Prevalence of Fecal Carriage of Extended-Spectrum-Lactamase-Producing Enterobacteriaceae during Nonoutbreak Situations in Spain. J. Clin. Microbiol. 2004, 42, 7. [Google Scholar] [CrossRef] [PubMed]
- Livermore, D.M. Current Epidemiology and Growing Resistance of Gram-Negative Pathogens. Korean J. Intern. Med. 2012, 27, 128. [Google Scholar] [CrossRef] [PubMed]
- Dahmen, S.; Bettaieb, D.; Mansour, W.; Boujaafar, N.; Bouallègue, O.; Arlet, G. Characterization and Molecular Epidemiology of Extended-Spectrum β-Lactamases in Clinical Isolates of Enterobacteriaceae in a Tunisian University Hospital. Microb. Drug Resist. 2010, 16, 163–170. [Google Scholar] [CrossRef]
- Mohamedalagamy, M.; Eldinashour, M.; Wiegand, I. First Description of CTX-M β-Lactamase-Producing Clinical Escherichia coli Isolates from Egypt. Int. J. Antimicrob. Agents 2006, 27, 545–548. [Google Scholar] [CrossRef]
- Chong, Y.; Ito, Y.; Kamimura, T. Genetic Evolution and Clinical Impact in Extended-Spectrum β-Lactamase-Producing Escherichia coli and Klebsiella pneumoniae. Infect. Genet. Evol. 2011, 11, 1499–1504. [Google Scholar] [CrossRef]
- Mathlouthi, N.; Al-Bayssari, C.; Bakour, S.; Rolain, J.M.; Chouchani, C. RETRACTED ARTICLE: Prevalence and Emergence of Carbapenemases-Producing Gram-Negative Bacteria in Mediterranean Basin. Crit. Rev. Microbiol. 2017, 43, 43–61. [Google Scholar] [CrossRef]
- Mairi, A.; Pantel, A.; Sotto, A.; Lavigne, J.-P.; Touati, A. OXA-48-like Carbapenemases Producing Enterobacteriaceae in Different Niches. Eur. J. Clin. Microbiol. Infect. Dis. 2018, 37, 587–604. [Google Scholar] [CrossRef]
- Loucif, L.; Chelaghma, W.; Helis, Y.; Sebaa, F.; Baoune, R.D.; Zaatout, W.; Rolain, J.-M. First Detection of OXA-48-Producing Klebsiella pneumoniae in Community-Acquired Urinary Tract Infection in Algeria. J. Glob. Antimicrob. Resist. 2018, 12, 115–116. [Google Scholar] [CrossRef] [PubMed]
- Bajaj, P.; Singh, N.S.; Virdi, J.S. Escherichia coli β-Lactamases: What Really Matters. Front. Microbiol. 2016, 7, 417. [Google Scholar] [CrossRef] [PubMed]
- Nabti, L.Z.; Sahli, F.; Radji, N.; Mezaghcha, W.; Semara, L.; Aberkane, S.; Lounnas, M.; Solassol, J.; Didelot, M.-N.; Jean-Pierre, H.; et al. High Prevalence of Multidrug-Resistant Escherichia coli in Urine Samples from Inpatients and Outpatients at a Tertiary Care Hospital in Sétif, Algeria. Microb. Drug Resist. 2019, 25, 386–393. [Google Scholar] [CrossRef] [PubMed]
- Nzalie, R.N.; Gonsu, H.K.; Koulla-Shiro, S. Bacterial Etiology and Antibiotic Resistance Profile of Community-Acquired Urinary Tract Infections in a Cameroonian City. Int. J. Microbiol. 2016, 2016, 3240268. [Google Scholar] [CrossRef] [PubMed]
- Odongo, C.O.; Anywar, D.A.; Luryamamoi, K.; Odongo, P. Antibiograms from Community-Acquired Uropathogens in Gulu, Northern Uganda—A Cross-Sectional Study. BMC Infect. Dis. 2013, 13, 193. [Google Scholar] [CrossRef]
- Smaoui, S.; Abdelhedi, K.; Marouane, C.; Kammoun, S.; Messadi-Akrout, F. Résistance aux antibiotiques des entérobactéries responsables d’infections urinaires communautaires à Sfax (Tunisie). Méd. Mal. Infect. 2015, 45, 335–337. [Google Scholar] [CrossRef]
- Guessennd, N.; Bremont, S.; Gbonon, V.; Kacou-NDouba, A.; Ekaza, E.; Lambert, T.; Dosso, M.; Courvalin, P. Résistance aux quinolones de type qnr chez les entérobactéries productrices de bêta-lactamases à spectre élargi à Abidjan en Côte d’Ivoire. Pathol. Biol. 2008, 56, 439–446. [Google Scholar] [CrossRef]
- Mouanga Ndzime, Y.; Onanga, R.; Kassa Kassa, R.F.; Bignoumba, M.; Mbehang Nguema, P.P.; Gafou, A.; Lendamba, R.W.; Mbombe Moghoa, K.; Bisseye, C. Epidemiology of Community Origin Escherichia coli and Klebsiella pneumoniae Uropathogenic Strains Resistant to Antibiotics in Franceville, Gabon. IDR 2021, 14, 585–594. [Google Scholar] [CrossRef]
- Hassuna, N.A.; Khairalla, A.S.; Farahat, E.M.; Hammad, A.M.; Abdel-Fattah, M. Molecular Characterization of Extended-Spectrum β Lactamase- Producing E. coli Recovered from Community-Acquired Urinary Tract Infections in Upper Egypt. Sci. Rep. 2020, 10, 2772. [Google Scholar] [CrossRef]
- Dromigny, J.A.; Nabeth, P.; Perrier Gros Claude, J.D. Distribution and Susceptibility of Bacterial Urinary Tract Infections in Dakar, Senegal. Int. J. Antimicrob. Agents 2002, 20, 339–347. [Google Scholar] [CrossRef]
- Naziri, Z.; Derakhshandeh, A.; Soltani Borchaloee, A.; Poormaleknia, M.; Azimzadeh, N. Treatment Failure in Urinary Tract Infections: A Warning Witness for Virulent Multi-Drug Resistant ESBL- Producing Escherichia coli. IDR 2020, 13, 1839–1850. [Google Scholar] [CrossRef] [PubMed]
- El Bouamri, M.C.; Arsalane, L.; Kamouni, Y.; Yahyaoui, H.; Bennouar, N.; Berraha, M.; Zouhair, S. Profil actuel de résistance aux antibiotiques des souches d’Escherichia coli uropathogènes et conséquences thérapeutiques. Prog. Urol. 2014, 24, 1058–1062. [Google Scholar] [CrossRef] [PubMed]
- Kahlmeter, G.; Poulsen, H.O. Antimicrobial Susceptibility of Escherichia coli from Community-Acquired Urinary Tract Infections in Europe: The ECO·SENS Study Revisited. Int. J. Antimicrob. Agents 2012, 39, 45–51. [Google Scholar] [CrossRef] [PubMed]
- Iranpour, D.; Hassanpour, M.; Ansari, H.; Tajbakhsh, S.; Khamisipour, G.; Najafi, A. Phylogenetic Groups of Escherichia coli Strains from Patients with Urinary Tract Infection in Iran Based on the New Clermont Phylotyping Method. BioMed Res. Int. 2015, 2015, 846219. [Google Scholar] [CrossRef] [PubMed]
- Gonçalves, L.F.; de Oliveira Martins-Júnior, P.; de Melo, A.B.F.; da Silva, R.C.R.M.; de Paulo Martins, V.; Pitondo-Silva, A.; de Campos, T.A. Multidrug Resistance Dissemination by Extended-Spectrum β-Lactamase-Producing Escherichia coli Causing Community-Acquired Urinary Tract Infection in the Central-Western Region, Brazil. J. Glob. Antimicrob. Resist. 2016, 6, 1–4. [Google Scholar] [CrossRef]
- Jaureguy, F.; Landraud, L.; Passet, V.; Diancourt, L.; Frapy, E.; Guigon, G.; Carbonnelle, E.; Lortholary, O.; Clermont, O.; Denamur, E.; et al. Phylogenetic and Genomic Diversity of Human Bacteremic Escherichia coli Strains. BMC Genom. 2008, 9, 560. [Google Scholar] [CrossRef]
- Mshana, S.E.; Imirzalioglu, C.; Hain, T.; Domann, E.; Lyamuya, E.F.; Chakraborty, T. Multiple ST Clonal Complexes, with a Predominance of ST131, of Escherichia coli Harbouring BlaCTX-M-15 in a Tertiary Hospital in Tanzania. Clin. Microbiol. Infect. 2011, 17, 1279–1282. [Google Scholar] [CrossRef]
- Aibinu, I.; Odugbemi, T.; Koenig, W.; Ghebremedhin, B. Sequence Type ST131 and ST10 Complex (ST617) Predominant among CTX-M-15-Producing Escherichia coli Isolates from Nigeria. Clin. Microbiol. Infect. 2012, 18, E49–E51. [Google Scholar] [CrossRef][Green Version]
- Coque, T.M.; Baquero, F.; Cantón, R. Increasing Prevalence of ESBL-Producing Enterobacteriaceae in Europe. Eurosurveillance 2008, 13, 19044. [Google Scholar] [CrossRef]
- Petty, N.K.; Ben Zakour, N.L.; Stanton-Cook, M.; Skippington, E.; Totsika, M.; Forde, B.M.; Phan, M.-D.; Gomes Moriel, D.; Peters, K.M.; Davies, M.; et al. Global Dissemination of a Multidrug Resistant Escherichia coli Clone. Proc. Natl. Acad. Sci. USA 2014, 111, 5694–5699. [Google Scholar] [CrossRef]
- Rogers, B.A.; Sidjabat, H.E.; Paterson, D.L. Escherichia coli O25b-ST131: A Pandemic, Multiresistant, Community-Associated Strain. J. Antimicrob. Chemother. 2011, 66, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Cattoir, V.; Poirel, L.; Rotimi, V.; Soussy, C.-J.; Nordmann, P. Multiplex PCR for Detection of Plasmid-Mediated Quinolone Resistance Qnr Genes in ESBL-Producing Enterobacterial Isolates. J. Antimicrob. Chemother. 2007, 60, 394–397. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Xu, B.; Yang, Y.; Liu, D.; Yang, M.; Wang, J.; Shen, H.; Zhou, X.; Ma, X. A High Throughput Multiplex PCR Assay for Simultaneous Detection of Seven Aminoglycoside-Resistance Genes in Enterobacteriaceae. BMC Microbiol. 2013, 13, 58. [Google Scholar] [CrossRef] [PubMed]
- Clermont, O.; Christenson, J.K.; Denamur, E.; Gordon, D.M. The Clermont Escherichia coli Phylo-Typing Method Revisited: Improvement of Specificity and Detection of New Phylo-Groups: A New E. coli Phylo-Typing Method. Environ. Microbiol. Rep. 2013, 5, 58–65. [Google Scholar] [CrossRef] [PubMed]
| Factors. | ESBL-Producing (n = 65) | non-ESBL-Producing (n = 77) |
|---|---|---|
| Age (years) | N (%) | N (%) |
| 15–24 years | 19 (13%) | 30 (21%) |
| 25–49 years | 32 (23%) | 24 (17%) |
| ≥50 years | 14 (10%) | 23 (16%) |
| Sex | ||
| Male | 21 (15%) | 40 (28%) |
| Female | 44 (31%) | 37 (26%) |
| Previous use of antibiotics (last 3 months) | 35 (54%) | 14 (18%) |
| Isolate | Beta-Lactamases | Carbapenemases | Phylogroup | Sequence Type | Resistance to Quinolones |
|---|---|---|---|---|---|
| E. coli13 | CTX-M-15+OXA-1 | B2 | ST-43 | qnrB | |
| E. coli7U | OXA48 | A | ST2450 | ||
| E. coli49 | CTX-M-15+OXA-1 | B2 | ST-43 | qnrA | |
| E. coli53 | CTX-M-15+OXA-1 | B2 | ST-43 | ||
| E. coli48 | CTX-M-15+OXA-1 | B2 | ST-43 | qnrA | |
| E. coli56 | CTX-M-15+TEM-1, OXA-1 | B2 | ST-43 | ||
| E. coli95 | CTX-M-15+OXA-1 | B2 | ST-43 | ||
| E. coli64 | CTX-M-15+TEM-1, OXA-1 | B2 | ST-43 | qnrD | |
| E. coli11 | CTX-M-15+TEM-1, OXA-1 | B2 | ST-43 | qnrC | |
| E. coli24 | CTX-M-15+OXA-1 | B2 | ST-43 | qnrC | |
| E. coli43 | CTX-M-15+OXA-1 | B2 | ST53 | qnrD | |
| E. coli55 | CTX-M-15+OXA-1 | F | ST2 | ||
| E. coli41 | CTX-M-15+TEM-1, OXA-1 | B2 | ST53 | oqxAB | |
| E. coli49 | CTX-M-14+OXA-1 | D | ST829 | oqxAB | |
| E. coli58 | CTX-M-15+OXA-1 | A | ST692 | qnrC | |
| E. coli95 | CTX-M-15+TEM-1, OXA-1 | A | ST698 | qnrA | |
| E. coli02 | CTX-M-14+OXA-1 | A | ST2 | oqxAB | |
| E. coli13 | CTX-M-15+TEM-1, OXA-1 | B1 | ST954 | qnrD | |
| E. coli75 | CTX-M-15+OXA-1 | A | ST690 | oqxAB | |
| E. coli98 | CTX-M-15+OXA-1 | D | ST44 | ||
| E. coli91 | CTX-M-15 | B2 | ST-43 | ||
| E. coli08 | CTX-M-15+TEM-1 | D | ST44 | qnrA | |
| E. coli39 | CTX-M-15+OXA-1 | B2 | ST53 | qnrA | |
| E. coli08 | CTX-M-15+TEM-1, OXA-1 | B2 | ST53 | ||
| E. coli64 | CTX-M-15+OXA-1 | B2 | ST53 | ||
| E. coli19 | CTX-M-15+TEM-1, OXA-1 | B2 | ST53 | qnrA | |
| E. coli14 | CTX-M-15+TEM-1 | F | ST2 | ||
| E. coli78 | CTX-M-15+OXA-1 | F | ST2 | ||
| E. coli49 | CTX-M-15 | B2 | ST-43 | ||
| E. coli28 | CTX-M-15+OXA-1 | B2 | ST-43 | qnrA | |
| E. coli21 | CTX-M-15+OXA-1 | B2 | ST-43 | qnrA | |
| E. coli37 | CTX-M-15+OXA-1 | B2 | ST-43 | ||
| E. coli28 | CTX-M-15+OXA-1 | B2 | ST-43 | ||
| E. coli05 | CTX-M-15+OXA-1 | B2 | ST-43 | qnrA | |
| E. coli81 | CTX-M-15+OXA-1 | B2 | ST-43 | ||
| E. coli88 | CTX-M-15+TEM-1, OXA-1 | B2 | ST-43 | ||
| E. coli11 | CTX-M-15+OXA-1 | B2 | ST-43 | ||
| E. coli67 | CTX-M-15+TEM-1, OXA-1 | B2 | ST-43 | qnrA | |
| E. coli32 | CTX-M-15+OXA-1 | A | ST500 | ||
| E. coli30 | CTX-M-15 | B1 | ST741 | ||
| E. coli31 | CTX-M-15+OXA-1 | B1 | ST741 | ||
| E. coli78 | CTX-M-15+OXA-1 | B1 | ST960 | ||
| E. coli53 | CTX-M-15+TEM-1, OXA-1 | B1 | ST960 | ||
| E. coli95 | CTX-M-15+OXA-1 | B1 | ST960 | ||
| E. coli4U | CTX-M-15+OXA-1 | OXA48 | C | ST410 | |
| E. coli10 | CTX-M-15+OXA-1 | A | ST692 | ||
| E. coli54 | CTX-M-15+TEM-1 | A | ST692 | ||
| E. coli06 | CTX-M-15+OXA-1 | A | ST692 | ||
| E. coli90 | CTX-M-15+OXA-1 | A | ST692 | ||
| E. coli30 | CTX-M-15+OXA-1 | A | ST692 | ||
| E. coli76 | CTX-M-15+TEM-1, OXA-1 | A | ST692 | ||
| E. coli 29 | CTX-M-15+OXA-1 | A | ST692 | ||
| E. coli75 | CTX-M-15+OXA-1 | A | ST692 | ||
| E. coli90 | CTX-M-15+TEM-1 | D | ST829 | ||
| E. coli26 | CTX-M-15+TEM-1 | D | ST829 | ||
| E. coli063 | CTX-M-15+TEM-1 | E | ST244 | ||
| E. coli63 | CTX-M-15+OXA-1 | E | ST244 | ||
| E. coli076 | CTX-M-15+TEM-1 | F | ST2 | ||
| K. pneumoniae20 | CTX-M-15+ OXA-1 | ST592 | |||
| K. pneumoniae54 | CTX-M-15+ OXA-1 | ST464 | |||
| K. pneumoniae03 | CTX-M-15+TEM-1 | ST29 | |||
| K. pneumoniae11 | CTX-M-15+ OXA-1 | ST889 | |||
| K. pneumoniae26 | ST732 | qnrA | |||
| K. pneumoniae43 | ST16 | ||||
| K. pneumoniae13 | CTX-M-15+ OXA-1 | ST485 | |||
| K. pneumoniae710 | CTX-M-15+ OXA-1 | ST889 | |||
| K. pneumoniae50 | CTX-M-15+ OXA-1 | ST464 |
| Phylogenetic Group | MDR-E N (%) | non-MDR-E N (%) | Total ESBL-EC N (%) |
|---|---|---|---|
| B2 | 24 (21%) | 26 (22%) | 50 (43%) |
| A | 16 (14%) | 14 (12%) | 30 (26%) |
| B1 | 10 (9%) | 8 (7%) | 18 (15%) |
| D | 4 (3%) | 4 (3%) | 8 (6%) |
| F | 3 (3%) | 4 (3%) | 7 (6%) |
| E | 2 (2%) | 2 (2%) | 4 (4%) |
| Total | 59 (5%) | 58 (50%) | 117 (100%) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mohamed, H.S.; Houmed Aboubaker, M.; Dumont, Y.; Didelot, M.-N.; Michon, A.-L.; Galal, L.; Jean-Pierre, H.; Godreuil, S. Multidrug-Resistant Enterobacterales in Community-Acquired Urinary Tract Infections in Djibouti, Republic of Djibouti. Antibiotics 2022, 11, 1740. https://doi.org/10.3390/antibiotics11121740
Mohamed HS, Houmed Aboubaker M, Dumont Y, Didelot M-N, Michon A-L, Galal L, Jean-Pierre H, Godreuil S. Multidrug-Resistant Enterobacterales in Community-Acquired Urinary Tract Infections in Djibouti, Republic of Djibouti. Antibiotics. 2022; 11(12):1740. https://doi.org/10.3390/antibiotics11121740
Chicago/Turabian StyleMohamed, Hasna Said, Mohamed Houmed Aboubaker, Yann Dumont, Marie-Noëlle Didelot, Anne-Laure Michon, Lokman Galal, Hélène Jean-Pierre, and Sylvain Godreuil. 2022. "Multidrug-Resistant Enterobacterales in Community-Acquired Urinary Tract Infections in Djibouti, Republic of Djibouti" Antibiotics 11, no. 12: 1740. https://doi.org/10.3390/antibiotics11121740
APA StyleMohamed, H. S., Houmed Aboubaker, M., Dumont, Y., Didelot, M.-N., Michon, A.-L., Galal, L., Jean-Pierre, H., & Godreuil, S. (2022). Multidrug-Resistant Enterobacterales in Community-Acquired Urinary Tract Infections in Djibouti, Republic of Djibouti. Antibiotics, 11(12), 1740. https://doi.org/10.3390/antibiotics11121740






