New Insights into Listeria monocytogenes Antimicrobial Resistance, Virulence Attributes and Their Prospective Correlation
Abstract
1. Introduction
2. Results
2.1. Prevalence of L. monocytogenes from Different Sources and Governorates
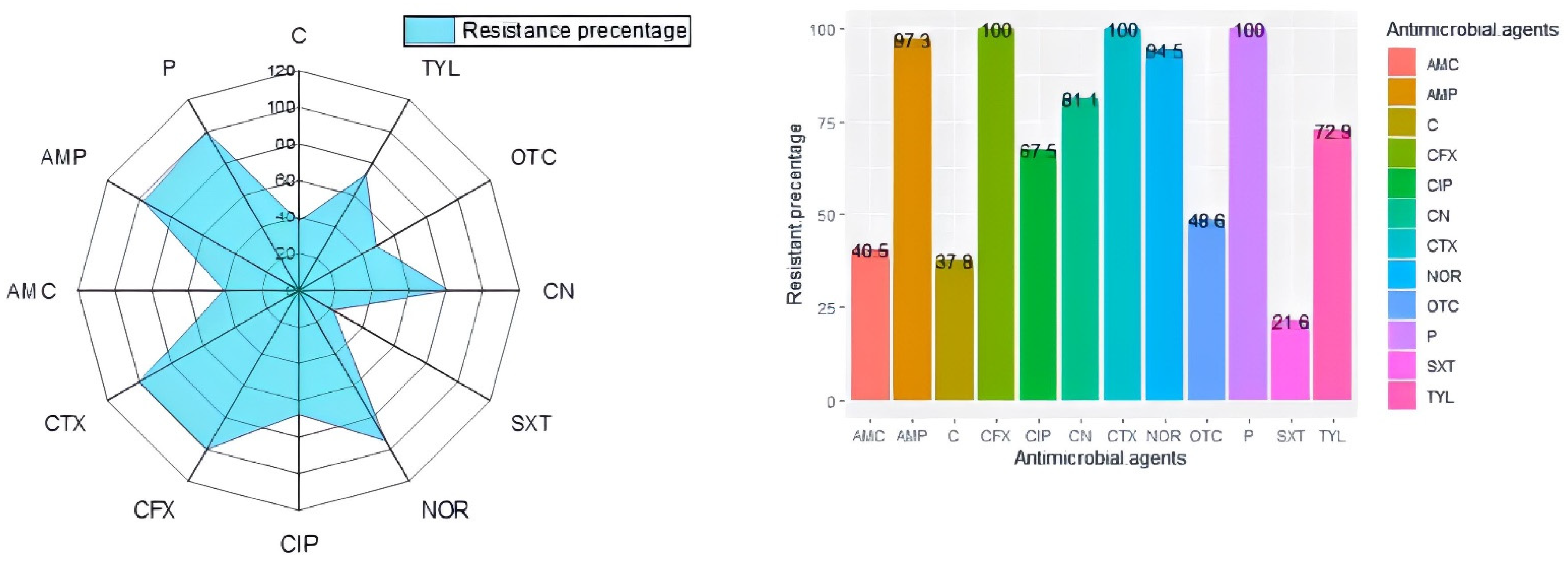
2.2. Antibiogram Typing and Virulence Gene Profiles of L. monocytogenes
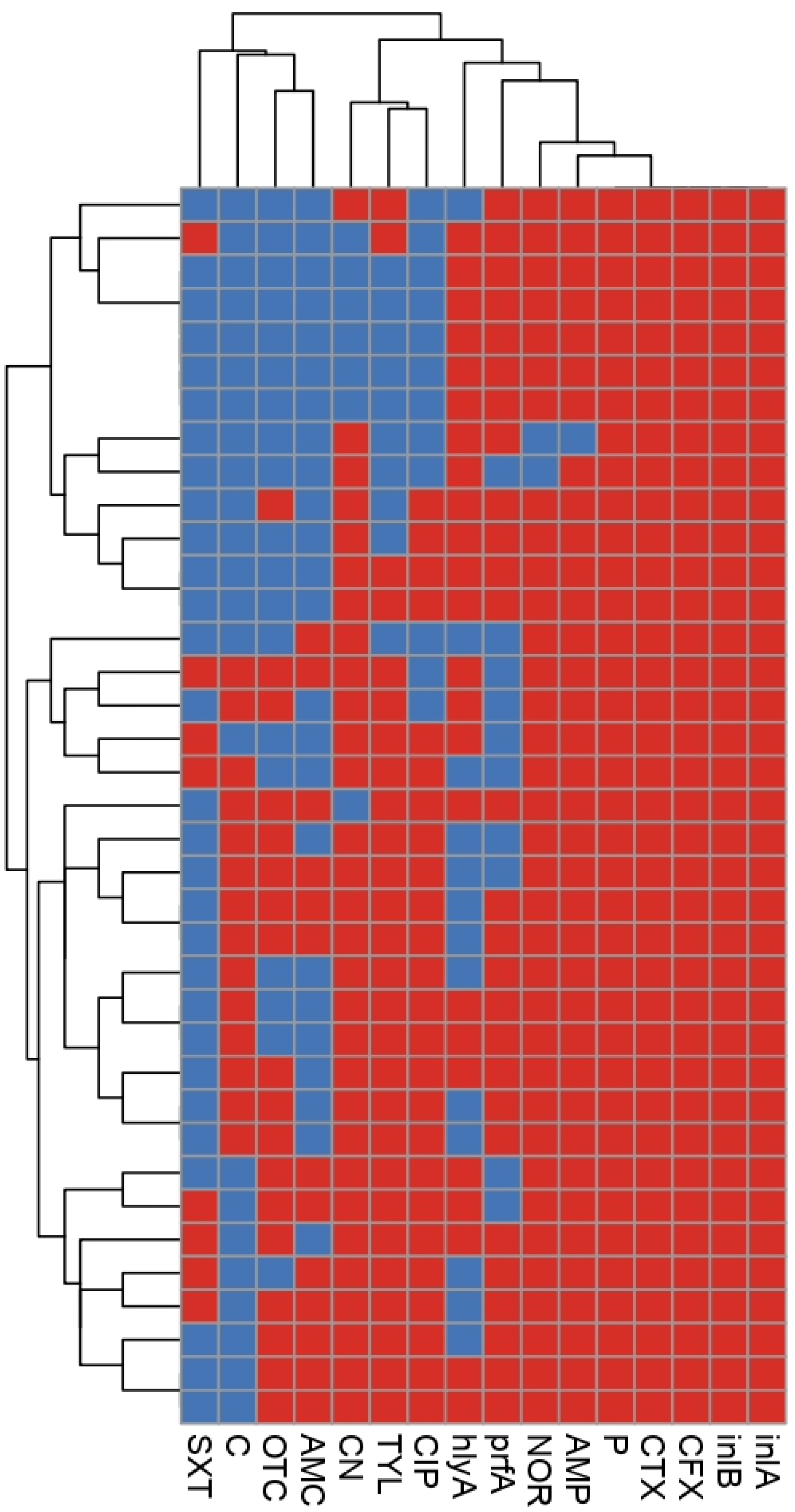
2.3. Phenotypic and Genetic Diversity of L. monocytogenes
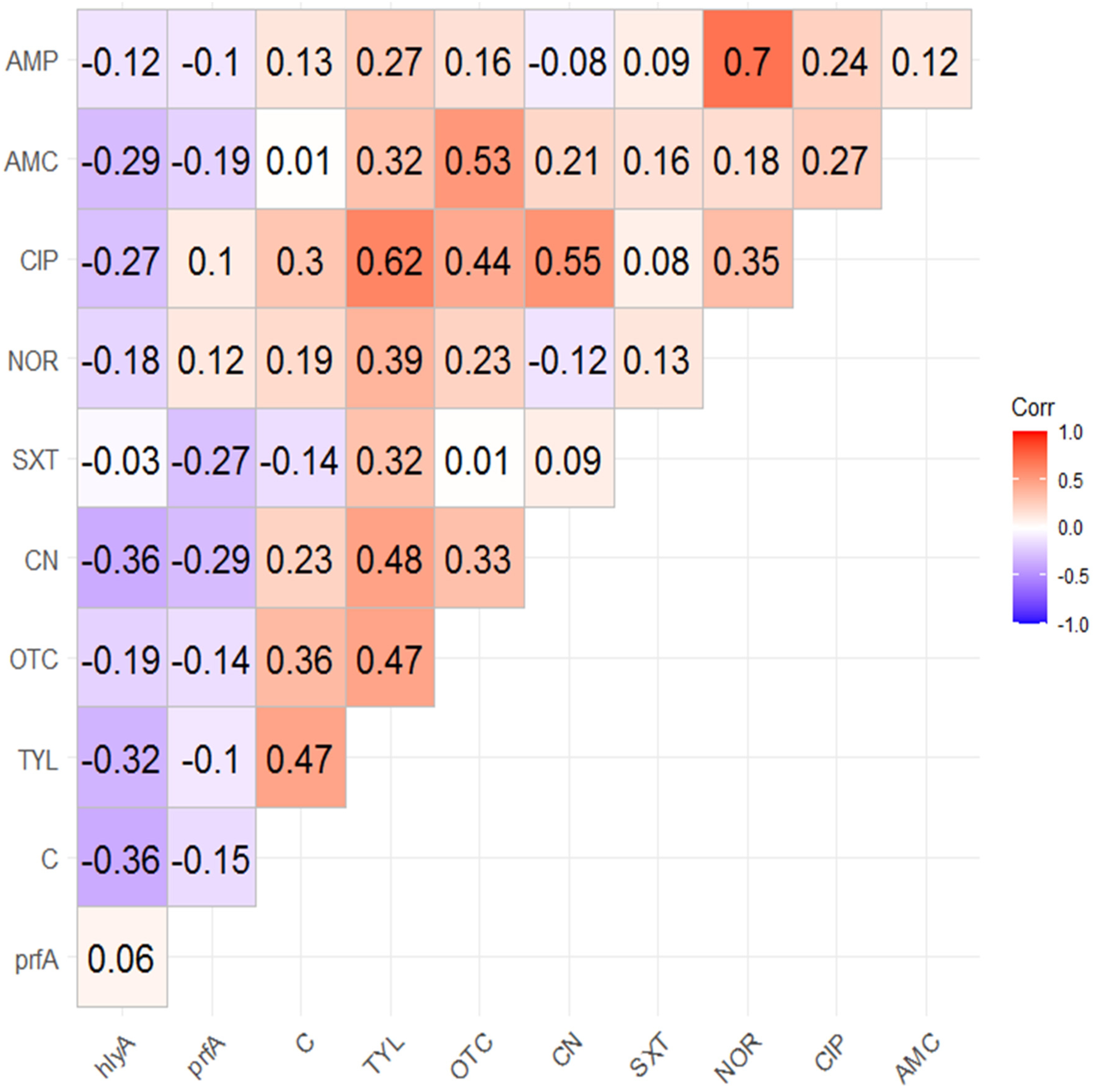
2.4. Correlation Analysis
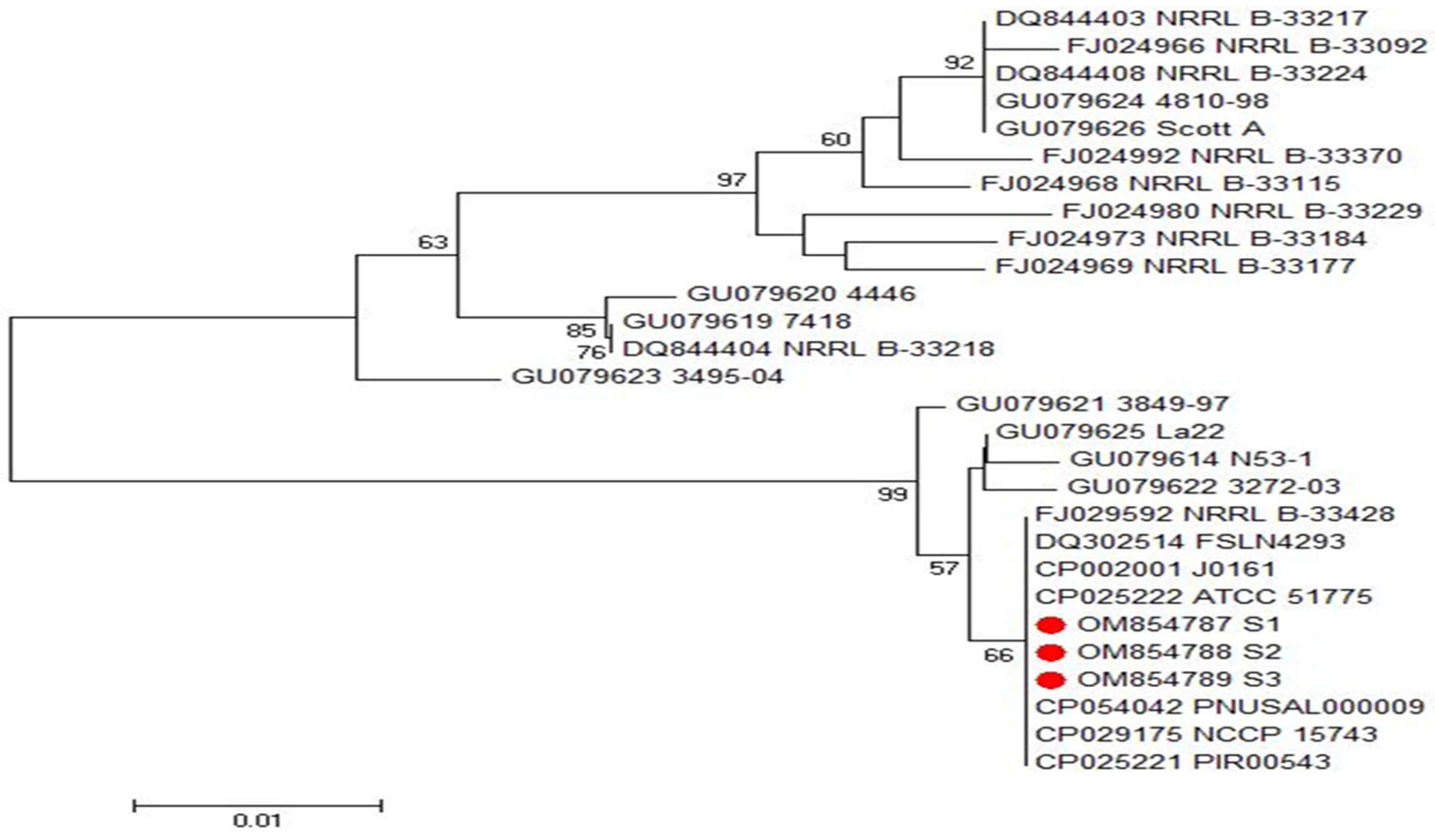
2.5. Phylogenetic Analyses of inlA and inlB Genes of L. monocytogenes
3. Discussion
4. Materials and Methods
4.1. Ethics Statement
4.2. Sampling
4.3. Isolation and Characterization of L. monocytogenes
4.4. Phenotypic Assessment of L. monocytogenes Virulence
4.4.1. Anton’s Test
4.4.2. CAMP (Christie–Atkins–Munch–Peterson) Test
4.4.3. Hemolytic Activity
4.5. Antibiogram Patterns of L. monocytogenes Isolates
4.6. Molecular Detection of L. monocytogenes Virulence Genes
4.7. Sequencing and Phylogenetic Analysis of L. monocytogenes inlA and inlB Genes
4.8. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bintsis, T. Foodborne pathogens. AIMS Microbiol. 2017, 3, 529–563. [Google Scholar] [CrossRef]
- Jalali, M.; Abedi, D. Prevalence of Listeria species in food products in Isfahan, Iran. Int. J. Food Microbiol. 2007, 122, 336–340. [Google Scholar] [CrossRef]
- Berrang, M.E.; Meinersmann, R.J.; Frank, J.F.; Smith, D.P.; Genzlinger, L.L. Distribution of Listeria monocytogenes subtypes within a poultry further processing plant. J. Food Prot. 2005, 68, 980–985. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Faour-Klingbeil, D.; Todd, E. Prevention and Control of Foodborne Diseases in Middle-East North African Countries: Review of National Control Systems. Int. J. Environ. Res. Public Health 2019, 17, 70. [Google Scholar] [CrossRef]
- Abd El-Hamid, M.I.; Sewid, A.H.; Samir, M.; Hegazy, W.; Bahnass, M.M.; Mosbah, R.A.; Ghaith, D.M.; Khalifa, E.; Ramadan, H.; Alshareef, W.A.; et al. Clonal Diversity and Epidemiological Characteristics of ST239-MRSA Strains. Front. Cell. Infect. Microbiol. 2022, 12, 782045. [Google Scholar] [CrossRef] [PubMed]
- Mosallam, F.M.; Helmy, E.A.; Bendary, M.M.; El-Batal, A.I. Potency of a novel synthesized Ag-eugenol nanoemulsion for treating some bacterial and fungal pathogens. J. Mater. Res. 2021, 36, 1524–1537. [Google Scholar] [CrossRef]
- Bendary, M.M.; Abd El-Hamid, M.I.; Alhomrani, M.; Alamri, A.S.; Elshimy, R.; Mosbah, R.A.; Bahnass, M.M.; Omar, N.N.; Al-Sanea, M.M.; Elmanakhly, A.R. What Is behind the Correlation Analysis of Diarrheagenic, E. coli Pathotypes? Biology 2022, 11, 1004. [Google Scholar] [CrossRef]
- Ghaly, M.F.; Nasr, Z.M.; Abousaty, A.I.; Seadawy, H.G.; Shaheen, M.; Albogami, S.; Al-Sanea, M.M.; Bendary, M.M. Alternative and Complementary Therapies against Foodborne Salmonella Infections. Antibiotics 2021, 10, 1453. [Google Scholar] [CrossRef]
- Moretro, T.; Langsrud, S. Listeria monocytogenes: Biofilm formation and persistence in food-processing environments. Biofilms 2004, 1, 107–121. [Google Scholar] [CrossRef]
- Samelis, J.; Metaxopoulos, J. Incidence and principal sources of Listeria spp. and Listeria monocytogenes contamination in processed meats and a meat processing plant. Food Microbiol. 1999, 16, 465–477. [Google Scholar] [CrossRef]
- Feng, Y.; Wu, S.; Varma, J.K.; Klena, J.D.; Angulo, F.J.; Ran, L. Systematic review of human listeriosis in China, 1964–2010. Trop. Med. Int. Health 2013, 18, 1248–1256. [Google Scholar] [CrossRef] [PubMed]
- Flamm, R.K.; Hinrichs, D.J.; Thomashow, M.F. Introduction of pAM beta 1 into Listeria monocytogenes by conjugation and homology between native L. monocytogenes plasmids. Infect. Immun. 1984, 44, 157–161. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Diaz, J.C.; Vicente, M.F.; Baquero, F. Plasmids in Listeria. Plasmid 1982, 8, 112–118. [Google Scholar] [CrossRef]
- Bendary, M.M.; Abd El-Hamid, M.I.; El-Tarabili, R.M.; Hefny, A.A.; Algendy, R.M.; Elzohairy, N.A.; Ghoneim, M.M.; Al-Sanea, M.M.; Nahari, M.H.; Moustafa, W.H. Clostridium perfringens Associated with Foodborne Infections of Animal Origins: Insights into Prevalence, Antimicrobial Resistance, Toxin Genes Profiles, and Toxinotypes. Biology 2022, 11, 551. [Google Scholar] [CrossRef] [PubMed]
- Ammar, A.M.; Abd El-Hamid, M.I.; El-Malt, R.M.S.; Azab, D.S.; Albogami, S.; Al-Sanea, M.M.; Soliman, W.E.; Ghoneim, M.M.; Bendary, M.M. Molecular Detection of Fluoroquinolone Resistance among Multidrug-, Extensively Drug-, and Pan-Drug-Resistant Campylobacter Species in Egypt. Antibiotics 2021, 10, 1342. [Google Scholar] [CrossRef]
- Elfaky, M.A.; Abdel-Hamid, M.I.; Khalifa, E.; Alshareef, W.A.; Mosbah, R.A.; Elazab, S.T.; Ghoneim, M.M.; Al-Sanea, M.M.; Bendary, M.M. Innovative next-generation therapies in combating multi-drug-resistant and multi-virulent Escherichia coli isolates: Insights from in vitro, in vivo, and molecular docking studies. Appl. Microbiol. Biotechnol. 2022, 106, 1691–1703. [Google Scholar] [CrossRef]
- Ghaly, M.F.; Shaheen, A.A.; Bouhy, A.M.; Bendary, M.M. Alternative therapy to manage otitis media caused by multidrug-resistant fungi. Arch. Microbiol. 2020, 202, 1231–1240. [Google Scholar] [CrossRef]
- Pagliano, P.; Ascione, T.; Boccia, G.; Caro, F.D.; Esposito, S. Listeria monocytogenes meningitis in the elderly: Epidemiological, clinical and therapeutic findings. Infez Med. 2016, 24, 105–111. [Google Scholar]
- Abdeen, E.E.; Mousa, W.S.; Harb, O.H.; Fath-Elbab, G.A.; Nooruzzaman, M.; Gaber, A.; Alsanie, W.F.; Abdeen, A. Prevalence, Antibiogram and Genetic Characterization of Listeria monocytogenes from Food Products in Egypt. Foods 2021, 10, 1381. [Google Scholar] [CrossRef]
- Osman, K.M.; Kappell, A.D.; Fox, E.M.; Orabi, A.; Samir, A. Prevalence, pathogenicity, virulence, antibiotic resistance, and phylogenetic analysis of biofilm-producing L. monocytogenes isolated from different ecological niches in Egypt. food, humans, animals, and environment. Pathogens 2020, 9, 5. [Google Scholar] [CrossRef]
- Gartley, S.; Anderson-Coughlin, B.; Sharma, M.; Kniel, K.E. Listeria monocytogenes in Irrigation Water: An Assessment of Outbreaks, Sources, Prevalence, and Persistence. Microorganisms 2022, 10, 1319. [Google Scholar] [CrossRef] [PubMed]
- Szczawiński, J.; Ewa Szczawińska, M.; Łobacz, A.; Tracz, M.; Jackowska-Tracz, A. Modelling the Growth Rate of Listeria Monocytogenes in Cooked Ham Stored at Different Temperatures. J. Vet. Res. 2017, 61, 45–51. [Google Scholar] [CrossRef]
- Aureli, P.; Ferrini, A.M.; Mannoni, V.; Hodzic, S.; Wedell-Weergaard, C.; Oliva, B. Susceptibility of Listeria monocytogenes isolated from food in Italy to antibiotics. Int. J. Food Microbiol. 2003, 83, 325–330. [Google Scholar] [CrossRef]
- Charpentier, E.; Gerbaud, G.; Jacquet, C.; Rocourt, J.; Courvalin, P. Incidence of antibiotic resistance in Listeria species. J. Infect. Dis. 1995, 172, 277–281. [Google Scholar] [CrossRef]
- Ahmed, H.A.; Tahoun, A.B.M.B.; Abou Elez, R.M.M.; Abd El-Hamid, M.I.; Abd Ellatif, S.S. Prevalence of Yersinia enterocolitica in milk and dairy products and the effects of storage temperatures on survival and virulence gene expression. Int. Dairy J. 2019, 94, 16–21. [Google Scholar] [CrossRef]
- Conter, M.; Paludi, D.; Zanardi, E.; Ghidini, S.; Vergara, A.; Ianieri, A. Characterization of antimicrobial resistance of foodborne Listeria monocytogenes. Int. J. Food Microbiol. 2009, 128, 497–500. [Google Scholar] [CrossRef]
- Ammar, A.M.; El-Naenaeey, E.-S.Y.; El-Malt, R.M.S.; El-Gedawy, A.A.; Khalifa, E.; Elnahriry, S.S.; Abd El-Hamid, M.I. Prevalence, antimicrobial susceptibility, virulence and genotyping of Campylobacter jejuni with a special reference to the anti-virulence potential of eugenol and beta-resorcylic acid on some multi-drug resistant isolates in Egypt. Animals 2021, 11, 3. [Google Scholar] [CrossRef]
- Hansen, J.M.; Gerner-Smidt, P.; Bruun, B. Antibiotic susceptibility of Listeria monocytogenes in Denmark 1958–2001. APMIS 2005, 113, 31–36. [Google Scholar] [CrossRef]
- Elsayed, M.M.; Elkenany, R.M.; Zakaria, A.I.; Badawy, B.M. Epidemiological study on Listeria monocytogenes in Egyptian dairy cattle farms’ insights into genetic diversity of multi-antibiotic-resistant strains by ERIC-PCR. Environ. Sci. Pollut. Res. 2022, 29, 54359–54377. [Google Scholar] [CrossRef]
- Akrami-Mohajeri, F.; Derakhshan, Z.; Ferrante, M.; Hamidiyan, N.; Soleymani, M.; Conti, G.O.; Tafti, R.D. The prevalence and antimicrobial resistance of Listeria spp in raw milk and traditional dairy products delivered in Yazd, Central Iran (2016). Food Chem. Toxicol. 2018, 114, 141–144. [Google Scholar] [CrossRef]
- Heger, W.; Dierich, M.P.; Allerberger, F. In vitro susceptibility of Listeria monocytogenes: Comparison of the E test with the agar dilution test. Chemotherapy 1997, 43, 303–310. [Google Scholar] [CrossRef] [PubMed]
- Poyart-Salmeron, C.; Trieu-Cuot, P.; Carlier, C.; Courvalin, P. Molecular characterization of two proteins involved in the excision of the conjugative transposon Tn1545: Homologies with other site-specific recombinases. EMBO J. 1989, 8, 2425–2433. [Google Scholar] [CrossRef]
- Godreuil, S.; Galimand, M.; Gerbaud, G.; Jacquet, C.; Courvalin, P. Efflux pump Lde is associated with fluoroquinolone resistance in Listeria monocytogenes. Antimicrob. Agents Chemother. 2003, 47, 704–708. [Google Scholar] [CrossRef]
- Charpentier, E.; Courvalin, P. Antibiotic resistance in Listeria spp. Antimicrob. Agents Chemother. 1999, 43, 2103–2108. [Google Scholar] [CrossRef] [PubMed]
- Hof, H. An update on the medical management of listeriosis. Expert Opin. Pharmacother. 2004, 5, 1727–1735. [Google Scholar] [CrossRef] [PubMed]
- Temple, M.E.; Nahata, M.C. Treatment of listeriosis. Ann. Pharmacother. 2000, 34, 656–661. [Google Scholar] [CrossRef]
- Vázquez-Boland, J.A.; Kuhn, M.; Berche, P.; Chakraborty, T.; Domínguez-Bernal, G.; Goebel, W.; González-Zorn, B.; Wehland, J.; Kreft, J. Listeria pathogenesis and molecular virulence determinants. Clin. Microbiol. Rev. 2001, 14, 584–640. [Google Scholar] [CrossRef]
- Quereda, J.J.; Morón-García, A.; Palacios-Gorba, C.; Dessaux, C.; García-Del, P.F.; Pucciarelli, M.G.; Ortega, A.D. Pathogenicity and virulence of Listeria monocytogenes: A trip from environmental to medical microbiology. Virulence 2021, 12, 2509–2545. [Google Scholar] [CrossRef]
- Berche, P.; Gaillard, J.L.; Sansonetti, P.J. Intracellular growth of Listeria monocytogenes as a prerequisite for in vivo induction of T cell-mediated immunity. J. Immunol. 1987, 138, 2266–2271. [Google Scholar]
- Busch, D.H.; Kerksiek, K.; Pamer, E.G. Processing of Listeria monocytogenes antigens and the in vivo T-cell response to bacterial infection. Immunol. Rev. 1999, 172, 163–169. [Google Scholar] [CrossRef]
- Braun, L.; Ghebrehiwet, B.; Cossart, P. gC1q-R/p32, a C1q-binding protein, is a receptor for the InlB invasion protein of Listeria monocytogenes. EMBO J. 2000, 19, 1458–1466. [Google Scholar] [CrossRef] [PubMed]
- Mengaud, J.; Ohayon, H.; Gounon, P.; Mege, R.M.; Cossart, P. E-Cadherin is the receptor for internalin, a surface protein required for entry of L. monocytogenes into epithelial cells. Cell 1996, 84, 923–932. [Google Scholar] [CrossRef]
- Morvan, A.; Moubareck, C.; Leclercq, A.; Hervé-Bazin, M.; Bremont, S.; Lecuit, M.; Courvalin, P.; Le Monnier, A. Antimicrobial resistance of Listeria monocytogenes strains isolated from humans in France. Antimicrob. Agents Chemother. 2010, 54, 2728–2731. [Google Scholar] [CrossRef] [PubMed]
- Scortti, M.; Han, L.; Alvarez, S.; Leclercq, A.; Moura, A.; Lecuit, M.; Vazquez-Boland, J. Epistatic control of intrinsic resistance by virulence genes in Listeria. PLoS Genet. 2018, 14, e1007525. [Google Scholar] [CrossRef] [PubMed]
- Beceiro, A.; Tomás, M.; Bou, G. Antimicrobial resistance and virulence: A successful or deleterious association in the bacterial world? Clin. Microbiol. Rev. 2013, 26, 185–230. [Google Scholar] [CrossRef]
- Bendary, M.M.; Solyman, S.M.; Azab, M.M.; Mahmoud, N.F.; Hanora, A.M. Genetic diversity of multidrug resistant Staphylococcus aureus isolated from clinical and non-clinical samples in Egypt. Cell. Mol. Biol. 2016, 62, 55–61. [Google Scholar]
- Bendary, M.M.; Solyman, S.M.; Azab, M.M.; Mahmoud, N.F.; Hanora, A.M. Characterization of Methicillin Resistant Staphylococcus aureus isolated from human and animal samples in Egypt. Cell. Mol. Biol. 2016, 62, 94–100. [Google Scholar]
- Diard, M.; Hardt, W.D. Evolution of bacterial virulence. FEMS Microbiol. Rev. 2017, 41, 679–697. [Google Scholar] [CrossRef]
- Chen, Y.; Chen, M.; Wang, J.; Wu, Q.; Cheng, J.; Zhang, J.; Sun, Q.; Xue, L.; Zeng, H.; Lei, T.; et al. Heterogeneity, Characteristics, and Public Health Implications of Listeria monocytogenes in Ready-to-Eat Foods and Pasteurized Milk in China. Front. Microbiol. 2020, 15, 642. [Google Scholar] [CrossRef]
- Nyarko, E.B.; Donnelly, C.W. Listeria monocytogenes: Strain Heterogeneity, Methods, and Challenges of Subtyping. J. Food Sci. 2015, 80, 2868–2878. [Google Scholar] [CrossRef]
- Hitchins, A.D.; Whiting, R.C. Food-borne Listeria monocytogenes risk assessment. Food Addit. Contam. 2001, 18, 1108–1117. [Google Scholar] [CrossRef] [PubMed]
- Markey, B.K.; Leonard, F.C.; Archambault, M.; Cullinane, A.; Maguire, D. Clinical Veterinary Microbiology, 2nd ed.; MOSBY. Elsevier Ltd.: Edinburgh, UK; London, UK; New York, NY, USA; Oxford, UK; Philadelphia, PA, USA; St. Louis, MI, USA; Sydney, Australia; Toronto, ON, Canada, 2013. [Google Scholar]
- Kumar, A.; Grover, S.; Batish, V.K. Exploring specific primers targeted against different genes for a multiplex PCR for detection of Listeria monocytogenes. 3 Biotech 2015, 5, 261–269. [Google Scholar] [CrossRef] [PubMed]
- Quinn, P.J.; Markey, B.K.; Carter, M.E.; Donnelly, W.J.C.; Leonard, F.C.; Maguire, D. Veterinary Microbiology and Microbial Disease; Blackwell Science: Oxford, UK, 2002; pp. 84–96. [Google Scholar]
- ISO 11290-1:1996; Microbiology of Food and Animal Feeding Stuffs–Horizontal Method for the Detection and Enumeration of Listeria Monocytogenes–Part 1: Detection Method. 1st ed. ISO: Geneva, Switzerland, 1996.
- Abd El-Hamid, M.I.; Ibrahim, S.M.; Eldemery, F.; El-Mandrawy, S.A.M.; Metwally, A.S.; Khalifa, E.; Elnahriry, S.S.; Ibrahim, D. Dietary cinnamaldehyde nanoemulsion boosts growth and transcriptomes of antioxidant and immune related genes to fight Streptococcus agalactiae infection in Nile tilapia (Oreochromis niloticus). Fish Shellfish Immunol. 2021, 113, 96–105. [Google Scholar] [CrossRef] [PubMed]
- Fujisawa, T.; Mori, M. Evaluation of media for determining hemolytic activity and that of API Listeria system for identifying strains of Listeria monocytogenes. J. Clin. Microbiol. 1994, 32, 1127–1129. [Google Scholar] [CrossRef]
- The European Committee on Antimicrobial Susceptibility Testing (ECoAS). Breakpoint Tables for Interpretation of MICs and Zone Diameters. 2018, Version 8.0. 2018. Available online: http://www.sciepub.com/reference/368572 (accessed on 1 October 2022).
- Magiorakos, A.P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef] [PubMed]
- Krumperman, P.H. Multiple antibiotic resistance indexing of Escherichia coli to identify high-risk sources of fecal contamination of foods. Appl. Environ. Microbiol. 1983, 46, 165–170. [Google Scholar] [CrossRef]
- Dickinson, J.H.; Kroll, R.G.; Grant, K.A. The direct application of the polymerase chain reaction to DNA extracted from foods. Lett. Appl. Microbiol. 1995, 20, 212–216. [Google Scholar] [CrossRef]
- Liu, D.; Lawrence, M.L.; Austin, F.W.; Ainsworth, A.J. A multiplex PCR for species- and virulence-specific determination of Listeria monocytogenes. J. Microbiol. Methods 2007, 71, 133–140. [Google Scholar] [CrossRef]
- Deneer, H.G.; Boychuk, I. Species-Specific Detection of Listeria monocytogenes by DNA Amplification. Appl. Environ. Microbiol. 1991, 57, 606–609. [Google Scholar] [CrossRef]
- Hunter, P.R.; Gaston, M.A. Numerical index of the discriminatory ability of typing systems: An application of Simpson’s index of diversity. J. Clin. Microbiol. 1988, 26, 2465–2466. [Google Scholar] [CrossRef]


| Target Gene | Oligonucleotide Primer Sequence (5′-3′) | Amplified Product (bp) | Reference |
|---|---|---|---|
| 16S rRNA | F: GGACCGGGGCTAATACCGAATGAT AA R: TTC ATGTAGGCGAGTTGCAGC CTA | 1200 | [53] |
| prfA | F: TCTCCGAGCAACCTCGGAACC R: TGGATTGACAAAATGGAACA | 1052 | [61] |
| inlA | F: ACG AGT AAC GGG ACA AAT GC R: CCC GAC AGT GGT GCT AGA TT | 800 | [62] |
| inlB | F: CTGGAAAGTTTGTATTTGGGAAA R: TTTCATAATCGCCATCATCACT | 343 | [62] |
| hlyA | F: GCATCTGCATTCAATAAAGA R: TGTCACTGCATCTCCGTGGT | 174 | [63] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Elsayed, M.E.; Abd El-Hamid, M.I.; El-Gedawy, A.; Bendary, M.M.; ELTarabili, R.M.; Alhomrani, M.; Alamri, A.S.; Alghamdi, S.A.; Arnout, M.; Binjawhar, D.N.; et al. New Insights into Listeria monocytogenes Antimicrobial Resistance, Virulence Attributes and Their Prospective Correlation. Antibiotics 2022, 11, 1447. https://doi.org/10.3390/antibiotics11101447
Elsayed ME, Abd El-Hamid MI, El-Gedawy A, Bendary MM, ELTarabili RM, Alhomrani M, Alamri AS, Alghamdi SA, Arnout M, Binjawhar DN, et al. New Insights into Listeria monocytogenes Antimicrobial Resistance, Virulence Attributes and Their Prospective Correlation. Antibiotics. 2022; 11(10):1447. https://doi.org/10.3390/antibiotics11101447
Chicago/Turabian StyleElsayed, Mahmoud E., Marwa I. Abd El-Hamid, Attia El-Gedawy, Mahmoud M. Bendary, Reham M. ELTarabili, Majid Alhomrani, Abdulhakeem S. Alamri, Saleh A. Alghamdi, Marwa Arnout, Dalal N. Binjawhar, and et al. 2022. "New Insights into Listeria monocytogenes Antimicrobial Resistance, Virulence Attributes and Their Prospective Correlation" Antibiotics 11, no. 10: 1447. https://doi.org/10.3390/antibiotics11101447
APA StyleElsayed, M. E., Abd El-Hamid, M. I., El-Gedawy, A., Bendary, M. M., ELTarabili, R. M., Alhomrani, M., Alamri, A. S., Alghamdi, S. A., Arnout, M., Binjawhar, D. N., Al-Sanea, M. M., & Abousaty, A. I. (2022). New Insights into Listeria monocytogenes Antimicrobial Resistance, Virulence Attributes and Their Prospective Correlation. Antibiotics, 11(10), 1447. https://doi.org/10.3390/antibiotics11101447







