Dissemination of Carbapenemases and MCR-1 Producing Gram-Negative Bacteria in Aquatic Environments in Batna, Algeria
Abstract
1. Introduction
2. Results
2.1. Identification
2.1.1. Tap Water and Well Water
2.1.2. Wastewater
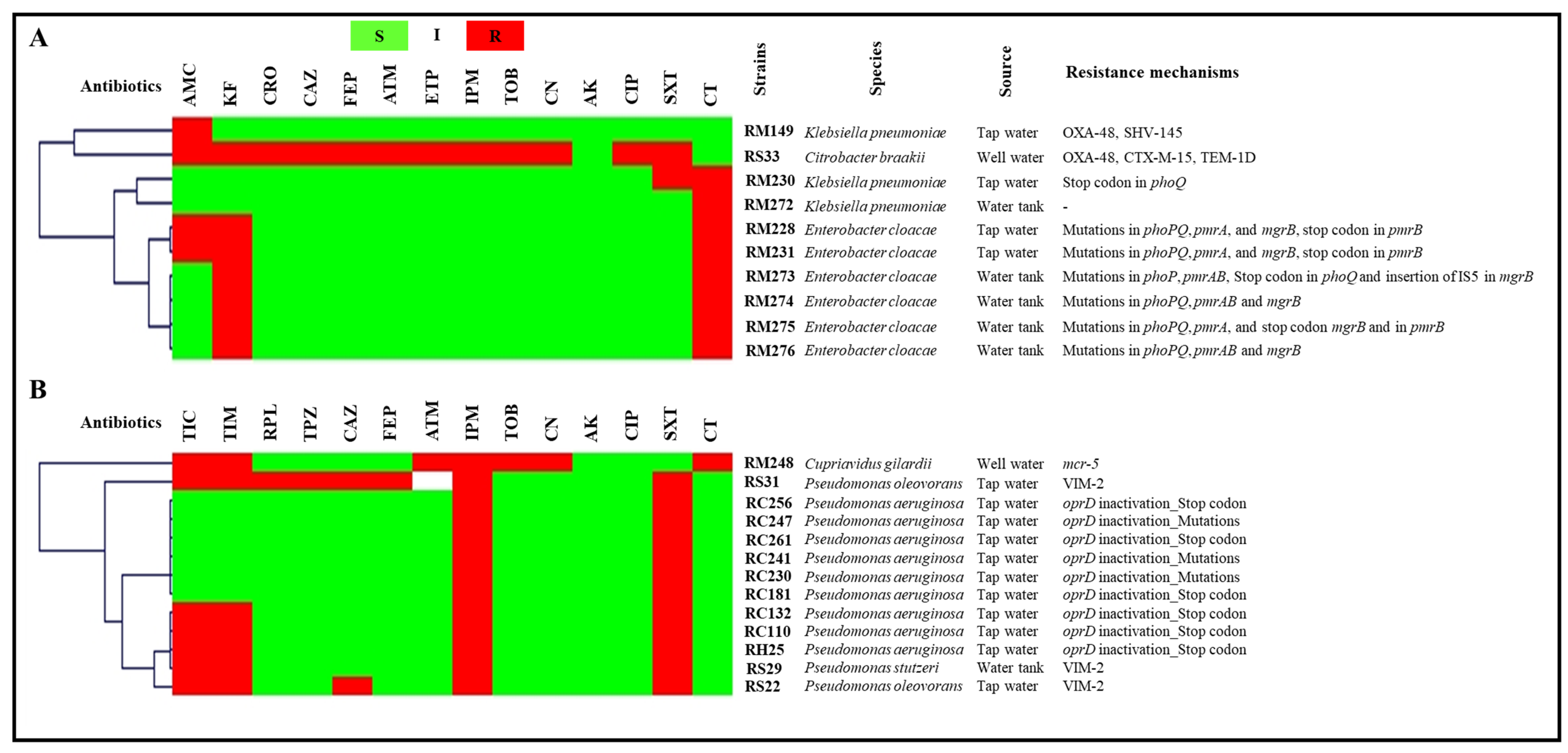
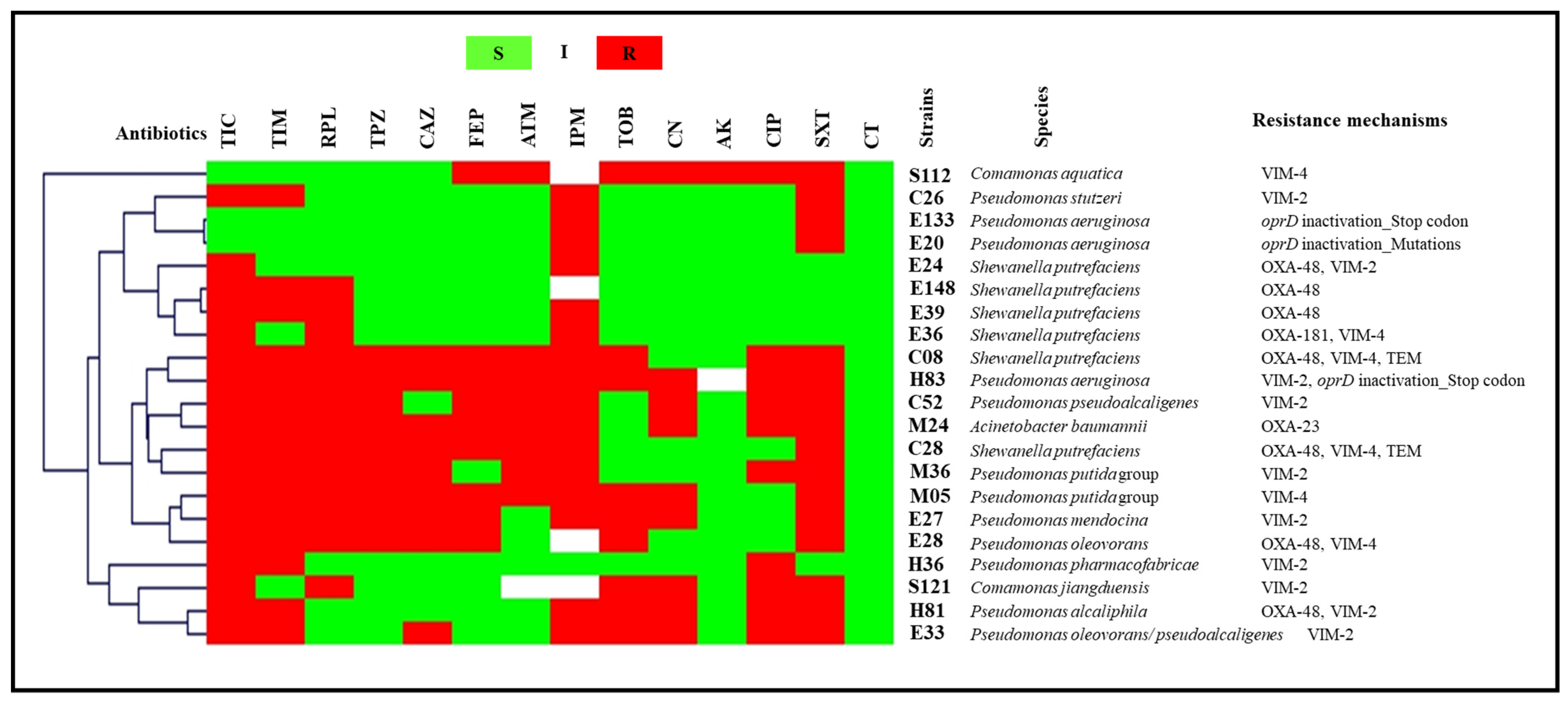
2.2. Antibiotic Susceptibility Testing
2.2.1. Tap Water and Well Water
2.2.2. Wastewater
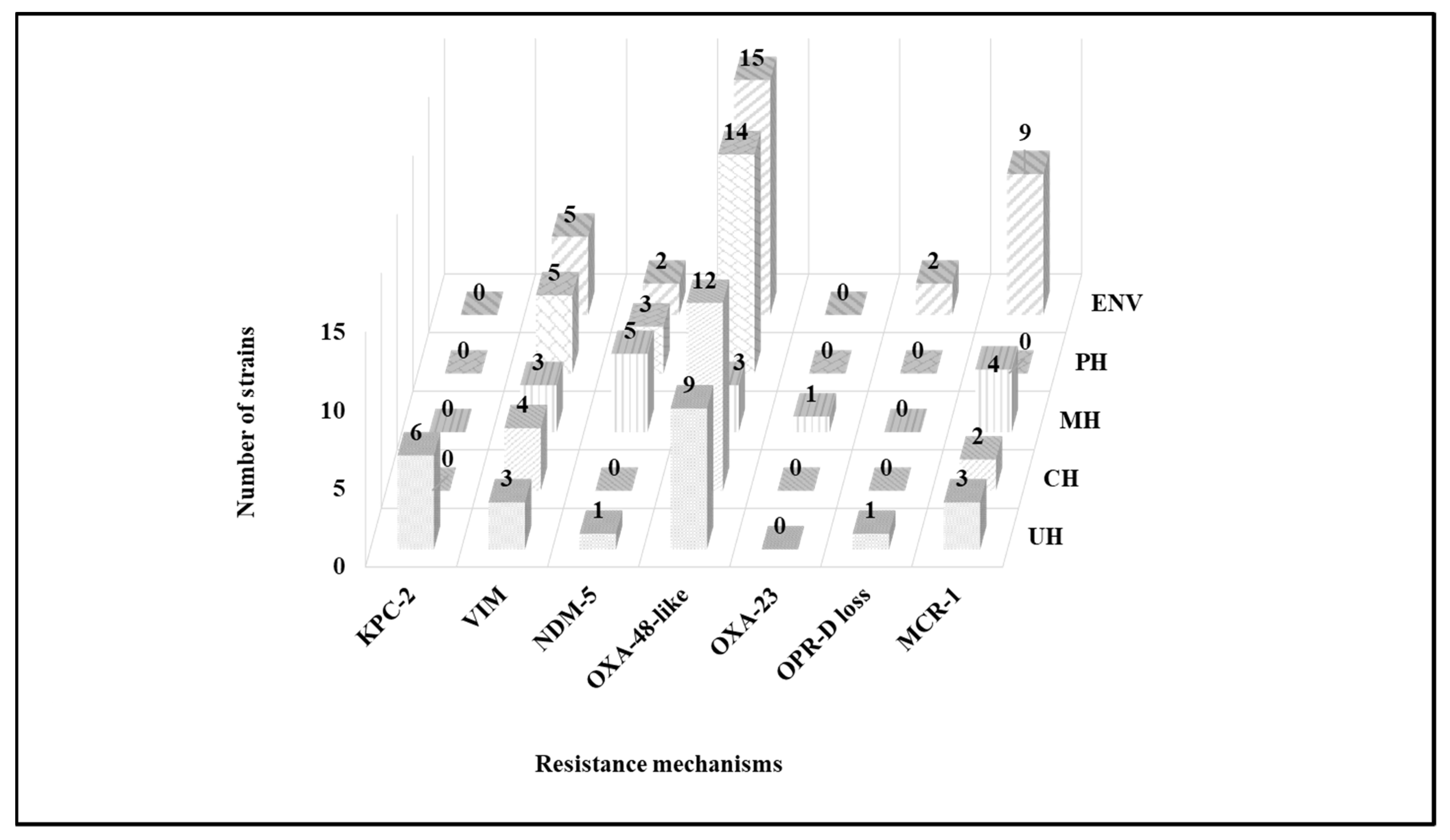
2.3. Molecular Characterization of Antibiotic Resistance Mechanisms
2.3.1. Tap Water and Well Water
2.3.2. Wastewater
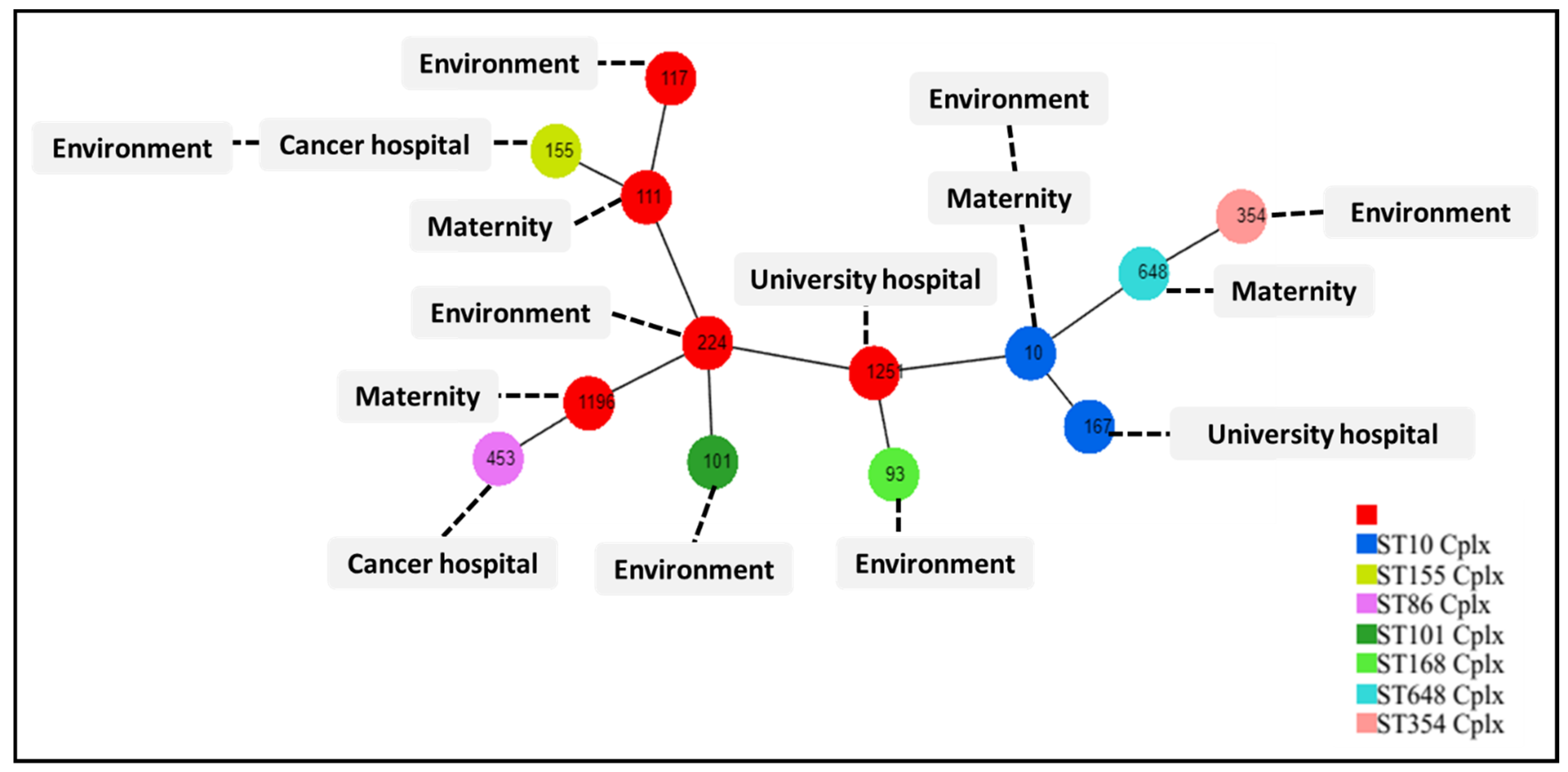
2.4. Multilocus Sequence Typing
3. Discussion
4. Materials and Methods
4.1. Sampling
4.2. Samples’ Processing and Bacterial Isolation
4.2.1. Wastewater
4.2.2. Well Water and Tap Water
4.3. Bacterial Identification
4.4. Antibiotic Susceptibility Testing and Phenotypic Detection of β-Lactamase Production
4.5. Molecular Characterization of β-Lactam- and Colistin Resistance Mechanisms
4.6. Multilocus Sequence Typing
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Maestre-Carballa, L.; Gomez, M.L.; Navarro, A.A.; Garcia-Heredia, I.; Martinez-Hernandez, F.; Martinez-Garcia, M. Insights into the antibiotic resistance dissemination in a wastewater effluent microbiome: Bacteria, viruses and vesicles matter. Environ. Microbiol. 2019, 21, 4582–4596. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Wei, W.; Huang, M.; Umar, Z.; Feng, Y. Definition of a Family of Nonmobile Colistin Resistance (NMCR-1) Determinants Suggests Aquatic Reservoirs for MCR-4. Adv. Sci. 2019, 6, 1900038. [Google Scholar] [CrossRef] [PubMed]
- Kelly, A.M.; Mathema, B.; Larson, E.L. Carbapenem-resistant Enterobacteriaceae in the community: A scoping review. Int. J. Antimicrob. Agents 2017, 50, 127–134. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Global Action Plan on Antimicrobial Resistance. 2015. Available online: http://www.wpro.who.int/entity/drug_resistance/resources/global_action_plan_eng.pdf (accessed on 15 December 2019).
- Baquero, F.; Martinez, J.L.; Cantón, R. Antibiotics and antibiotic resistance in water environments. Curr. Opin. Biotechnol. 2008, 19, 260–265. [Google Scholar] [CrossRef] [PubMed]
- Manaia, C.M.; Macedo, G.; Fatta-Kassinos, D.; Nunes, O.C. Antibiotic resistance in urban aquatic environments: Can it be controlled? Appl. Microbiol. Biotechnol. 2016, 100, 1543–1557. [Google Scholar] [CrossRef] [PubMed]
- Cherak, Z.; Loucif, L.; Moussi, A.; Rolain, J.-M. Epidemiology of mobile colistin resistance (mcr) genes in aquatic environments. J. Glob. Antimicrob. Resist. 2021, 27, 51–62. [Google Scholar] [CrossRef] [PubMed]
- Caniaux, I.; Van Belkum, A.; Zambardi, G.; Poirel, L.; Gros, M.F. MCR: Modern colistin resistance. Eur. J. Clin. Microbiol. Infect. Dis. 2017, 36, 415–420. [Google Scholar] [CrossRef]
- Diene, S.M.; Rolain, J.M. Carbapenemase genes and genetic platforms in Gram-negative bacilli: Enterobacteriaceae, Pseudomonas and Acinetobacter species. Clin. Microbiol. Infect. 2014, 20, 831–838. [Google Scholar] [CrossRef] [PubMed]
- Iovleva, A.; Doi, Y. Carbapenem-Resistant Enterobacteriaceae. Clin. Lab. Med. 2017, 37, 303–315. [Google Scholar] [CrossRef] [PubMed]
- Baron, S.; Hadjadj, L.; Rolain, J.-M.; Olaitan, A.O. Molecular mechanisms of polymyxin resistance: Knowns and unknowns. Int. J. Antimicrob. Agents 2016, 48, 583–591. [Google Scholar] [CrossRef] [PubMed]
- Cherak, Z.; Loucif, L.; Ben Khedher, M.; Moussi, A.; Benbouza, A.; Baron, S.A.; Rolain, J.-M. MCR-5-Producing Colistin-Resistant Cupriavidus gilardii Strain from Well Water in Batna, Algeria. mSphere 2021, 6, e0057521. [Google Scholar] [CrossRef] [PubMed]
- Hernando-Amado, S.; Coque, T.M.; Baquero, F.; Martínez, J.L. Defining and combating antibiotic resistance from One Health and Global Health perspectives. Nat. Microbiol. 2019, 4, 1432–1442. [Google Scholar] [CrossRef]
- Cherak, Z.; Loucif, L.; Moussi, A.; Rolain, J.-M. Carbapenemase-producing Gram-negative bacteria in aquatic environments: A review. J. Glob. Antimicrob. Resist. 2021, 25, 287–309. [Google Scholar] [CrossRef]
- Huijbers, P.M.; Flach, C.-F.; Larsson, D.J. A conceptual framework for the environmental surveillance of antibiotics and antibiotic resistance. Environ. Int. 2019, 130, 104880. [Google Scholar] [CrossRef]
- Poirel, L.; Potron, A.; Nordmann, P. OXA-48-like carbapenemases: The phantom menace. J. Antimicrob. Chemother. 2012, 67, 1597–1606. [Google Scholar] [CrossRef] [PubMed]
- Touati, A.; Mairi, A. Carbapenemase-Producing Enterobacterales in Algeria: A Systematic Review. Microb. Drug Resist. 2019, 26, 475–482. [Google Scholar] [CrossRef]
- Loucif, L.; Kassah-Laouar, A.; Saidi, M.; Messala, A.; Chelaghma, W.; Rolain, J.-M. Outbreak of OXA-48-Producing Klebsiella pneumoniae Involving a Sequence Type 101 Clone in Batna University Hospital, Algeria. Antimicrob. Agents Chemother. 2016, 60, 7494–7497. [Google Scholar] [CrossRef] [PubMed]
- Loucif, L.; Cherak, Z.; Chamlal, N.; Bendjama, E.; Gacemi-Kirane, D.; Grainat, N.; Rolain, J.-M. First Detection of VIM-2 Metallo-β-Lactamase-Producing Pseudomonas putida in Blattella germanica Cockroaches in an Algerian Hospital. Antimicrob. Agents Chemother. 2017, 61, e00357–e00417. [Google Scholar] [CrossRef]
- Loucif, L.; Gacemi-Kirane, D.; Cherak, Z.; Chamlal, N.; Grainat, N.; Rolain, J.-M. First Report of German Cockroaches (Blattella germanica) as Reservoirs of CTX-M-15 Extended-Spectrum-β-Lactamase- and OXA-48 Carbapenemase-Producing Enterobacteriaceae in Batna University Hospital, Algeria. Antimicrob. Agents Chemother. 2016, 60, 6377–6380. [Google Scholar] [CrossRef]
- Loucif, L.; Chelaghma, W.; Helis, Y.; Sebaa, F.; Baoune, R.D.; Zaatout, W.; Rolain, J.-M. First detection of OXA-48-producing Klebsiella pneumoniae in community-acquired urinary tract infection in Algeria. J. Glob. Antimicrob. Resist. 2018, 12, 115–116. [Google Scholar] [CrossRef] [PubMed]
- Bouaziz, A.; Loucif, L.; Ayachi, A.; Guehaz, K.; Bendjama, E.; Rolain, J.-M. Migratory White Stork (Ciconia ciconia): A Potential Vector of the OXA-48-Producing Escherichia coli ST38 Clone in Algeria. Microb. Drug Resist. 2018, 24, 461–468. [Google Scholar] [CrossRef] [PubMed]
- Loucif, L.; Chelaghma, W.; Cherak, Z.; Bendjama, E.; Beroual, F.; Rolain, J.-M. Detection of NDM-5 and MCR-1 antibiotic resistance encoding genes in Enterobacterales in long-distance migratory bird species Ciconia ciconia, Algeria. Sci. Total Environ. 2022, 814, 152861. [Google Scholar] [CrossRef] [PubMed]
- Bendjama, E.; Loucif, L.; Chelaghma, W.; Attal, C.; Bellakh, F.Z.; Benaldjia, R.; Kahlat, I.; Meddour, A.; Rolain, J.-M. First detection of an OXA-48-producing Enterobacter cloacae isolate from currency coins in Algeria. J. Glob. Antimicrob. Resist. 2020, 23, 162–166. [Google Scholar] [CrossRef] [PubMed]
- Loucif, L.; Chelaghma, W.; Bendjama, E.; Bouaziz, A.; Cherak, Z.; Serrar, W.; Mohammed, F.S.; Rolain, J.-M. Urban pigeons as a reservoir of carbapenem resistant Enterobacterales: First report of OXA-48-producing Klebsiella pneumoniae. New Microbes New Infect. 2022, 47, 100981. [Google Scholar] [CrossRef] [PubMed]
- Loucif, L.; Chelaghma, W.; Bendjama, E.; Cherak, Z.; Khellaf, M.; Khemri, A.; Rolain, J.-M. Detection of bla(OXA-48) and mcr-1 Genes in Escherichia coli Isolates from Pigeon (Columba livia) in Algeria. Microorganisms 2022, 10, 975. [Google Scholar] [CrossRef]
- Chelaghma, W.; Loucif, L.; Bendjama, E.; Cherak, Z.; Bendahou, M.; Rolain, J.-M. Occurrence of Extended Spectrum Cephalosporin-, Carbapenem- and Colistin-Resistant Gram-Negative Bacteria in Fresh Vegetables, an Increasing Human Health Concern in Algeria. Antibiotics 2022, 11, 988. [Google Scholar] [CrossRef]
- Cherak, Z.; Loucif, L.; Moussi, A.; Bendjama, E.; Benbouza, A.; Rolain, J.-M. Emergence of Metallo-β-Lactamases and OXA-48 Carbapenemase Producing Gram-Negative Bacteria in Hospital Wastewater in Algeria: A Potential Dissemination Pathway into the Environment. Microb. Drug Resist. 2021, 28, 23–30. [Google Scholar] [CrossRef]
- Merradi, M.; Kassah-Laouar, A.; Ayachi, A.; Heleili, N.; Menasria, T.; Hocquet, D.; Cholley, P.; Sauget, M. Occurrence of VIM-4 metallo-β-lactamase-producing Pseudomonas aeruginosa in an Algerian hospital. J. Infect. Dev. Ctries. 2019, 13, 284–290. [Google Scholar] [CrossRef]
- Shen, Y.; Wu, Z.; Wang, Y.; Zhang, R.; Zhou, H.-W.; Wang, S.; Lei, L.; Li, M.; Cai, J.; Tyrrell, J.; et al. Heterogeneous and Flexible Transmission of mcr-1 in Hospital-Associated Escherichia coli. mBio 2018, 9, e00943–e01018. [Google Scholar] [CrossRef]
- Bok, E.; Kożańska, A.; Mazurek-Popczyk, J.; Wojciech, M.; Baldy-Chudzik, K. Extended Phylogeny and Extraintestinal Virulence Potential of Commensal Escherichia coli from Piglets and Sows. Int. J. Environ. Res. Public Health 2020, 17, 366. [Google Scholar] [CrossRef]
- Huang, J.; Hu, X.; Zhao, Y.; Shi, Y.; Ding, H.; Wu, R.; Zhao, Z.; Ji, J. Comparative Analysis of bla(KPC) Expression in Tn 4401 Transposons and the Tn3 -Tn4401 Chimera. Antimicrob. Agents Chemother. 2019, 63, e02434–e02518. [Google Scholar] [CrossRef] [PubMed]
- Cury, A.P.; Girardello, R.; Duarte, A.J.D.S.; Rossi, F. KPC-producing Enterobacterales with uncommon carbapenem susceptibility profile in Vitek 2 system. Int. J. Infect. Dis. 2020, 93, 118–120. [Google Scholar] [CrossRef] [PubMed]
- Franklin, C.; Liolios, L.; Peleg, A.Y. Phenotypic Detection of Carbapenem-Susceptible Metallo-β-Lactamase-Producing Gram-Negative Bacilli in the Clinical Laboratory. J. Clin. Microbiol. 2006, 44, 3139–3144. [Google Scholar] [CrossRef]
- Corbella, L.; Fernández-Ruiz, M.; Ruiz-Ruigómez, M.; Rodríguez-Goncer, I.; Silva, J.T.; Hernández-Jiménez, P.; López-Medrano, F.; Lizasoain, M.; Villa, J.; Carretero, O.; et al. Prognostic factors of OXA-48 carbapenemase-producing Klebsiella pneumoniae infection in a tertiary-care Spanish hospital: A retrospective single-center cohort study. Int. J. Infect. Dis. 2022, 119, 59–68. [Google Scholar] [CrossRef] [PubMed]
- Baaloudj, O.; Assadi, I.; Nasrallah, N.; El Jery, A.; Khezami, L.; Assadi, A.A. Simultaneous removal of antibiotics and inactivation of antibiotic-resistant bacteria by photocatalysis: A review. J. Water Process Eng. 2021, 42, 102089. [Google Scholar] [CrossRef]
- Pruden, A.; Pei, R.; Storteboom, H.; Carlson, K.H. Antibiotic Resistance Genes as Emerging Contaminants: Studies in Northern Colorado. Environ. Sci. Technol. 2006, 40, 7445–7450. [Google Scholar] [CrossRef] [PubMed]
- Gothwal, R.; Thatikonda, S. Modeling transport of antibiotic resistant bacteria in aquatic environment using stochastic differential equations. Sci. Rep. 2020, 10, 15081. [Google Scholar] [CrossRef]
- Sanganyado, E.; Gwenzi, W. Antibiotic resistance in drinking water systems: Occurrence, removal, and human health risks. Sci. Total Environ. 2019, 669, 785–797. [Google Scholar] [CrossRef]
- Seng, P.; Drancourt, M.; Gouriet, F.; La Scola, B.; Fournier, P.-E.; Rolain, J.M.; Raoult, D. Ongoing Revolution in Bacteriology: Routine Identification of Bacteria by Matrix-Assisted Laser Desorption Ionization Time-of-Flight Mass Spectrometry. Clin. Infect. Dis. 2009, 49, 543–551. [Google Scholar] [CrossRef]
- Weisburg, W.G.; Barns, S.M.; Pelletier, D.A.; Lane, D.J. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 1991, 173, 697–703. [Google Scholar] [CrossRef]
- Bru, J.P.; Caron, F.; Cattoen, C.; Cattoir, V.; Dubreuil, L.; Lina, G.; Merens, A.; Plesiat, P.; Ploy, M.C.; Soussy, C.J.; et al. Antibiogram Committee of the French Microbiology Society. Second edition. Available online: https://www.sfm-microbiologie.org/wp-content/uploads/2020/04/CASFM2020_Avril2020_V1.1.pdf (accessed on 27 August 2022).
- Jarlier, V.; Nicolas, M.H.; Fournier, G.; Philippon, A. Extended broad-spectrum beta-lactamases conferring transferable resistance to newer beta-lactam agents in Enterobacteriaceae: Hospital prevalence and susceptibility patterns. Rev. Infect. Dis. 1988, 10, 867–878. [Google Scholar] [CrossRef] [PubMed]
- Bakour, S.; Garcia, V.; Loucif, L.; Brunel, J.-M.; Gharout-Sait, A.; Touati, A.; Rolain, J.-M. Rapid identification of carbapenemase-producing Enterobacteriaceae, Pseudomonas aeruginosa and Acinetobacter baumannii using a modified Carba NP test. New Microbes New Infect. 2015, 7, 89–93. [Google Scholar] [CrossRef]
- Touati, M.; Hadjadj, L.; Berrazeg, M.; Baron, S.A.; Rolain, J.-M. Emergence of Escherichia coli harbouring mcr-1 and mcr-3 genes in North West Algerian farmlands. J. Glob. Antimicrob. Resist. 2020, 21, 132–137. [Google Scholar] [CrossRef] [PubMed]
- Nabti, L.Z.; Sahli, F.; Ngaiganam, E.P.; Radji, N.; Mezaghcha, W.; Lupande-Mwenebitu, D.; Baron, S.A.; Rolain, J.-M.; Diene, S.M. Development of real-time PCR assay allowed describing the first clinical Klebsiella pneumoniae isolate harboring plasmid-mediated colistin resistance mcr-8 gene in Algeria. J. Glob. Antimicrob. Resist. 2020, 20, 266–271. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.K.; Padmanabhan, B.R.; Diene, S.M.; Lopez-Rojas, R.; Kempf, M.; Landraud, L.; Rolain, J.-M. ARG-ANNOT, a New Bioinformatic Tool to Discover Antibiotic Resistance Genes in Bacterial Genomes. Antimicrob. Agents Chemother. 2014, 58, 212–220. [Google Scholar] [CrossRef] [PubMed]
- Hong, J.S.; Yoon, E.-J.; Lee, H.; Jeong, S.H.; Lee, K. Clonal Dissemination of Pseudomonas aeruginosa Sequence Type 235 Isolates Carrying blaIMP-6 and Emergence of blaGES-24 and blaIMP-10 on Novel Genomic Islands PAGI-15 and -16 in South Korea. Antimicrob. Agents Chemother. 2016, 60, 7216–7223. [Google Scholar] [CrossRef]
- Borowiak, M.; Fischer, J.; Hammerl, J.A.; Hendriksen, R.S.; Szabo, I.; Malorny, B. Identification of a novel transposon-associated phosphoethanolamine transferase gene, mcr-5, conferring colistin resistance in d-tartrate fermenting Salmonella enterica subsp. enterica serovar Paratyphi B. J. Antimicrob. Chemother. 2017, 72, 3317–3324. [Google Scholar] [CrossRef] [PubMed]
- Chabou, S.; Leangapichart, T.; Okdah, L.; Le Page, S.; Hadjadj, L.; Rolain, J.M. Real-time quantitative PCR assay with Taqman® probe for rapid detection of MCR-1 plasmid-mediated colistin resistance. N. Microbes N. Infect. 2016, 13, 71–74. [Google Scholar] [CrossRef] [PubMed]
- Olaitan, A.O.; Diene, S.M.; Kempf, M.; Berrazeg, M.; Bakour, S.; Gupta, S.K.; Thongmalayvong, B.; Akkhavong, K.; Somphavong, S.; Paboriboune, P.; et al. Worldwide emergence of colistin resistance in Klebsiella pneumoniae from healthy humans and patients in Lao PDR, Thailand, Israel, Nigeria and France owing to inactivation of the PhoP/PhoQ regulator mgrB: an epidemiological and molecular study. Int. J. Antimicrob. Agents 2014, 44, 500–507. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cherak, Z.; Loucif, L.; Bendjama, E.; Moussi, A.; Benbouza, A.; Grainat, N.; Rolain, J.-M. Dissemination of Carbapenemases and MCR-1 Producing Gram-Negative Bacteria in Aquatic Environments in Batna, Algeria. Antibiotics 2022, 11, 1314. https://doi.org/10.3390/antibiotics11101314
Cherak Z, Loucif L, Bendjama E, Moussi A, Benbouza A, Grainat N, Rolain J-M. Dissemination of Carbapenemases and MCR-1 Producing Gram-Negative Bacteria in Aquatic Environments in Batna, Algeria. Antibiotics. 2022; 11(10):1314. https://doi.org/10.3390/antibiotics11101314
Chicago/Turabian StyleCherak, Zineb, Lotfi Loucif, Esma Bendjama, Abdelhamid Moussi, Amel Benbouza, Nadia Grainat, and Jean-Marc Rolain. 2022. "Dissemination of Carbapenemases and MCR-1 Producing Gram-Negative Bacteria in Aquatic Environments in Batna, Algeria" Antibiotics 11, no. 10: 1314. https://doi.org/10.3390/antibiotics11101314
APA StyleCherak, Z., Loucif, L., Bendjama, E., Moussi, A., Benbouza, A., Grainat, N., & Rolain, J.-M. (2022). Dissemination of Carbapenemases and MCR-1 Producing Gram-Negative Bacteria in Aquatic Environments in Batna, Algeria. Antibiotics, 11(10), 1314. https://doi.org/10.3390/antibiotics11101314




