Extended-Spectrum Beta-Lactamases Producing Enterobacteriaceae in the USA Dairy Cattle Farms and Implications for Public Health
Abstract
1. Introduction
2. Mechanisms of Resistance to Beta-Lactam Antibiotics
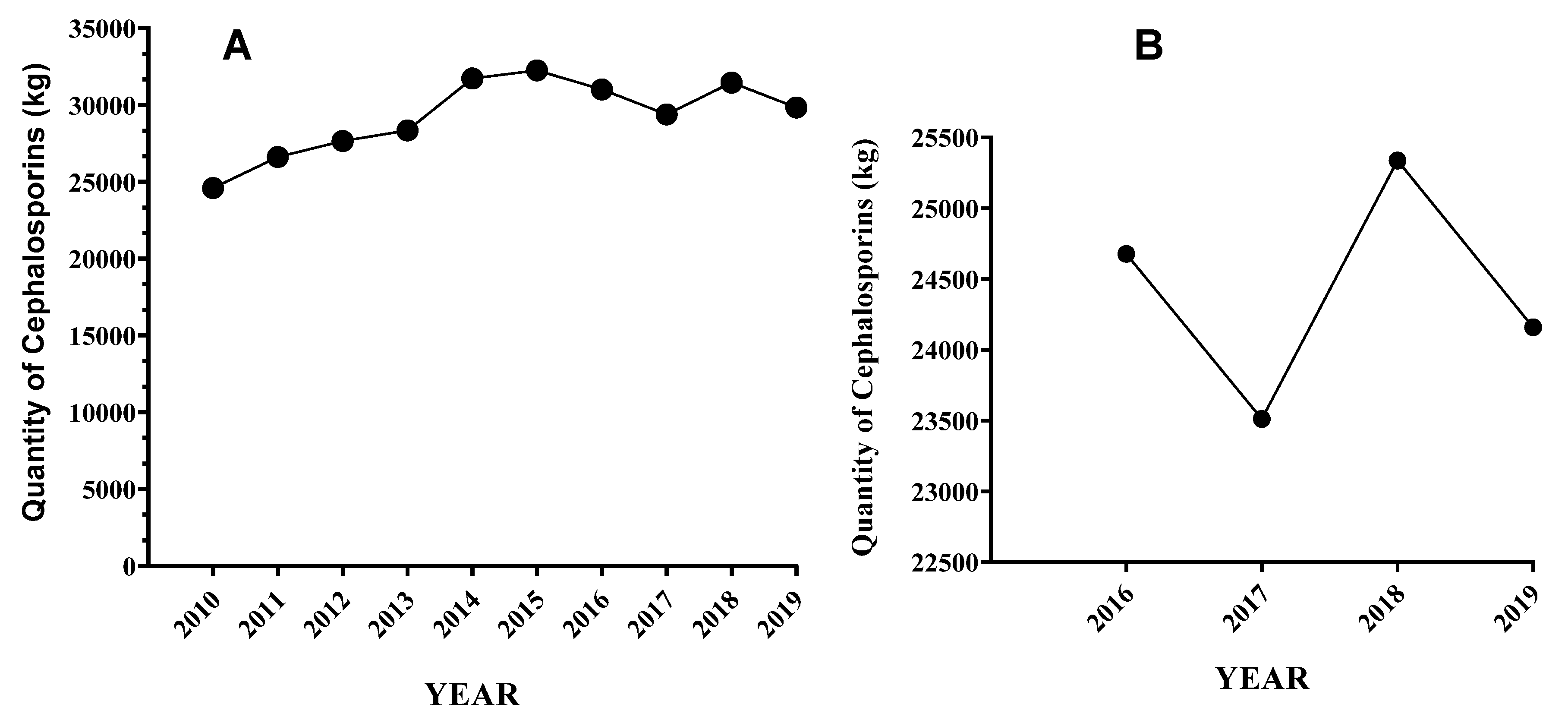
3. Use of Beta-Lactam Antibiotics in the USA Dairy Cattle Farms
4. Molecular Epidemiology of Extended-Spectrum Beta-Lactamases Producing Enterobacteriaceae in the USA Dairy Farms
5. Emergence and Status of Extended-Spectrum Beta-Lactamases Producing Enterobacteriaceae in the USA Dairy Farms
6. Public Health Implications of the Rise in Extended-Spectrum Beta-Lactamases in Dairy Cattle Farms
6.1. Unpasteurized Milk and Undercooked Beef as Possible Sources of Extended-Spectrum Beta-Lactamases Producing Enterobacteriaceae
6.2. Direct Contact (Hand to Mouth) with Dairy Cattle or Their Excretions (Feces, Urine, Milk)
6.3. Fresh Vegetables, Fruits, and Crops
7. Priority Research Gaps That Need to Be Addressed
- A major weakness of current studies is the lack of reliable data on the amount of beta-lactam antibiotics, especially 3GCs used in dairy farms. For example, cephalosporins sales data is often used as an indicator for cephalosporins use which is an unreliable indicator of its use. Furthermore, the sale data does not separately show the amount sold for use in dairy and beef cattle productions. In the absence of these data, it is difficult to assess the impact of their use, develop appropriate interventions, and evaluate the impact of interventions (e.g., the effect of reducing the use of cephalosporins on the prevalence of resistance to cephalosporins). Thus, improving surveillance data on preexisting (baseline) resistance to 3GCs and their use and resistance dynamics after their use is crucial to understanding how antibiotic use may influence antibiotic resistance.
- Currently, the prevalence of ESBLs-Ent in the USA dairy farms is mostly unknown. For instance, despite the veterinary and public health importance of Klebsiella spp., information on its prevalence and the variants of ESBL genes carried by Klebsiella spp. isolates from dairy farms are mostly unknown. Further research should address the status of ESBLs-Ent in dairy farms and their potential risk to human health.
- Among the significant ESBL genes, blaCTX-M encoding lineages are establishing themselves as dominant ESBL in Enterobacteriaceae, particularly among E. coli in the USA dairy farms and across the globe. However, the driver of the successful dissemination of this gene variant is not understood beyond speculation. Understanding the mechanisms for its rapid dissemination in E. coli and other members of Enterobacteriaceae may help to reduce the emergence and spread of antibiotic resistant commensals and pathogenic strains. Thus, further study is critically important to unravel the mechanisms for widespread dissemination of blaCTX-M encoding genes and the bacteria hosting these genes
- Recently, cases of community-acquired ESBL-Ent infection have been rising in the USA. Despite the widespread speculation, there is a lack of adequate scientific data on the level of ESBLs-Ent transmission from dairy cattle and their farm environments to humans. A further detailed investigation is needed to address the potential transmission of ESBLs-Ent from dairy farms to humans using high-resolution genome sequencing technologies such as WGS in epidemiologically linked settings in a system-based one-health approach. This will help to develop a prudent usage plan and antimicrobial stewardship and infection control policies through one health approach consisting of animal, human and environments.
- Factors such as antimicrobial usage and farm management practices that may drive the increased prevalence, spread, persistence, and diversity of ESBL-Ent in dairy farms are not adequately investigated in the USA dairy farms. Such studies are needed to enhance our understanding of factors that influence the occurrence and spread of ESBLs-Ent so that evidenced-based control measures can be devised.
- Archived and contemporary isolates of the members of Enterobacteriaceae should be tested to track any temporal changes in the trends (changes) of phenotypic and genotypic resistance to 3GCs over time in the USA dairy farms.
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Antimicrobial Resistance Collaborators. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef]
- O’Neill, J. Antimicrobial Resistance: Tackling a Crisis for the Health and Wealth of Nations; Wellcome Trust: London, UK, 2014. [Google Scholar]
- CDC. Antimicrobial Resistance Threats Report. 2019. Available online: https://www.cdc.gov/DrugResistance/Biggest-Threats.html#cam (accessed on 25 January 2022).
- Holmes, A.H.; Moore, L.S.; Sundsfjord, A.; Steinbakk, M.; Regmi, S.; Karkey, A.; Guerin, P.J.; Piddock, L.J. Understanding the mechanisms and drivers of antimicrobial resistance. Lancet 2016, 387, 176–187. [Google Scholar] [CrossRef]
- Redding, L.E.; Bender, J.; Baker, L. Quantification of antibiotic use on dairy farms in Pennsylvania. J. Dairy Sci. 2019, 102, 1494–1507. [Google Scholar] [CrossRef] [PubMed]
- FDA. Summary Report on Antimicrobials Sold or Distributed for Use in Food-Producing Animals; FDA: Rockville, MD, USA, 2019.
- WHO. Critically Important Antimicrobials for Human Medicine. In Proceedings of the 6th Revision 2018, Ranking of medically Important Antimicrobials for Risk Management of Antimicrobial Resistance Due to non-Human Use. 2019. Available online: https://www.who.int/publications/i/item/9789241515528 (accessed on 28 August 2022).
- Taylor, E.A.; Ossa-Trujillo, C.; Vinasco, J.; Jordan, E.R.; Buitrago, G.J.A.; Hagevoort, K.N.; Norman, S.D.; Lawhon, R.; Pineiro, J.M.; Levent, G.; et al. Use of critically important antimicrobial classes early in life may adversely impact bacterial resistance profiles during adult years: Potential co-selection for plasmid-borne fluoroquinolone and macrolide resistance via extendedspectrum beta-lactam use in dairy cattle. Lett. Appl. Microbiol. 2020, 72, 220–224. [Google Scholar] [PubMed]
- Bahrani-Mougeot, F.K.; Ansonetti, P.J.S. Encyclopedia of Microbiology, 3rd ed.; Elsevier: Amsterdam, The Netherlands, 2009. [Google Scholar]
- U.S. Department of Health and Human Services Food and Drug Administration Center for Veterinary Medicine. #152 Guidance for Industry, Evaluating the Safety of Antimicrobial New Animal Drugs with Regard to Their Microbiological Effects on Bacteria of Human Health Concern. 2003. Available online: https://www.fda.gov/files/animal%20&%20veterinary/published/CVM-GFI--152-Evaluating-the-Safety-of-Antimicrobial-New-Animal-Drugs-with-Regard-to-Their-Microbiological-Effects-on-Bacteria-of-Human-Health-Concern.pdf (accessed on 28 August 2022).
- Wichmann, F.; Udikovic-Kolic, N.; Andrew, S.; Handelsman, J. Diverse antibiotic resistance genes in dairy cow manure. MBio 2014, 5, e01017. [Google Scholar] [CrossRef] [PubMed]
- Catry, B.; Dewulf, J.; Maes, D.; Pardon, B.; Callens, B.; Vanrobaeys, M.; Opsomer, G.; De Kruif, A.; Haesebrouck, F. Effect of Antimicrobial Consumption and Production Type on Antibacterial Resistance in the Bovine Respiratory and Digestive Tract. PLoS ONE 2016, 11, e0146488. [Google Scholar] [CrossRef] [PubMed]
- O’Hara, A.M.; Shanahan, F. The gut flora as a forgotten organ. Embo Rep. 2006, 7, 688–693. [Google Scholar] [CrossRef]
- Bennani, H.; Mateus, A.; Mays, N.; Eastmure, E.; Stärk, K.D.C.; Häsler, B. Overview of Evidence of Antimicrobial Use and Antimicrobial Resistance in the Food Chain. Antibiotics 2020, 9, 49. [Google Scholar] [CrossRef]
- Thaden, J.T.; Fowler, V.G.; Sexton, D.J.; Anderson, D.J. Increasing Incidence of Extended-Spectrum beta-Lactamase-Producing Escherichia coli in Community Hospitals throughout the Southeastern United States. Infect. Control Hosp. Epidemiol. 2016, 37, 49–54. [Google Scholar] [CrossRef]
- Lee, S.; Mir, R.A.; Park, S.H.; Kim, D.; Kim, H.Y.; Boughton, R.K.; Morris, J.G., Jr.; Jeong, K.C. Prevalence of extended-spectrum b-lactamases in the local farm environment and livestock: Challenges to mitigate antimicrobial resistance. Crit. Rev. Microbiol. 2020, 46, 1–14. [Google Scholar] [CrossRef]
- USDA. Dairy 14 Part III. Health and Management Practices on U.S. Dairy Operations, 2014; USDA-APHIS-VS-CEAHNAHMS, Fort Collins, CO, USA; USDA: Washington, DC, USA, 2018.
- Gelalcha, B.D.; Ensermu, D.B.; Agga, G.E.; Vancuren, M.; Gillespie, B.E.; D’Souza, D.H.; Okafor, C.C.; Kerro Dego, O. Prevalence of Antimicrobial Resistant and Extended-Spectrum Beta-Lactamase-producing Escherichia coli in Dairy Cattle Farms in East Tennessee. Foodborne Pathog. Dis 2022, 19, 408–416. [Google Scholar] [CrossRef]
- Rawat, D.; Nair, D. Extended-spectrum beta-lactamases in Gram Negative Bacteria. J. Glob. Infect. Dis. 2010, 2, 263–274. [Google Scholar] [CrossRef]
- Lalak, A.; Wasyl, D.; Zając, M.; Skarżyńska, M.; Hoszowski, A.; Samcik, I.; Woźniakowski, G.; Szulowski, K. Mechanisms of cephalosporin resistance in indicator Escherichia coli isolated from food animals. Vet. Microbiol. 2016, 194, 69–73. [Google Scholar] [CrossRef]
- Collis, R.M.; Burgess, S.A.; Biggs, P.J.; Midwinter, A.C.; French, N.P.; Toombs-Ruane, L.; Cookson, A.L. Extended-Spectrum Beta-Lactamase-Producing Enterobacteriaceae in Dairy Farm Environments: A New Zealand Perspective. Foodborne Pathog. Dis. 2019, 16, 5–22. [Google Scholar] [CrossRef]
- Bush, K. Proliferation and significance of clinically relevant beta-lactamases. Ann. N. Y. Acad. Sci. 2013, 1277, 84–90. [Google Scholar] [CrossRef]
- de Been, M.; Lanza, V.F.; de Toro, M.; Scharringa, J.; Dohmen, W.; Du, Y.; Hu, J.; Lei, Y.; Li, N.; Tooming-Klunderud, A.; et al. Dissemination of Cephalosporin Resistance Genes between Escherichia coli Strains from Farm Animals and Humans by Specific Plasmid Lineages. PLoS Genet. 2014, 10, e1004776. [Google Scholar] [CrossRef]
- Afema, J.A.; Ahmed, S.; Besser, T.E.; Jones, L.P.; Sischo, W.M.; Davis, M.A. Molecular Epidemiology of Dairy Cattle-Associated Escherichia coli Carrying blaCTX-M Genes in Washington State. Appl. Environ. Microbiol. 2018, 84, e02430-17. [Google Scholar] [CrossRef]
- Stutman, H.R. Salmonella, Shigella, and Campylobacter: Common bacterial causes of infectious diarrhea. Pediatr. Ann. 1994, 23, 538–543. [Google Scholar] [CrossRef]
- Stoycheva, M.V.; Murdjeva, M.A. Antimicrobial therapy of salmonelloses--current state and perspectives. Folia Med. 2006, 48, 5–10. [Google Scholar]
- Wittum, T.E.; Mollenkopf, D.F.; Daniels, J.B.; Parkinson, A.E.; Mathews, J.L.; Fry, P.R.; Abley, M.J.; Gebreyes, W.A. CTX-M-type extended-spectrum beta-lactamases present in Escherichia coli from the feces of cattle in Ohio, United States. Foodborne Pathog. Dis. 2010, 7, 1575–1579. [Google Scholar] [CrossRef]
- Heider, L.C.; Funk, J.A.; Hoet, A.E.; Meiring, R.W.; Gebreyes, W.A.; Wittum, T.E. Identification of Escherichia coli and Salmonella enterica organisms with reduced susceptibility to ceftriaxone from fecal samples of cows in dairy herds. Am. J. Vet. Res. 2009, 70, 389–393. [Google Scholar] [CrossRef]
- Dunne, E.F.; Fey, P.D.; Kludt, P.; Reporter, R.; Mostashari, F.; Shillam, P.; Wicklund, J.; Miller, C.; Holland, B.; Stamey, K.; et al. Emergence of domestically acquired ceftriaxone-resistant Salmonella infections associated with AmpC beta-lactamase. JAMA 2000, 284, 3151–3156. [Google Scholar] [CrossRef]
- Doi, Y.; Iovleva, A.; Bonomo, R.A. The ecology of extended-spectrum beta-lactamases (ESBLs) in the developed world. J. Travel. Med. 2017, 24 (Suppl. 1), S44–S51. [Google Scholar] [CrossRef]
- Sauvage, E.; Kerff, F.; Terrak, M.; Ayala, J.A.; Charlier, P. The penicillin-binding proteins: Structure and role in peptidoglycan biosynthesis. FEMS Microbiol. Rev. 2008, 32, 234–258. [Google Scholar] [CrossRef]
- Bush, K.; Bradford, P.A. beta-Lactams and beta-Lactamase Inhibitors: An Overview. Cold Spring Harb. Perspect. Med. 2016, 6, a025247c. [Google Scholar] [CrossRef]
- Seiffert, S.N.; Hilty, M.; Perreten, V.; Endimiani, A. Extended-spectrum cephalosporin-resistant Gram-negative organisms in livestock: An emerging problem for human health? Drug Resist. Updates 2013, 16, 22–45. [Google Scholar] [CrossRef]
- Letourneau, A.R. Beta-Lactam Antibiotics: Mechanisms of Action and Resistance and Adverse Effects. 2021. Available online: https://www.uptodate.com/contents/beta-lactam-antibiotics-mechanisms-of-action-and-resistance-and-adverse-effects (accessed on 28 August 2022).
- King, D.T.; Sobhanifar, S.; Strynadka, N.C.J. The Mechanisms of Resistance to β-Lactam Antibiotics. In Handbook of Antimicrobial Resistance; Springer: New York, NY, USA, 2017; pp. 177–201. [Google Scholar]
- Bush, K.; Jacoby, G.A. Updated functional classification of beta-lactamases. Antimicrob. Agents Chemother. 2010, 54, 969–976. [Google Scholar] [CrossRef] [PubMed]
- Caroff, N.; Espaze, E.; Gautreau, D.; Richet, H.; Reynaud, A. Analysis of the effects of −42 and −32 ampC promoter mutations in clinical isolates of Escherichia coli hyperproducing ampC. J. Antimicrob. Chemother. 2000, 45, 783–788. [Google Scholar] [CrossRef] [PubMed]
- Bevan, E.R.; Jones, A.M.; Hawkey, P.M. Global epidemiology of CTX-M beta-lactamases: Temporal and geographical shifts in genotype. J. Antimicrob. Chemother. 2017, 72, 2145–2155. [Google Scholar] [CrossRef] [PubMed]
- Bradford, P.A. Extended-spectrum beta-lactamases in the 21st century: Characterization, epidemiology, and detection of this important resistance threat. Clin. Microbiol. Rev. 2001, 14, 933–951. [Google Scholar] [CrossRef] [PubMed]
- Paterson, D.L.; Bonomo, R.A. Extended-spectrum beta-lactamases: A clinical update. Clin. Microbiol. Rev. 2005, 18, 657–686. [Google Scholar] [CrossRef]
- Castanheira, M.; Simner, P.J.; Bradford, P.A. Extended-spectrum β-lactamases: An update on their characteristics, epidemiology and detection. JAC Antimicrob. Resist. 2021, 16, dlab092. [Google Scholar] [CrossRef]
- Bradford, P.A.; Bonomo, R.A.; Bush, K.; Carattoli, A.; Feldgarden, M.; Haft, D.H.; Ishii, Y.; Jacoby, G.A.; Klimke, W.; Palzkill, T.; et al. Consensus on beta-Lactamase Nomenclature. Antimicrob. Agents Chemother. 2022, 66, e0033322. [Google Scholar] [CrossRef]
- Medeiros, A.A. β-LACTAMASES. Br. Med. Bull. 1984, 40, 18–27. [Google Scholar] [CrossRef]
- Bush, K. The ABCD’s of beta-lactamase nomenclature. J. Infect. Chemother. 2013, 19, 549–559. [Google Scholar] [CrossRef]
- Livermore, D.M. Defining an extended-spectrum beta-lactamase. Clin. Microbiol. Infect. 2008, 14 (Suppl. 1), 3–10. [Google Scholar] [CrossRef]
- Gniadkowski, M. Evolution of extended-spectrum beta-lactamases by mutation. Clin. Microbiol. Infect. 2008, 14 (Suppl. 1), 11–32. [Google Scholar] [CrossRef]
- Matthew, M.; Hedges, R.W.; Smith, J.T. Types of beta-lactamase determined by plasmids in gram-negative bacteria. J. Bacteriol. 1979, 138, 657–662. [Google Scholar] [CrossRef]
- Pitout, J.D. Extraintestinal pathogenic Escherichia coli: An update on antimicrobial resistance, laboratory diagnosis and treatment. Expert Rev. Anti Infect 2012, 10, 1165–1176. [Google Scholar] [CrossRef]
- NCBI. Reference Gene Catalog. Available online: https://www.ncbi.nlm.nih.gov/pathogens/refgene/#gene_family:(blaSHV/blaTEM (accessed on 5 July 2022).
- Pitout, J.D.; Hossain, A.; Hanson, N.D. Phenotypic and molecular detection of CTX-M-beta-lactamases produced by Escherichia coli and Klebsiella spp. J. Clin. Microbiol. 2004, 42, 5715–5721. [Google Scholar] [CrossRef]
- Hussain, H.I.; Aqib, A.I.; Seleem, M.N.; Shabbir, M.A.; Hao, H.; Iqbal, Z.; Kulyar, M.F.; Zaheer, T.; Li, K. Genetic basis of molecular mechanisms in beta-lactam resistant gram-negative bacteria. Microb. Pathog. 2021, 158, 105040. [Google Scholar] [CrossRef]
- Matsumoto, Y.; Ikeda FKamimura, T.; Yokota, Y.; Mine, Y. Novel plasmid-mediated Beta-lactamase from Escherichia coli that inactivates oxyimino-cephalosporins. Antimicrob. Agents Chemother. 1988, 32, 1243–1246. [Google Scholar] [CrossRef]
- Canton, R.; Gonzalez-Alba, J.M.; Galan, J.C. CTX-M Enzymes: Origin and Diffusion. Front. Microbiol. 2012, 3, 110. [Google Scholar] [CrossRef]
- Peirano, G.; Sang, J.H.; Pitondo-Silva, A.; Laupland, K.B.; Pitout, J.D. Molecular epidemiology of extended-spectrum-beta-lactamase-producing Klebsiella pneumoniae over a 10 year period in Calgary, Canada. J. Antimicrob. Chemother. 2012, 67, 1114–1120. [Google Scholar] [CrossRef] [PubMed]
- Pitout, J.D.; Laupland, K.B. Extended-spectrum beta-lactamase-producing Enterobacteriaceae: An emerging public-health concern. Lancet Infect. Dis. 2008, 8, 159–166. [Google Scholar] [CrossRef]
- Bonnet, R. Growing group of extended-spectrum beta-lactamases: The CTX-M enzymes. Antimicrob. Agents Chemother. 2004, 48, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Davis, M.A.; Sischo, W.M.; Jones, L.P.; Moore, D.A.; Ahmed, S.; Short, D.M.; Besser, T.E. Recent Emergence of Escherichia coli with Cephalosporin Resistance Conferred by bla(CTX-M) on Washington State Dairy Farms. Appl. Environ. Microbiol. 2015, 81, 4403–4410. [Google Scholar] [CrossRef]
- D’Andrea, M.M.; Arena, F.; Pallecchi, L.; Rossolini, G.M. CTX-M-type beta-lactamases: A successful story of antibiotic resistance. Int. J. Med. Microbiol. 2013, 303, 305–317. [Google Scholar] [CrossRef]
- NCBI. Reference Gene Catalog. Available online: https://www.ncbi.nlm.nih.gov/pathogens/refgene/#gene_family:(blaCTX-M) (accessed on 5 July 2022).
- USDA(United States Department of Agriculture). Dairy 2014, Milk Quality, Milking Procedures, and Mastitis in the United States, 2014; USDA–APHIS–VS–CEAH–NAHMS; USDA: Fort Collins, CO, USA, 2016.
- Schrag, N.F.; Godden, S.M.; Apley, M.D.; Singer, R.S.; Lubbers, B.V. Antimicrobial Use Quantification in Adult Dairy Cows—Part 3—Use Measured by Standardized Regimens and Grams on 29 Dairies in the United States. Zoonoses Public Health 2020, 67 (Suppl. 1), 82–93. [Google Scholar] [CrossRef]
- FDA. Animal Drugs. In: U.S. Food & Drug Administration. In Animal Drugs. Available online: https://animaldrugsatfda.fda.gov/adafda/views/#/search (accessed on 28 August 2022).
- FDA. Extralabel Use and Antimicrobials. Available online: https://www.fda.gov/animal-veterinary/antimicrobial-resistance/extralabel-use-and-antimicrobials (accessed on 30 June 2022).
- FDA. Cephalosporin Order of Prohibition Questions and Answers. Available online: https://www.fda.gov/animal-veterinary/antimicrobial-resistance/cephalosporin-order-prohibition-questions-and-answers (accessed on 3 July 2022).
- Arnold, M. The U.S. Food and Drug Administration (FDA) and Cephalosporin Use: How Will This New Rule Affect KY Dairy Producers? Available online: https://afs.ca.uky.edu/dairy/us-food-and-drug-administration-fda-and-cephalosporin-use-how-will-new-rule-affect-ky-dairy (accessed on 3 July 2022).
- FDA. Cephalosporin use in cattle. Available online: https://www.mda.state.mn.us/sites/default/files/inline-files/Cephalosporin.pdf (accessed on 15 July 2022).
- Zoetis Services, LLC Zoetis United States Website. Available online: https://www.zoetisus.com/ (accessed on 3 July 2022).
- Ekakoro, J.E.; Caldwell, M.; Strand, E.B.; Okafor, C.C. Drivers of Antimicrobial Use Practices among Tennessee Dairy Cattle Producers. Vet. Med. Int. 2018, 2018, 1836836. [Google Scholar] [CrossRef]
- Schrag, N.F.; Apley, M.D.; Godden, S.M.; Lubbers, B.V.; Singer, R.S. Antimicrobial use quantification in adult dairy cows—Part 1—Standardized regimens as a method for describing antimicrobial use. Zoonoses Public Health 2020, 67, 51–68. [Google Scholar] [CrossRef]
- Pol, M.; Ruegg, P.L. Treatment practices and quantification of antimicrobial drug usage in conventional and organic dairy farms in Wisconsin. J. Dairy Sci. 2007, 90, 249–261. [Google Scholar] [CrossRef]
- Kumar, N.; Manimaran, A.; Kumaresan, A.; Sreela, L.; Patbandha, T.K.; Tiwari, S.; Chandra, S. Episodes of clinical mastitis and its relationship with duration of treatment and seasonality in crossbred cows maintained in organized dairy farm. Vet. World 2016, 9, 75–79. [Google Scholar] [CrossRef]
- Sawant, A.A.; Sordillo, L.M.; Jayarao, B.M. A survey on antibiotic usage in dairy herds in Pennsylvania. J. Dairy Sci. 2005, 88, 2991–2999. [Google Scholar] [CrossRef]
- U.S. Food & Drug Administration. Antimicrobial Use and Resistance in Animal Agriculture in the United States 2016–2019; 2022. Available online: https://www.fda.gov/media/159544/download (accessed on 28 August 2022).
- Fey, P.D.; Safranek, T.J.; Rupp, M.E.; Dunne, E.F.; Ribot, E.; Iwen, P.C.; Bradford, P.A.; Angulo, F.J.; Hinrichs, S.H. Ceftriaxone-resistant salmonella infection acquired by a child from cattle. N. Engl. J. Med. 2000, 342, 1242–1249. [Google Scholar] [CrossRef]
- Food and Drug Administration, Center for Veteinary Medicine. 2020 Summary Report on Animicrobials sold or Distributed for Use in Food-Producing Animals. 2021. Available online: https://www.fda.gov/media/154820/download (accessed on 28 August 2022).
- Fuenzalida, M.J.; Furmaga, E.; Aulik, N. Antimicrobial resistance in Klebsiella species from milk specimens submitted for bovine mastitis testing at the Wisconsin Veterinary Diagnostic Laboratory, 2008–2019. JDS Commun. 2021, 2, 148–152. [Google Scholar] [CrossRef]
- Aarestrup, F.M.; Seyfarth, A.M.; Emborg, H.D.; Pedersen, K.; Hendriksen, R.S.; Bager, F. Effect of abolishment of the use of antimicrobial agents for growth promotion on occurrence of antimicrobial resistance in fecal enterococci from food animals in Denmark. Antimicrob. Agents Chemother. 2001, 45, 2054–2059. [Google Scholar] [CrossRef]
- Berge, A.C.B.; Atwill, E.R.; Sischo, W.M. Animal and farm influences on the dynamics of antibiotic resistance in faecal Escherichia coli in young calves. Prev. Vet. Med. 2005, 69, 25–38. [Google Scholar] [CrossRef]
- Poppe, C.; Martin, L.C.; Gyles, C.L.; Reid-Smith, R.; Boerlin, P.; McEwen, S.A.; Prescott, J.F.; Forward, K.R. Acquisition of resistance to extended-spectrum cephalosporins by Salmonella enterica subsp. enterica serovar Newport and Escherichia coli in the turkey poult intestinal tract. Appl. Environ. Microbiol. 2005, 71, 1184–1192. [Google Scholar] [CrossRef]
- Fan, S.; Foster, D.; Miller, W.; Osborne, J.; Kathariou, S. Impact of Ceftiofur Administration in Steers on the Prevalence and Antimicrobial Resistance of Campylobacter spp. Microorganisms 2021, 9, 318. [Google Scholar] [CrossRef]
- Beyi, A.F.; Brito-Goulart, D.; Hawbecker, T.; Ruddell, B.; Hassall, A.; Dewell, R.; Dewell, G.; Sahin, O.; Zhang, Q.; Plummer, P.J. Enrofloxacin Alters Fecal Microbiota and Resistome Irrespective of Its Dose in Calves. Microorganisms 2021, 9, 2162. [Google Scholar] [CrossRef]
- Beyi, A.F.; Brito-Goulart, D.; Hawbecker, T.; Slagel, C.; Ruddell, B.; Hassall, A.; Dewell, R.; Dewell, G.; Sahin, O.; Zhang, Q.; et al. Danofloxacin Treatment Alters the Diversity and Resistome Profile of Gut Microbiota in Calves. Microorganisms 2021, 9, 2023. [Google Scholar] [CrossRef]
- Guri, A.; Flaks-Manov, N.; Ghilai, A.; Hoshen, M.; Rimon, O.F.; Ciobotaro, P.; Zimhony, O. Risk factors for third-generation cephalosporin resistant Enterobacteriaceae in gestational urine cultures: A retrospective cohort study based on centralized electronic health records. PLoS ONE 2020, 15, e0226515. [Google Scholar] [CrossRef]
- Foster, D.M.; Jacob, M.E.; Farmer, K.A.; Callahan, B.; Theriot, C.M.; Kathariou, S.; Cernicchiaro, N.; Prange, T.; Papich, M.G. Ceftiofur formulation differentially affects the intestinal drug concentration, resistance of fecal Escherichia coli, and the microbiome of steers. PLoS ONE 2019, 14, e0223378. [Google Scholar] [CrossRef]
- Oliveira, L.; Ruegg, P.L. Treatments of clinical mastitis occurring in cows on 51 large dairy herds in Wisconsin. J. Dairy Sci. 2014, 97, 5426–5436. [Google Scholar] [CrossRef]
- Makovec, J.A.; Ruegg, P.L. Antimicrobial resistance of bacteria isolated from dairy cow milk samples submitted for bacterial culture: 8,905 samples (1994–2001). J. Am. Vet. Med. Assoc. 2003, 222, 1582–1589. [Google Scholar] [CrossRef]
- Oliver, S.P.; Murinda, S.E.; Jayarao, B.M. Impact of Antibiotic Use in Adult Dairy Cows on Antimicrobial Resistance of Veterinary and Human Pathogens: A Comprehensive Review. Foodborne Pathog. Dis. 2011, 8, 337–355. [Google Scholar] [CrossRef]
- Abdi, R.D.; Gillespie, B.E.; Vaughn, J.; Merrill, C.; Headrick, S.I.; Ensermu, D.B.; D’Souza, D.H.; Agga, G.E.; Almeida, R.A.; Oliver, S.P.; et al. Antimicrobial Resistance of Staphylococcus aureus Isolates from Dairy Cows and Genetic Diversity of Resistant Isolates. Foodborne Pathog. Dis. 2018, 15, 449–458. [Google Scholar] [CrossRef]
- Abdi, R.D.; Gillespie, B.E.; Ivey, S.; Pighetti, G.M.; Almeida, R.A.; Dego, O.K. Antimicrobial Resistance of Major Bacterial Pathogens from Dairy Cows with High Somatic Cell Count and Clinical Mastitis. Animals 2021, 11, 131. [Google Scholar] [CrossRef]
- Oliver, J.P.; Gooch, C.A.; Lansing, S.; Schueler, J.; Hurst, J.J.; Sassoubre, L.; Crossette, E.M.; Aga, D.S. Invited review: Fate of antibiotic residues, antibiotic-resistant bacteria, and antibiotic resistance genes in US dairy manure management systems. J. Dairy Sci. 2020, 103, 1051–1071. [Google Scholar] [CrossRef]
- Gelalcha, B.D.; Agga, G.E.; Dego, O.K. Antimicrobial Usage for the Management of Mastitis in the USA: Impacts on Antimicrobial Resistance and Potential Alternative Approaches. In Mastitis in Dairy Cattle, Sheep and Goats; Dego, O.K., Ed.; Intech Open: London, UK, 2021; pp. 1–21. [Google Scholar]
- Gonggrijp, M.A.; Santman-Berends, I.; Heuvelink, A.E.; Buter, G.J.; van Schaik, G.; Hage, J.J.; Lam, T. Prevalence and risk factors for extended-spectrum beta-lactamase- and AmpC-producing Escherichia coli in dairy farms. J. Dairy Sci. 2016, 99, 9001–9013. [Google Scholar] [CrossRef] [PubMed]
- Gomes, F.; Henriques, M. Control of Bovine Mastitis: Old and Recent Therapeutic Approaches. Curr. Microbiol. 2016, 72, 377–382. [Google Scholar] [CrossRef] [PubMed]
- Rabiee, A.R.; Lean, I.J. The effect of internal teat sealant products (Teatseal and Orbeseal) on intramammary infection, clinical mastitis, and somatic cell counts in lactating dairy cows: A meta-analysis. J. Dairy Sci. 2013, 96, 6915–6931. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.H.; Hu, Z.Q. Epidemiology and genetics of CTX-M extended-spectrum beta-lactamases in Gram-negative bacteria. Crit. Rev. Microbiol. 2013, 39, 79–101. [Google Scholar] [CrossRef]
- Li, Q.; Chang, W.; Zhang, H.; Hu, D.; Wang, X. The Role of Plasmids in the Multiple Antibiotic Resistance Transfer in ESBLs-Producing Escherichia coli Isolated From Wastewater Treatment Plants. Front. Microbiol. 2019, 10, 633. [Google Scholar] [CrossRef]
- Partridge, S.R.; Kwong, S.M.; Firth, N.; Jensen, S.O. MMobile Genetic Elements Associated with Antimicrobial Resistance. Clin. Microbiol. Rev. 2018, 31, e00088-17. [Google Scholar] [CrossRef]
- Peirano, G.; Pitout, J. Extended-Spectrum β-Lactamase-Producing Enterobacteriaceae: Update on Molecular Epidemiology and Treatment Options. Drugs 2019, 79, 1529–1541. [Google Scholar] [CrossRef]
- Poirel, L.; Lartigue, M.F.; Decousser, J.W.; Nordmann, P. ISEcp1B-mediated transposition of blaCTX-M in Escherichia coli. Antimicrob. Agents Chemother. 2005, 49, 447–450. [Google Scholar] [CrossRef]
- Lartigue, M.F.; Poirel, L.; Aubert, D.; Nordmann, P. In vitro analysis of ISEcp1B-mediated mobilization of naturally occurring beta-lactamase gene blaCTX-M of Kluyvera ascorbata. Antimicrob. Agents Chemother. 2006, 50, 1282–1286. [Google Scholar] [CrossRef]
- Mathers, A.J.; Peirano, G.; Pitout, J.D. The role of epidemic resistance plasmids and international high-risk clones in the spread of multidrug-resistant Enterobacteriaceae. Clin. Microbiol. Rev. 2015, 28, 565–591. [Google Scholar] [CrossRef]
- Nordmann, P.; Poirel, L. The difficult-to-control spread of carbapenemase producers among Enterobacteriaceae worldwide. Clinical microbiology and infection. Eur. Soc. Clin. Microbiol. Infect. Dis. 2014, 20, 821–830. [Google Scholar] [CrossRef]
- Nicolas-Chanoine, M.-H.; Blanco, J.; Leflon-Guibout, V.; Demarty, R.; Alonso, M.P.; Caniça, M.M.; Park, Y.-J.; Lavigne, J.-P.; Pitout, J.; Johnson, J.R. Intercontinental emergence of Escherichia coli clone O25: H4-ST131 producing CTX-M-15. J. Antimicrob. Chemother. 2008, 61, 273–281. [Google Scholar] [CrossRef]
- Irrgang, A.; Falgenhauer, L.; Fischer, J.; Ghosh, H.; Guiral, E.; Guerra, B.; Schmoger, S.; Imirzalioglu, C.; Chakraborty, T.; Hammerl, J.A.; et al. CTX-M-15-Producing E. coli Isolates from Food Products in Germany Are Mainly Associated with an IncF-Type Plasmid and Belong to Two Predominant Clonal E. coli Lineages. Front. Microbiol. 2017, 8, 2318. [Google Scholar] [CrossRef]
- Carattoli, A. Plasmids and the spread of resistance. Int. J. Med. Microbiol. 2013, 303, 298–304. [Google Scholar] [CrossRef]
- Gozi, K.S.; Froes, J.R.; Deus Ajude, L.P.T.; da Silva, C.R.; Baptista, R.S.; Peiro, J.R.; Marinho, M.; Mendes LC, N.; Nogueira MC, L.; Casella, T. Dissemination of Multidrug-Resistant Commensal Escherichia coli in Feedlot Lambs in Southeastern Brazil. Front. Microbiol. 2019, 10, 1394. [Google Scholar] [CrossRef]
- Villa, L.; García-Fernández, A.; Fortini, D.; Carattoli, A. Replicon sequence typing of IncF plasmids carrying virulence and resistance determinants. J. Antimicrob. Chemother. 2010, 65, 2518–2529. [Google Scholar] [CrossRef]
- Pitout, J.D.; DeVinney, R. Escherichia coli ST131: A multidrug-resistant clone primed for global domination. F1000Research 2017, 6, 195. [Google Scholar] [CrossRef]
- Shin, J.; Choi, M.J.; Ko, K.S. Replicon sequence typing of IncF plasmids and the genetic environments of blaCTX-M-15 indicate multiple acquisitions of blaCTX-M-15 in Escherichia coli and Klebsiella pneumoniae isolates from South Korea. J. Antimicrob. Chemother. 2012, 67, 1853–1857. [Google Scholar] [CrossRef]
- Canton, R.; Coque, T.M. The CTX-M beta-lactamase pandemic. Curr. Opin. Microbiol. 2006, 9, 466–475. [Google Scholar] [CrossRef]
- Donaldson, S.C.; Straley, B.A.; Hegde, N.V.; Sawant, A.A.; DebRoy, C.; Jayarao, B.M. Molecular epidemiology of ceftiofur-resistant Escherichia coli isolates from dairy calves. Appl. Env. Microbiol. 2006, 72, 3940–3948. [Google Scholar] [CrossRef]
- Tamta, S.; Kumar, O.; Singh, S.V.; Pruthvishree, B.S.; Karthikeyan, R.; Rupner, R.; Sinha, D.K.; Singh, B.R. Antimicrobial resistance pattern of extended-spectrum beta-lactamase-producing Escherichia coli isolated from fecal samples of piglets and pig farm workers of selected organized farms of India. Vet. World 2020, 13, 360–363. [Google Scholar] [CrossRef]
- Liu, X.; Liu, H.; Wang, L.; Peng, Q.; Li, Y.; Zhou, H.; Li, Q. Molecular Characterization of Extended-Spectrum beta-Lactamase-Producing Multidrug Resistant Escherichia coli From Swine in Northwest China. Front. Microbiol. 2018, 9, 1756. [Google Scholar] [CrossRef]
- Badr, H.; Reda, R.M.; Hagag, N.M.; Kamel, E.; Elnomrosy, S.M.; Mansour, A.I.; Shahein, M.A.; Ali, S.F.; Ali, H.R. Multidrug-Resistant and Genetic Characterization of Extended-Spectrum Beta-Lactamase-Producing E. coli Recovered from Chickens and Humans in Egypt. Animals 2022, 12, 346. [Google Scholar] [CrossRef]
- Carey, A.M.; Capik, S.F.; Giebel, S.; Nickodem, C.; Piñeiro, J.M.; Scott, H.M.; Vinasco, J.; Norman, K.N. Prevalence and Profiles of Antibiotic Resistance Genes mph(A) and qnrB in Extended-Spectrum Beta-Lactamase (ESBL)-Producing Escherichia coli Isolated from Dairy Calf Feces. Microorganisms 2022, 10, 411. [Google Scholar] [CrossRef]
- Winokur, P.L.; Brueggemann, A.; DeSalvo, D.L.; Hoffmann, L.; Apley, M.D.; Uhlenhopp, E.K.; Pfaller, M.A.; Doern, G.V. Animal and human multidrug-resistant, cephalosporin-resistant salmonella isolates expressing a plasmid-mediated CMY-2 AmpC beta-lactamase. Antimicrob. Agents Chemother. 2000, 44, 2777–2783. [Google Scholar] [CrossRef]
- Weber, D.A.; Sanders, C.C.; Bakken, J.S.; Quinn, J.P. A novel chromosomal TEM derivative and alterations in outer membrane proteins together mediate selective ceftazidime resistance in Escherichia coli. J. Infect. Dis. 1990, 162, 460–465. [Google Scholar] [CrossRef]
- Jacoby, G.A.; Medeiros, A.A. More extended-spectrum beta-lactamases. Antimicrob. Agents Chemother. 1991, 35, 1697–1704. [Google Scholar] [CrossRef]
- Moland, E.S.; Black, J.A.; Hossain, A.; Hanson, N.D.; Thomson, K.S.; Pottumarthy, S. Discovery of CTX-M-like extended-spectrum beta-lactamases in Escherichia coli isolates from five US States. Antimicrob. Agents Chemother. 2003, 47, 2382–2383. [Google Scholar] [CrossRef]
- Agga, G.E.; Schmidt, J.W.; Arthur, T.M. Antimicrobial-Resistant Fecal Bacteria from Ceftiofur-Treated and Nonantimicrobial-Treated Comingled Beef Cows at a Cow-Calf Operation. Microb. Drug Resist. 2016, 22, 598–608. [Google Scholar] [CrossRef]
- Berge, A.C.B.; Adaska, J.M.; Sischo, W.M. Use of antibiotic susceptibility patterns and pulsed-field gel electrophoresis to compare historic and contemporary isolates of multi-drug-resistant Salmonella enterica subsp enterica serovar Newport. Appl. Environ. Microbiol. 2004, 70, 318–323. [Google Scholar] [CrossRef]
- Yang, Y.; Higgins, C.H.; Rehman, I.; Galvao, K.N.; Brito, I.L.; Bicalho, M.L.; Song, J.; Wang, H.; Bicalho, R.C. Genomic Diversity, Virulence, and Antimicrobial Resistance of Klebsiella pneumoniae Strains from Cows and Humans. Appl. Environ. Microbiol. 2019, 85, e02654-18. [Google Scholar] [CrossRef] [PubMed]
- Josman Dantas Palmeira, H.M.N.F. Extended-spectrum beta-lactamase (ESBL)-producing Enterobacteriaceae in cattle production—A threat around the world. Heliyon 2020, 6, e03206. [Google Scholar] [CrossRef] [PubMed]
- Hutchinson, H.; Finney, S.; Muñoz-Vargas, L.; Feicht, S.; Masterson, M.; Habing, G. Prevalence and Transmission of Antimicrobial Resistance in a Vertically Integrated Veal Calf Production System. Foodborne Pathog. Dis. 2017, 14, 711–718. [Google Scholar] [CrossRef] [PubMed]
- Davidson, K.E.; Byrne, B.A.; Pires, A.F.A.; Magdesian, K.G.; Pereira, R.V. Antimicrobial resistance trends in fecal Salmonella isolates from northern California dairy cattle admitted to a veterinary teaching hospital, 2002–2016. PLoS ONE 2018, 13, e0199928. [Google Scholar] [CrossRef] [PubMed]
- Mollenkopf, D.F.; Weeman, M.F.; Daniels, J.B.; Abley, M.J.; Mathews, J.L.; Gebreyes, W.A.; Wittum, T.E. Variable within- and between-Herd Diversity of CTX-M Cephalosporinase-Bearing Escherichia coli Isolates from Dairy Cattle. Appl. Environ. Microbiol. 2012, 78, 4552–4560. [Google Scholar] [CrossRef] [PubMed]
- Frye, J.G.; Fedorka-Cray, P.J. Prevalence, distribution and characterisation of ceftiofur resistance in Salmonella enterica isolated from animals in the USA from 1999 to 2003. Int. J. Antimicrob. Agents 2007, 30, 134–142. [Google Scholar] [CrossRef]
- Li, X.; Aly, S.S.; Su, Z.; Pereira, R.V.; Williams, D.R.; Rossitto, P.; Champagne, J.D.; Chase, J.; Nguyen, T.; Atwill, E.R. Phenotypic Antimicrobial Resistance Profiles of E. coli and Enterococcus from Dairy Cattle in Different Management Units on a Central California Dairy. Clin. Microbiol. Infect. 2018, 7, 2. [Google Scholar] [CrossRef]
- Cao, H. Antimicrobial Resistance of Salmonella and E. coli from Pennsylvania Dairy Herds; University of Maryland: College Park, MD, USA, 2015. [Google Scholar]
- Cummings, K.; Warnick, L.; Alexander, K.; Cripps, C.; Gröhn, Y.; McDonough, P.; Nydam, D.; Reed, K. The incidence of salmonellosis among dairy herds in the northeastern United States. J. Dairy Sci. 2009, 92, 3766–3774. [Google Scholar] [CrossRef]
- Edrington, T.S.; Callaway, T.R.; Anderson, R.C.; Nisbet, D.J. Prevalence of multidrug-resistant Salmonella on commercial dairies utilizing a single heifer raising facility. J. Food Prot. 2008, 71, 27–34. [Google Scholar] [CrossRef]
- Ray, K.; Warnick, L.; Mitchell, R.; Kaneene, J.; Ruegg, P.; Wells, S.; Fossler, C.; Halbert, L.; May, K. Prevalence of antimicrobial resistance among Salmonella on midwest and northeast USA dairy farms. Prev. Vet. Med. 2007, 79, 204–223. [Google Scholar] [CrossRef]
- Davis, M.A.; Besser, T.E.; Orfe, L.H.; Baker, K.N.K.; Lanier, A.S.; Broschat, S.L.; New, D.; Call, D.R. Genotypic-Phenotypic Discrepancies between Antibiotic Resistance Characteristics of Escherichia coli Isolates from Calves in Management Settings with High and Low Antibiotic Use. Appl. Environ. Microbiol. 2011, 77, 3293–3299. [Google Scholar] [CrossRef]
- NARMS. Global Resistome Data. Available online: https://www.fda.gov/animal-veterinary/national-antimicrobial-resistance-monitoring-system/global-resistome-data (accessed on 20 July 2022).
- Chang, Q.; Wang, W.; Regev-Yochay, G.; Lipsitch, M.; Hanage, W.P. Antibiotics in agriculture and the risk to human health: How worried should we be? Evol. Appl. 2015, 8, 240–247. [Google Scholar] [CrossRef]
- Price, L.B.; Graham, J.P.; Lackey, L.G.; Roess, A.; Vailes, R.; Silbergeld, E. Elevated risk of carrying gentamicin-resistant Escherichia coli among U.S. poultry workers. Environ. Health Perspect. 2007, 115, 1738–1742. [Google Scholar] [CrossRef]
- Oliveira, L.; Hulland, C.; Ruegg, P.L. Characterization of clinical mastitis occurring in cows on 50 large dairy herds in Wisconsin. J. Dairy Sci. 2013, 96, 7538–7549. [Google Scholar] [CrossRef]
- Locatelli, C.; Scaccabarozzi, L.; Pisoni, G.; Moroni, P. CTX-M1 ESBL-producing Klebsiella pneumoniae subsp. pneumoniae isolated from cases of bovine mastitis. J. Clin. Microbiol. 2010, 48, 3822–3823. [Google Scholar] [CrossRef]
- Wang, G.; Huang, T.; Surendraiah, P.K.; Wang, K.; Komal, R.; Zhuge, J.; Chern, C.R.; Kryszuk, A.A.; King, C.; Wormser, G.P. CTX-M β-Lactamase–producing Klebsiella pneumoniae in Suburban New York City, New York, USA. Emerg. Infect. Dis. 2013, 19, 1803. [Google Scholar] [CrossRef]
- McDanel, J.; Schweizer, M.; Crabb, V.; Nelson, R.; Samore, M.; Khader, K.; Blevins, A.E.; Diekema, D.; Chiang, H.-Y.; Nair, R.; et al. Incidence of Extended-Spectrum beta-Lactamase (ESBL)-Producing Escherichia coli and Klebsiella Infections in the United States: A Systematic Literature Review. Infect. Control Hosp. Epidemiol. 2017, 38, 1209–1215. [Google Scholar] [CrossRef]
- Kassakian, S.Z.; Mermel, L.A. Changing epidemiology of infections due to extended spectrum beta-lactamase producing bacteria. Antimicrob. Resist. Infect. Control 2014, 3, 9. [Google Scholar] [CrossRef]
- Allen, K.J.; Poppe, C. Occurrence and characterization of resistance to extended-spectrum cephalosporins mediated by beta-lactamase CMY-2 in Salmonella isolated from food-producing animals in Canada. Can. J. Vet. Res. 2002, 66, 137–144. [Google Scholar]
- Levy, S.B.; FitzGerald, G.B.; Macone, A.B. Changes in intestinal flora of farm personnel after introduction of a tetracycline-supplemented feed on a farm. N. Engl. J. Med. 1976, 295, 583–588. [Google Scholar] [CrossRef]
- Smith, K.E.; Besser, J.M.; Hedberg, C.W.; Leano, F.T.; Bender, J.B.; Wicklund, J.H.; Johnson, B.P.; Moore, K.A.; Osterholm, M.T. Quinolone-resistant Campylobacter jejuni infections in Minnesota, 1992–1998 Investigation Team. N. Engl. J. Med. 1999, 340, 1525–1532. [Google Scholar] [CrossRef]
- Winokur, P.L.; Vonstein, D.L.; Hoffman, L.J.; Uhlenhopp, E.K.; Doern, G.V. Evidence for transfer of CMY-2 AmpC beta-lactamase plasmids between Escherichia coli and Salmonella isolates from food animals and humans. Antimicrob. Agents Chemother. 2001, 45, 2716–2722. [Google Scholar] [CrossRef]
- Iwamoto, M.; Reynolds, J.; Karp, B.E.; Tate, H.; Fedorka-Cray, P.J.; Plumblee, J.R.; Hoekstra, R.M.; Whichard, J.M.; Mahon, B.E. Ceftriaxone-Resistant Nontyphoidal Salmonella from Humans, Retail Meats, and Food Animals in the United States, 1996–2013. Foodborne Pathog. Dis. 2017, 14, 74–83. [Google Scholar] [CrossRef]
- Reeves, P.R.; Liu, B.; Zhou, Z.; Li, D.; Guo, D.; Ren, Y.; Clabots, C.; Lan, R.; Johnson, J.R.; Wang, L. Rates of mutation and host transmission for an Escherichia coli clone over 3 years. PLoS ONE 2011, 6, e26907. [Google Scholar] [CrossRef]
- Kantele, A.; Kuenzli, E.; Dunn, S.J.; Dance, D.A.; Newton, P.N.; Davong, V.; Mero, S.; Pakkanen, S.H.; Neumayr, A.; Hatz, C.; et al. Real-time sampling of travelers shows intestinal colonization by multidrug-resistant bacteria to be a dynamic process with multiple transient acquisitions. bioRxiv 2022, 827915. [Google Scholar] [CrossRef]
- Wee, B.A.; Muloi, D.M.; Bunnik, A.D. Quantifying the transmission of antimicrobial resistance at the human and livestock interface with genomics. Clin. Microbiol. Infect. 2020, 26, 1612–1616. [Google Scholar] [CrossRef]
- Davis, G.S.; Price, L.B. Recent Research Examining Links Among Klebsiella pneumoniae from Food, Food Animals, and Human Extraintestinal Infections. Curr. Environ. Health Rep. 2016, 3, 128–135. [Google Scholar] [CrossRef]
- Madigan, T.; Johnson, J.R.; Clabots, C.; Johnston, B.D.; Porter, S.B.; Slater, B.S.; Banerjee, R. Extensive Household Outbreak of Urinary Tract Infection and Intestinal Colonization due to Extended-Spectrum β-Lactamase-Producing Escherichia coli Sequence Type 131. Clin. Infect. Dis. 2015, 61, e5–e12. [Google Scholar] [CrossRef] [PubMed]
- Huddleston, J.R. Horizontal gene transfer in the human gastrointestinal tract: Potential spread of antibiotic resistance genes. Infect. Drug Resist. 2014, 7, 167–176. [Google Scholar] [CrossRef] [PubMed]
- Oliver, S.P.; Jayarao, B.M.; Almeida, R.A. Foodborne pathogens in milk and the dairy farm environment: Food safety and public health implications. Foodborne Pathog. Dis. 2005, 2, 115–129. [Google Scholar] [CrossRef] [PubMed]
- Oliver, S.P.; Boor, K.J.; Murphy, S.C.; Murinda, S.E. Food safety hazards associated with consumption of raw milk. Foodborne Pathog. Dis. 2009, 6, 793–806. [Google Scholar] [CrossRef]
- Gelalcha, B.D.; Brown, S.M.; Crocker, H.E.; Agga, G.E.; Dego, O.K. Regulation Mechanisms of Virulence Genes in Enterohemorrhagic Escherichia coli. Foodborne Pathog. Dis. 2022, 19, 598–612. [Google Scholar] [CrossRef]
- Costard, S.; Espejo, L.; Groenendaal, H.; Zagmutt, F.J. Outbreak-Related Disease Burden Associated with Consumption of Unpasteurized Cow’s Milk and Cheese, United States, 2009–2014. Emerg. Infect. Dis. 2017, 23, 957–964. [Google Scholar] [CrossRef]
- Langer, A.J.; Ayers, T.; Grass, J.; Lynch, M.; Angulo, F.J.; Mahon, B.E. Nonpasteurized dairy products, disease outbreaks, and state laws—United States, 1993–2006. Emerg Infect. Dis. 2012, 18, 385–391. [Google Scholar] [CrossRef]
- Manyi-Loh, C.; Mamphweli, S.; Meyer, E.; Okoh, A. Antibiotic Use in Agriculture and Its Consequential Resistance in Environmental Sources: Potential Public Health Implications. Molecules 2018, 23, 795. [Google Scholar] [CrossRef]
- Straley, B.; Donaldson, S.; Hedge, N.; Sawant, A.; Srinivasan, V.; Oliver, S.; Jayarao, B. Public Health Significance of Antimicrobial-Resistant Gram-Negative Bacteria in Raw Bulk Tank Milk. Foodborne Pathog. Dis. 2006, 3, 222–233. [Google Scholar] [CrossRef]
- Kuehn, B. Drug-Resistant Infections from Raw Milk. JAMA 2018, 319, 1191. [Google Scholar] [CrossRef]
- Mungai, E.A.; Behravesh, C.B.; Gould, L.H. Increased outbreaks associated with nonpasteurized milk, United States, 2007–2012. Emerg. Infect. Dis. 2015, 21, 119–122. [Google Scholar] [CrossRef]
- Keene, W.E.; Hedberg, K.; Herriott, D.E.; Hancock, D.D.; McKay, R.W.; Barrett, T.J.; Fleming, D.W. A prolonged outbreak of Escherichia coli O157:H7 infections caused by commercially distributed raw milk. J. Infect. Dis. 1997, 176, 815–818. [Google Scholar] [CrossRef]
- Lim, J.Y.; Yoon, J.; Hovde, C.J. A brief overview of Escherichia coli O157:H7 and its plasmid O157. J. Microbiol. Biotechnol. 2010, 20, 5–14. [Google Scholar] [CrossRef]
- Herry, V.; Gitton, C.; Tabouret, G.; Répérant, M.; Forge, L.; Tasca, C.; Gilbert, F.B.; Guitton, E.; Barc, C.; Staub, C.; et al. Local immunization impacts the response of dairy cows to Escherichia coli mastitis, Local immunization impacts the response of dairy cows to Escherichia coli mastitis. Sci. Rep. 2017, 7, 3441. [Google Scholar] [CrossRef]
- Cho, Y.I.; Yoon, K.J. An overview of calf diarrhea—Infectious etiology, diagnosis, and intervention. J. Vet. Sci. 2014, 15, 1–17. [Google Scholar] [CrossRef]
- Scripps Local Media. Some Dairy Farmers Are Turning to Unpasteurized Milk. 2020. Available online: https://fox13now.com/2020/01/27/some-dairy-farmers-are-turning-to-unpasteurized-milk/ (accessed on 28 August 2022).
- Cody, S.H.; Abbott, S.L.; Marfin, A.A.; Schulz, B.; Wagner, P.; Robbins, K.; Mohle-Boetani, J.C.; Vugia, D.J. Two outbreaks of multidrug-resistant Salmonella serotype typhimurium DT104 infections linked to raw-milk cheese in Northern California. JAMA 1999, 281, 1805–1810. [Google Scholar] [CrossRef]
- United States Department of Agriculture, Animal and Plant Health Inspection Service. Economic Opportunities for Dairy Cow Culling Management Options; U.S. Department of Agriculture, Animal and Plant Health Inspection Service: Fort Collins, CO, USA, 1996.
- National Agricultural Statistics Service (NASS), Agricultural Statistics Board, United States Department of Agriculture (USDA). Livestock Slaughter. 2022. Available online: https://www.nass.usda.gov/Publications/Todays_Reports/reports/lstk0122.pdf (accessed on 28 August 2022).
- Troutt, H.F.; Osburn, B.I. Meat from dairy cows: Possible microbiological hazards and risks. Rev. Sci. Tech. 1997, 16, 405–414. [Google Scholar] [CrossRef]
- Marshall, K.E.H.; Tewell, M.; Tecle, S.; Leeper, M.; Sinatra, J.; Kissler, B.; Fung, A.; Brown, K.; Wagner, D.; Trees, E.; et al. Protracted Outbreak of Salmonella Newport Infections Linked to Ground Beef: Possible Role of Dairy Cows—21 States, 2016–2017. MMWR Morb. Mortal. Wkly. Rep. 2018, 67, 443–446. [Google Scholar] [CrossRef]
- Cobbold, R.N.; Rice, D.H.; Davis, M.A.; Besser, T.E.; Hancock, D.D. Long-term persistence of multi-drug-resistant Salmonella enterica serovar Newport in two dairy herds. J. Am. Vet. Med. Assoc. 2006, 228, 585–591. [Google Scholar] [CrossRef]
- Spika, J.S.; Waterman, S.H.; Hoo, G.W.; St Louis, M.E.; Pacer, R.E.; James, S.M.; Bissett, M.L.; Mayer, L.W.; Chiu, J.Y.; Hall, B. Chloramphenicol-Resistant Salmonella-Newport Traced through Hamburger to Dairy Farms—A Major Persisting Source of Human Salmonellosis in California. N. Engl. J. Med. 1987, 316, 565–570. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention (CDC). Outbreak of E. coli Infections Linked to Ground Beef; CDC: Atlanta, GA, USA, 2019.
- Centers for Disease Control and Prevention (CDC). Multistate Outbreak of E. coli O157:H7 Infections Linked to Ground Beef from Kroger/Nebraska Ltd.; (FINAL UPDATE); CDC: Atlanta, GA, USA, 2008.
- Centers for Disease Control and Prevention (CDC). Multistate Outbreak of E. coli O157:H7 Infections Associated with Beef from Fairbanks Farms; (FINAL UPDATE); CDC: Atlanta, GA, USA, 2009.
- Centers for Disease Control and Prevention (CDC). Multistate Outbreak of Shiga Toxin-Producing Escherichia coli O157:H7 Infections Linked to Ground Beef; CDC: Atlanta, GA, USA, 2014.
- Centers for Disease Control and Prevention (CDC). Multistate Outbreak of Shiga toxin-producing Escherichia coli O157:H7 Infections Linked to Beef Products Produced by Adams Farm; CDC: Atlanta, GA, USA, 2016.
- Centers for Disease Control and Prevention (CDC). Outbreak of E. coli Infections Linked to Ground Beef, 2018; CDC: Atlanta, GA, USA, 2018.
- Schwaber, M.J.; Navon-Venezia, S.; Schwartz, D.; Carmeli, Y. High Levels of Antimicrobial Coresistance among Extended-Spectrum- beta-Lactamase-Producing Enterobacteriaceae. Antimicrob. Agents Chemother. 2005, 49, 2137–2139. [Google Scholar] [CrossRef]
- Salah, F.D.; Soubeiga, S.T.; Ouattara, A.K.; Sadji, A.Y.; Metuor-Dabire, A.; Obiri-Yeboah, D.; Banla-Kere, A.; Karou, S.; Simpore, J. Distribution of quinolone resistance gene (qnr) in ESBL-producing Escherichia coli and Klebsiella spp. in Lome, Togo. Antimicrob. Resist. Infect. Control 2019, 8, 104. [Google Scholar] [CrossRef]
- Pereira, R.; Siler, J.; Ng, J.; Davis, M.; Grohn, Y.; Warnick, L. Effect of on-farm use of antimicrobial drugs on resistance in fecal Escherichia coli of preweaned dairy calves. J. Dairy Sci. 2014, 97, 7644–7654. [Google Scholar] [CrossRef]
- Jayarao, B.M.; Henning, D.R. Prevalence of foodborne pathogens in bulk tank milk. J. Dairy Sci. 2001, 84, 2157–2162. [Google Scholar] [CrossRef]
- Jayarao, B.M.; Wang, L. A study on the prevalence of gram-negative bacteria in bulk tank milk. J. Dairy Sci. 1999, 82, 2620–2624. [Google Scholar] [CrossRef]
- Jayarao, B.; Donaldson, S.; Straley, B.; Sawant, A.; Hegde, N.; Brown, J. A survey of foodborne pathogens in bulk tank milk and raw milk consumption among farm families in Pennsylvania. J. Dairy Sci. 2006, 89, 2451–2458. [Google Scholar] [CrossRef]
- Plumb, I.D.; Schwensohn, C.A.; Gieraltowski, L.; Tecle, S.; Schneider, Z.D.; Freiman, J.; Cote, A.; Noveroske, D.; Kolsin, J.; Brandenburg, J.; et al. Outbreak of Salmonella Newport Infections with Decreased Susceptibility to Azithromycin Linked to Beef Obtained in the United States and Soft Cheese Obtained in Mexico—United States, 2018–2019. MMWR Morb. Mortal. Wkly. Rep. 2019, 68, 713–717. [Google Scholar] [CrossRef] [PubMed]
- Del Collo, L.P.; Karns, J.S.; Biswas, D.; Lombard, J.E.; Haley, B.J.; Kristensen, R.C.; Kopral, C.A.; Fossler, C.P.; Van Kessel, J.A.S. Prevalence, antimicrobial resistance, and molecular characterization of Campylobacter spp. in bulk tank milk and milk filters from US dairies. J. Dairy Sci. 2017, 100, 3470–3479. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention (CDC). In Multistate Outbreak of Multidrug-Resistant Salmonella Heidelberg Infections Linked to Contact with Dairy Calves; (Final Update); 2017. Available online: https://www.cdc.gov/salmonella/heidelberg-11-16/index.html (accessed on 28 August 2022).
- Gupta, A.; Fontana, J.; Crowe, C.; Bolstorff, B.; Stout, A.; Van Duyne, S.; Hoekstra, M.P.; Whichard, J.M.; Barrett, T.J.; Angulo, F.J.; et al. Emergence of multidrug-resistant Salmonella enterica serotype Newport infections resistant to expanded-spectrum cephalosporins in the United States. J. Infect. Dis. 2003, 188, 1707–1716. [Google Scholar] [CrossRef]
- Bhutani, N.; Muraleedharan, C.; Talreja, D.; Rana, S.W.; Walia, S.; Kumar, A.; Walia, S.K. Occurrence of Multidrug Resistant Extended Spectrum Beta-Lactamase-Producing Bacteria on Iceberg Lettuce Retailed for Human Consumption. Biomed. Res. Int. 2015, 2015, 547547. [Google Scholar] [CrossRef]
- Kim, H.S.; Chon, J.W.; Kim, Y.J.; Kim, D.H.; Kim, M.S.; Seo, K.H. Prevalence and characterization of extended-spectrum-beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae in ready-to-eat vegetables. Int. J. Food Microbiol. 2015, 207, 83–86. [Google Scholar] [CrossRef]
- Boehme, S.; Werner, G.; Klare, I.; Reissbrodt, R.; Witte, W. Occurrence of antibiotic-resistant enterobacteria in agricultural foodstuffs. Mol. Nutr. Food. Res. 2004, 48, 522–531. [Google Scholar] [CrossRef]
- Liao, N.; Borges, C.A.; Rubin, J.; Hu, Y.; Ramirez, H.A.; Chen, J.; Zhou, B.; Zhang, Y.; Zhang, R.; Jiang, J.; et al. Prevalence of beta-Lactam Drug-Resistance Genes in Escherichia coli Contaminating Ready-to-Eat Lettuce. Foodborne Pathog. Dis. 2020, 17, 739–742. [Google Scholar] [CrossRef]
- Richter, L.; Du Plessis, E.M.; Duvenage, S.; Korsten, L. Occurrence, Identification, and Antimicrobial Resistance Profiles of Extended-Spectrum and AmpC beta-Lactamase-Producing Enterobacteriaceae from Fresh Vegetables Retailed in Gauteng Province, South Africa. Foodborne Pathog. Dis. 2019, 16, 421–427. [Google Scholar] [CrossRef]
- Bottichio, L.; Keaton, A.; Thomas, D.; Fulton, T.; Tiffany, A.; Frick, A.; Mattioli, M.; Kahler, A.; Murphy, J.; Otto, M.; et al. Shiga Toxin-Producing Escherichia coli Infections Associated With Romaine Lettuce-United States, 2018. Clin. Infect. Dis. 2020, 71, e323–e330. [Google Scholar] [CrossRef]
- Ribeiro, T.G.; Novais, Â.; Peixe, L.; Machado, E. Atypical epidemiology of CTX-M-15 among Enterobacteriaceae from a high diversity of non-clinical niches in Angola. J. Antimicrob. Chemother. 2016, 71, 1169–1177. [Google Scholar] [CrossRef]
- Wendel, A.M.; Johnson, D.H.; Sharapov, U.; Grant, J.; Archer, J.R.; Monson, T.; Koschmann, C.; Davis, J.P. Multistate Outbreak of Escherichia coli O157:H7 Infection Associated with Consumption of Packaged Spinach, August-September 2006: The Wisconsin Investigation. Clin. Infect. Dis. 2009, 48, 1079–1086. [Google Scholar] [CrossRef]
- Taylor, E.V.; Nguyen, T.A.; Machesky, K.D.; Koch, E.; Sotir, M.J.; Bohm, S.R.; Folster, J.P.; Bokanyi, R.; Kupper, A.; Bidol, S.A.; et al. Multistate outbreak of Escherichia coli O145 infections associated with romaine lettuce consumption, 2010. J. Food Prot. 2013, 76, 939–944. [Google Scholar] [CrossRef]
- Ackers, M.; Mahon, B.E.; Leahy, E.; Goode, B.; Damrow, T.; Hayes, P.S.; Bibb, W.F.; Rice, D.H.; Barrett, T.J.; Hutwagner, L.; et al. An outbreak of Escherichia coli O157:H7 infections associated with leaf lettuce consumption. J. Infect. Dis. 1998, 177, 1588–1593. [Google Scholar] [CrossRef]
- Sharapov, U.M.; Wendel, A.M.; Davis, J.P.; Keene, W.E.; Farrar, J.; Sodha, S.; Hyytia-Trees, E.; Leeper, M.; Gerner-Smidt, P.; Griffin, P.M.; et al. Multistate Outbreak of Escherichia coli O157:H7 Infections Associated with Consumption of Fresh Spinach: United States, 2006. J. Food Prot. 2016, 79, 2024–2030. [Google Scholar] [CrossRef]
- Raphael, E.; Wong, L.K.; Riley, L.W. Extended-spectrum Beta-lactamase gene sequences in gram-negative saprophytes on retail organic and nonorganic spinach. Appl. Env. Microbiol. 2011, 77, 1601–1607. [Google Scholar] [CrossRef]
- Berman, H.F.; Riley, L.W. Identification of novel antimicrobial resistance genes from microbiota on retail spinach. BMC Microbiol. 2013, 13, 272. [Google Scholar] [CrossRef]
- Marshall, K.E.; Hexemer, A.; Seelman, S.L.; Fatica, M.K.; Blessington, T.; Hajmeer, M.; Kisselburgh, H.; Atkinson, R.; Hill, K.; Sharma, D.; et al. Lessons Learned from a Decade of Investigations of Shiga Toxin-Producing Escherichia coli Outbreaks Linked to Leafy Greens, United States and Canada. Emerg. Infect. Dis. 2020, 26, 2319–2328. [Google Scholar] [CrossRef]
- Tadesse, D.A.; Li, C.; Mukherjee, S.; Hsu, C.-H.; Bodeis Jones, S.; Gaines, S.A.; Kabera, C.; Loneragan, G.H.; Torrence, M.; Harhay, D.M.; et al. Whole-Genome Sequence Analysis of CTX-M Containing Escherichia coli Isolates from Retail Meats and Cattle in the United States. Microb. Drug Resist. 2018, 24, 939–948. [Google Scholar] [CrossRef]
- Walia, S.; Rana, S.W.; Maue, D.; Rana, J.; Kumar, A.; Walia, S.K. Prevalence of multiple antibiotic-resistant Gram-negative bacteria on bagged, ready-to-eat baby spinach. Int. J. Environ. Health Res. 2013, 23, 108–118. [Google Scholar] [CrossRef]


| Sample from Conventional Farm | Method | Study Design/Population | Pathogens/Prevalence | State/ Region | Reference |
|---|---|---|---|---|---|
| Manure, bulk tank milk, manure fertilized soil | CAM, CSM, PCR of ESBL genes | A cross-sectional study on four dairy farms | Prevalence of CTXr E. coli was 20.5%, about 36% of BTM isolates were CTXr Over 83% of CTXr isolates carried ESBL genes | TN | [18] |
| Feces, swabs (pre-evisceration and carcass) | CSM, PCR of ESBL genes | Prospective study on veal calves from four cohorts (farms) | CTXr E. coli were 91%, 34% & 19% in feces, pre-evisceration and final carcass swabs, respectively. ESBL genes were detected in 89% of CTXr E. coli | OH | [124] |
| Feces | CSM, WGS | Matched-pair longitudinal study in CEF-treated and non-treated cows | More than 19 CEFr E. coli isolates and multiple ESBL genes found | TX, NM | [8] |
| Feces | Culture | A longitudinal study on cattle with clinical signs of salmonellosis and asymptomatic ones | The proportion of CEFr and CTRr Salmonella were 16.5% and 16%, respectively | CA | [125] |
| Feces | CSM, PCR of ESBL genes | A cross-sectional study on 747 dairy cattle from 25 conveniently selected dairy farms | More than 9% of E. coli isolates were CEFr, CTXr, and CPDr. All the 70 E. coli isolates carried the ESBL genes | OH | [126] |
| Feces | CSM, PCR of ESBL genes | On-farm from healthy dairy cattle and dairy cattle submitted for diagnostic purposes | Prevalence of CEFr Salmonella isolates were 35.8% and 1.8% among diagnostic and on-farm isolates, respectively | [127] | |
| Feces, lagoons, and milk filters | CSM, PCR of ESBL genes | A retrospective study on E. coli isolates banked from a previous survey of 30 dairy farms | The proportions of E. coli with ESBL genes were 53.5%, 57.1%, and 50.0% in feces, lagoon, and milk filters from 28 farms, respectively | WA | [57] |
| Feces | CSM, PCR of ESBL genes | A longitudinal study on 20 dairy heifer calves monthly for five months | About 93% of heifers harbored CEFr E. coli. The proportion of CEFr E. coli was 100%. ESBL and cephamycinase genes detected | PA | [111] |
| Milk | Culture, WGS | A cross-sectional study on milk from cows with mastitis from four farms | The prevalence of CEFr K. pneumoniae was 2.8%. ESBL genes detected | NY | [122] |
| Milk | Culture, AST | A retrospective study on 483 Klebsiella isolates from milk submitted for testing mastitis | The prevalence of CEFr Klebsiella spp. was 6.6% | WI | [76] |
| Feces | Culture, AST | A cross-sectional study on healthy and sick dairy cattle under different management systems | About 95% and 93% of E. coli isolates were CEFr and CTRr, respectively | CA | [128] |
| Composite manure | Culture, AST, and PCR of ESBL genes | A cross-sectional study on 80 dairy farms | CEFr and CTRr E. coli were identified in 31.2% and 36.4% of calves, respectively. Similarly, 6.2% and 5% of cows had CTRr, and CEFr E. coli isolates, respectively. E. coli carrying blaCTX-M was identified in about 5% of the farms | PA | [129] |
| Feces | Culture and AST | A prospective study on Salmonella suspected cases over eight months from 2,565 dairy cattle in 412 farms | The prevalence of CEFr Salmonella spp. was 60.4% | NY, PA, VT, MA, CT | [130] |
| Feces from pen floors | CSM and AST | A cross-sectional study on healthy and sick cows from four large-sized dairy farms | More than 51% of Salmonella isolates were CEFr, and all were susceptible to CTR | SW | [131] |
| Feces | CSM and AST | A longitudinal study on 110 dairy herds with five times sampling at a two-month interval | Prevalences of CEFr Salmonella isolates were 2.4%, 10%, and 10.8% in healthy cows, sick cows, and calves, respectively | NY, MI, MN, WI | [132] |
| ESBL Gene Type | Bacteria | State/Region | Sample | Reference |
|---|---|---|---|---|
| CTX-M-1 | E. coli | OH, WA, SW | Fecal | [8,29,130] |
| K. pneumoniae | NY | Mastitic milk | [122] | |
| CTX-M-12 | E. coli | WA | Fecal | [133] |
| CTX-14 | E. coli | OH, WA | Fecal | [24,126] |
| CTX-15 | E. coli | OH, WA, SW | Fecal | [8,24,126] |
| CTX-M-24 | E. coli | WA | Fecal | [24] |
| CTX-M-27 | E. coli | WA, SW | Fecal | [8,24] |
| CTX-M-32 | E. coli | SW | Fecal | [8] |
| CTX-M-55 | E. coli | WA, SW | Fecal | [8,24] |
| CTX-M-65 | E. coli | WA, SW | Fecal | [8,24] |
| CTX-M-79 | E. coli | OH | Fecal | [27] |
| CTX-M | Salmonella spp. | Not available | Feces from clinical case | [127] |
| E. coli | WA | Fecal | [57] | |
| E. coli | TN | Fecal & BTM | [18] | |
| E. coli | OH | Fecal & carcass swabs | [124] | |
| E. coli | PA | Fecal | [129] | |
| SHV | Salmonella spp. | Not available | Feces from clinical case | [127] |
| E. coli | WA | Fecal | [57] | |
| K. pneumoniae | NY | Mastitic milk | [122] | |
| TEM | E. coli | OH, WA | Fecal | [126,133] |
| E. coli | PA | Fecal | [111] | |
| Salmonella spp. | Not available | Fecal | [127] | |
| OXA-27 | E. coli | WA | Fecal | [133] |
| Beta-Lactam Antibiotic Resistance Gene | Host Bacterium | State | The Proportion of Total Beta-Lactam ARGs of Dairy Source | Time (year) |
|---|---|---|---|---|
| blaCTXM-27 | E. coli | WA, TX, OH and SD | 7.2% (7/97) | 2017, 2019 and 2021 |
| blaCTXM-55 | E. coli | TX, ND | 2.1% (2/97) | 2019 and 2020 |
| blaCXM-14 | E. coli | PA | 1% (1/97) | 2019 |
| blaCTXM-15 | E. coli | TX | 1% (1/97) | 2018 |
| blaCTXM-65 | E. coli | FL | 1% (1/97) | 2020 |
| blaTEM-1 | E. coli | TX, UT, WI, TN, WA, NY, OH, KS, MI, SD, CA, AZ, ID, PA and NE | 69.1% (67/97) | 2014–2020 |
| blaCMY-2 | E. coli | SD, MD, CA, PA, MI, ID, WA and WI | 14.4% (14/97) | 2018 |
| blaAmpC | E. coli | WI | 3.2% (1/31) | 2018 |
| blaOXA-2 | E. coli | MI | 1/97 | 2019 |
| blaCARB-2 | E. coli | CA | 1/97 | 2018 |
| blaSHV-12 | Salmonella | CA | 1/95 | 2018 |
| blaCMY-2 | Salmonella | UT, WA, WI, CA, TX, ID, UT, CO, AZ, TN and SC | 72.6% (69/95) | 2015–2020 |
| blaTEM-1 | Salmonella | WI, ID, CA, SD, WA, TX, UT, RI, IA and GA | 22.11% (21/95) | 2015–2019 |
| blaCARB-2 | Salmonella | WA, CA | 4.21% (4/95) | 2014 and 2016 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gelalcha, B.D.; Kerro Dego, O. Extended-Spectrum Beta-Lactamases Producing Enterobacteriaceae in the USA Dairy Cattle Farms and Implications for Public Health. Antibiotics 2022, 11, 1313. https://doi.org/10.3390/antibiotics11101313
Gelalcha BD, Kerro Dego O. Extended-Spectrum Beta-Lactamases Producing Enterobacteriaceae in the USA Dairy Cattle Farms and Implications for Public Health. Antibiotics. 2022; 11(10):1313. https://doi.org/10.3390/antibiotics11101313
Chicago/Turabian StyleGelalcha, Benti Deresa, and Oudessa Kerro Dego. 2022. "Extended-Spectrum Beta-Lactamases Producing Enterobacteriaceae in the USA Dairy Cattle Farms and Implications for Public Health" Antibiotics 11, no. 10: 1313. https://doi.org/10.3390/antibiotics11101313
APA StyleGelalcha, B. D., & Kerro Dego, O. (2022). Extended-Spectrum Beta-Lactamases Producing Enterobacteriaceae in the USA Dairy Cattle Farms and Implications for Public Health. Antibiotics, 11(10), 1313. https://doi.org/10.3390/antibiotics11101313






