Abstract
Staphylococcal cassette chromosome mec (SCCmec) typing was established in the 2000s and has been employed as a tool for the molecular epidemiology of methicillin-resistant Staphylococcus aureus, as well as the evolution investigation of Staphylococcus species. Molecular cloning and the conventional sequencing of SCCmec have been adopted to verify the presence and structure of a novel SCCmec type, while convenient PCR-based SCCmec identification methods have been used in practical settings for many years. In addition, whole-genome sequencing has been widely used, and various SCCmec and similar structures have been recently identified in various species. The current status of the SCCmec types, SCCmec subtypes, rules for nomenclature, and multiple methods for identifying SCCmec types and subtypes were summarized in this review, according to the perspective of the International Working Group on the Classification of Staphylococcal Cassette Chromosome Elements.
1. Introduction
Methicillin, a semisynthetic penicillin that specifically targets beta-lactamase-producing staphylococci, was introduced to the medical field in 1960. However, in the same year (1960), methicillin-resistant Staphylococcus aureus (MRSA) was identified [1,2]. Since then, MRSA has been the most prevalent and well-known antimicrobial-resistant bacteria for more than 60 years. In the first report of MRSA, it was described that MRSA existed before starting the use of methicillin but was rarely isolated in clinical settings, but after that, the prevalence of MRSA in healthcare settings increased gradually. For example, in the UK, the proportion of isolates of S. aureus from blood cultures that were methicillin-resistant increased in the 1990s and reached approximately 40% around 2000, though it decreased to less than 10% in the 2010s [3,4]. A high prevalence of antimicrobial resistance not only to methicillin but also to other antimicrobials was thought to have correlations with the inappropriate use of antimicrobials, so the prevalence of MRSA is still used as an indicator of good infection control and prevention practices and appropriateness of antimicrobial usage [5,6].
MRSA is known to produce an additional penicillin-binding protein designated as PBP2’ (PBP2a). PBP2’ exhibits a low affinity to most semisynthetic penicillin, such as methicillin, nafcillin, and oxacillin, as well as most cephalosporins. With its low affinity to beta-lactams and encoding gene mecA, PBP2’ was first reported in the mid-1980s [7,8,9]. Later, it was revealed that mecA is surrounded by genes that control its expression, mecR1 (encoding the signal transducer protein mecR1), and mecI (encoding the repressor protein mecI) [10,11]. When mecA was determined to be widely disseminated among multiple staphylococcal species, it was hypothesized that mecA could be carried on a mobile genetic element that could be transferred among staphylococcal species (Figure 1).
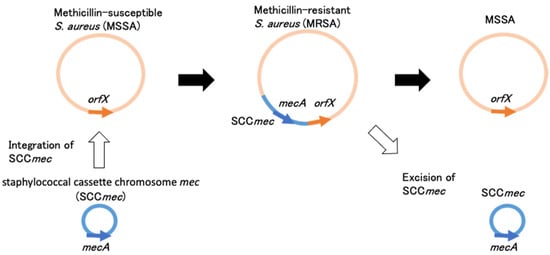
Figure 1.
Schematic representation of SCCmec excision and integration in S. aureus.
Mobile genetic elements containing mecA and their regulatory genes were designated as staphylococcal cassette chromosome mec (SCCmec) and were first reported in strain N315 (Sequence type 5, NY/JAPAN clone), of which the whole-genome sequence was subsequently revealed, followed by other strains, NCTC10442 and 85/2082 [12,13,14]. The SCCmec in NCTC10442, N315, and 85/2082 were designated as SCCmec I, SCCmec II, and SCCmec III, respectively [13].
The precise excision of SCCmec and integration of plasmid containing ccrA and ccrB was proven by experiments within a 24-h incubation period using N315 having SCCmec II [12]. However, the excision and integration of SCCmec from/to the S. aureus isolate is thought to happen in a complexed process in real clinical settings. It was observed that the excision and integration of small SCCmec such as SCCmec I could happen, but the integration of SCCmec II and V, which were larger than SCCmec I (>45 kb), could not happen even in vitro [15]. For large SCCmec structures, additional processes may be necessary to integrate into S. aureus, such as conjugative plasmids as a carrier or subsequent multiple recombination events. As an example, it was reported that the two isolates from a patient with chronic S. aureus infection of an intracardiac device changed from MSSA to MRSA during antimicrobial treatment, and the latter isolate possessed SCCmec II [16]. Some inversions and extra genetic fragments, which were not found in the original SCCmec II and its surrounding regions of N315, suggested that SCCmec II in the latter isolate did not directly integrate from N315, and additional processes might happen for integration.
2. International Working Group on the Classification of Staphylococcal Cassette Chromosome Elements (IWG-SCC)
Since the first reports of SCCmec I, II, and III in the early 2000s, various SCCmec elements have been reported by different researchers worldwide, and in addition to being adopted as a molecular epidemiology tool in healthcare settings, these elements have been utilized in the research on the evolution of staphylococci. These studies have also raised concerns about the confusion of the SCCmec nomenclature and the loss of its value. Therefore, the IWG-SCC released a report on the “Classification of Staphylococcal Cassette Chromosome mec (SCCmec): Guidelines for Reporting Novel SCCmec Elements” in 2009, followed by the Guidelines for Reporting Novel mecA Gene Homologues [17,18].
Recently, the IWG-SCC is updating its website, which contains information about IWG-SCC members, curators, requesting representative isolates of SCCmec types, current SCCmec types and subtypes approved by IWG-SCC, and IWG-SCC recommendations for designating and reporting new SCCmec elements (https://www.sccmec.org, under revision, accessed on: 14 December 2021).
3. Structure of SCCmec
The description provided in this section is based on the IWG-SCC recommendations, and additional information after the publication of these recommendations is presented [17,18].
3.1. Requirement for Defining SCCmec
According to the IWG-SCC recommendations, SCCmec is mainly characterized by four factors: (i) carriage of mecA in a mec gene complex, (ii) carriage of a ccr gene(s) (ccrAB and/or ccrC) in the ccr gene complex, (iii) integration at a specific site in the staphylococcal chromosome, designated as the integration site sequence (ISS) for SCC, which serves as a target for ccr-mediated recombination, and (iv) the presence of flanking direct repeat sequences containing the ISS [17]. As an example, a comparison of the structures of SCCmec I–V is presented in Figure 2. Several homologs of mecA have been reported recently, and instead of mecA, mecC is present in SCCmec IX. Details of the mecA homologs are described later in this review.

Figure 2.
Comparison of the structures of SCCmec I–V in S. aureus. SCCmec should contain the mec gene complex and ccr gene (complex), with integration site sequence and direct repeats at both ends. Adapted with permission from Ref. [17]. 2009, American Society for Microbiology.
3.2. ccr Gene Complex
The ccr gene complex comprises the ccr gene(s) and surrounding open reading frames, several of which have unknown functions. Currently, three phylogenetically distinct ccr genes, ccrA, ccrB, and ccrC, have been identified in S. aureus with DNA sequence similarities of less than 50%. Although the ccr sequences present in staphylococci other than S. aureus are remarkably diverse, novel ccr genes should be defined based on DNA sequence similarities of <50%, while novel allotypes of ccr genes should be designated if their DNA sequence similarity identities are between 50% and 85% [17]. Figure 3 presents a summary of the ccrA, ccrB, and ccrC genes and their allotypes. Wide implementations of whole-genome sequencing in the field of bacteriology may identify more diverse ccr genes from multiple species other than staphylococci; however, presently, this convention should be followed when naming novel ccr genes.
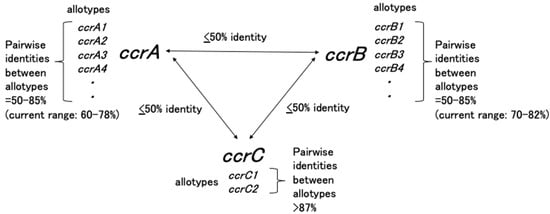
Figure 3.
Representation of the naming conventions for ccr genes in S. aureus. Adapted with permission from Ref. [17]. 2009, American Society for Microbiology.
The ccr gene complexes were numbered according to the timing of their descriptions. To date, two distinct groups have been reported: one carrying two adjacent ccr genes (ccrA and ccrB) and the other carrying only ccrC. For example, the ccr gene complexes identified in S. aureus include type 1 (carrying ccrA1B1), type 2 (carrying ccrA2B2), type 3 (carrying ccrA3B3), type 4 (carrying ccrA4B4), and type 5 (carrying ccrC).
3.3. mec Gene Complex
The mec gene complex comprises mecA, its regulatory genes, and the associated insertion sequences. Currently, three classes of the mec gene complex are recognized [17]. The class A mec gene complex (class A mec) is a prototype contained in SCCmec II. The class A mec gene complex contains mecA, the complete mecR1 and mecI regulatory genes upstream of mecA, and the hypervariable region (HVR) and insertion sequence IS431 downstream of mecA. The class B mec gene complex (class B mec) comprises mecA, a truncated mecR1 resulting from the insertion of IS1272 upstream of mecA, and HVR and IS431 downstream of mecA. The class C mec gene complex (class C mec) contains mecA and truncated mecR1 via the insertion of IS431 upstream of mecA and the HVR and IS431 downstream of mecA.
In the 21st century, multiple mecA homologs have been identified from staphylococci other than S. aureus and Macrococcus caseolyticus. Hence, the IWG-SCC released a commentary on the “Guidelines for Reporting Novel mecA Gene Homologues” in 2012 (Table 1) [18].

Table 1.
List of mecA homologs shown in the IWG-SCC guidelines. Adapted with permission from Ref. [18]. 2012, American Society for Microbiology.
The mecA genes with nucleotide sequence identities similar to the original mecA gene by 95% are designated as mecA, thus indicating that they are members of the allotype represented by the original mecA gene found in S. aureus N315. Those with nucleotide sequence identities to the original mecA of 70–95% are regarded as belonging to other allotypes of mecA. Accordingly, the mecA homologs detected in Staphylococcus sciuri, with a nucleotide sequence identity to the original mecA gene of approximately 80%, are designated as mecA1. The mecA homologs in Staphylococcus vitulinus, with nucleotide sequence identities similar to the original mecA gene of approximately 90%, are designated as mecA2.
In addition, the mec gene types are divided into allotypes, where each allotype encompasses a group of mec genes that share 70–95% nucleotide sequence identities to mecA of S. aureus N315 and mecB of Macrococcus caseolyticus JCSC5402, which was originally reported as mecAm, or mecC of S. aureus LGA251, which was isolated from bulk milk, daily cattle, and humans and originally designated as mecALGA251 [19,20]. Again, wide implementation of whole-genome sequencing may identify more diverse mec genes from multiple species other than staphylococci; however, presently, this convention should be followed when naming novel mec genes.
3.4. J Regions
In addition to the mec and ccr gene complexes, the SCCmec element also contains three J regions (previously called “junkyard regions”), which constitute nonessential components of the chromosome cassette. However, J regions may carry additional antimicrobial resistance determinants, and variations in the J regions within the same mec-ccr complex are adopted to define SCCmec subtypes. As illustrated in Figure 2, the J1 region (formerly called L-C) is located between the right chromosomal junction and the ccr complex, J2 (formerly called C-M) is between the ccr gene complex and the mec gene complex, and J3 (formerly called I-R) is between the mec complex and the left chromosomal junction.
4. Nomenclature of SCCmec and Its Subtypes
SCCmec elements are classified into types and subtypes by hierarchical systems. Currently, two methods are used to describe SCCmec types (Figure 4). These SSCCmec types are described by the combination of the mec gene complex class and the ccr gene complex type. SCCmec types can also be described using Roman numerals, such as SCCmec types I, II, and III. Generally, these two descriptions are reported and written together, for example, SCCmec type I (1B), SCCmec type II (2A), and SCCmec type III (3A).
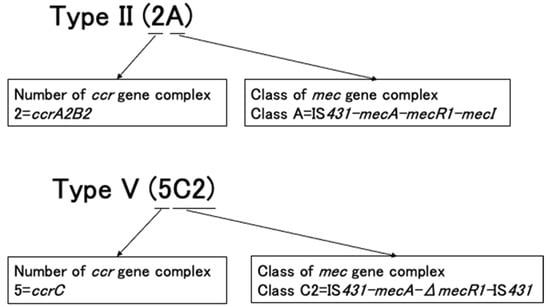
Figure 4.
Description of SCCmec under two methods.
SCCmec subtypes can be classified by differences in the J regions. The J regions contain characteristic genes, pseudogenes, noncoding regions, and mobile genetic elements such as insertion sequences and plasmids or transposons, which are used to define SCCmec subtypes. In the 2000s, some SCCmec subtypes could be reported with additional sequencing results of the J regions; however, this approach has a potential risk for subtype misclassification. Therefore, it is necessary to identify and report new SCCmec subtypes based on the entire nucleotide sequence. The following three methods have been adopted to describe subtypes of SCCmec: (i) expressing the J region differences as small letters, e.g., IVa, IVb, and IVc; (ii) expressing the differences due to the presence or absence of mobile genetic elements as capital letters, e.g., IA, IIA, and IVA; and (iii) expressing the differences in each J1, J2, and J3 region in Arabic numbers, which are designated according to the order of discovery, for example, II.1.1.1, II.1.1.2, and II.2.1.1. Currently, expressing J region differences as small letters is the most widely used to describe SCCmec subtypes. J1 region differences are mainly used to differentiate subtypes; however, some subtypes are characterized by unique contents in the J3 region.
5. Current SCCmec Types and Subtypes
The latest list of SCCmec types is presented in Table 2 and Figure 5, with the first official reports as references. Table 2 describes the details of the current SCCmec types, including the combinations of the ccr complex and mec complex and representative strains and their source, country, and reported year. Figure 5 shows the comparison of the structures of SCCmec listed in Table 2. To date, 14 SCCmec types have been officially reported and approved by the IWG-SCC.

Table 2.
Current SCCmec types (14 December 2021).
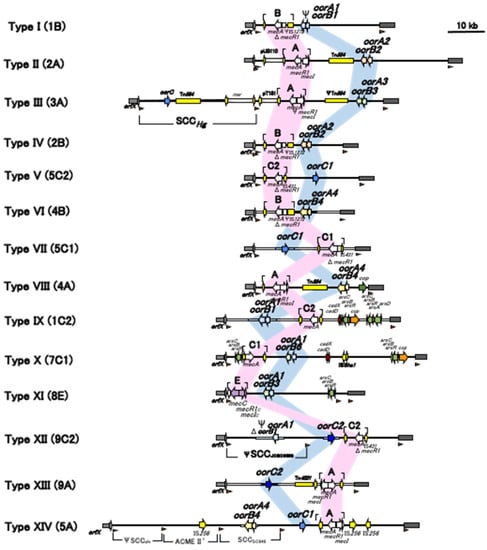
Figure 5.
Schematic comparison of the current SCCmec types (14 December 2021). Adapted with permissions from Ref. [17]. 2009, American Society for Microbiology, and Ref. [28]. 2018, Dr. Marc Stegger, and Ref. [29]. 2020, Dr. Noriko Urushibara.
SCCmec subtypes I, II, IV, and V are also presented in Table 3, together with their references. Table 3 shows the J1 region-based subtypes described by adding small letters, with one exception in SCCmec subtype IVn, which differs from the SCCmec subtype IVc in the J3 region. In addition to the J1 region-based subtypes, SCCmec subtypes IIB, IVE, and IVF were designated using a different method to express the differences due to the presence or absence of mobile genetic elements in the entire SCCmec structure as capital letters [30].

Table 3.
Current SCCmec I, II, IV and V subtypes (14 December 2021).
6. Methods to Identify SCCmec Types and Subtypes
6.1. PCR-Based Methods
PCR-based methods have been widely used to detect important components to differentiate SCCmec types and subtypes because of their relatively easy access to reagents and equipment. Single PCR and multiplex PCR are used to identify SCCmec types. Single PCR for each genomic component in SCCmec is a basic, generally sensitive, and specific method. The SCCmec type is identified by the combination of the detected components. However, it is difficult to guess the SCCmec type before analysis, and numerous different single PCRs are necessary for identification of the SCCmec type. Therefore, multiplex PCR methods have been reported as useful tools to detect multiple components in SCCmec and identify the SCCmec types and subtypes efficiently in one analysis, even though the sensitivity and specificity to detect each component were sometimes less than single PCRs. The multiplex PCR methods to which the IWG-SCC members contributed and were widely used are introduced here in this review. Dr. Zhang reported a multiplex PCR assay for characterization and concomitant subtyping of SCCmec types I, II, III, VIa, b, c, d, and V in 2005 and its updated version in 2012 [43,44]. Dr. Kondo formulated a combination of multiplex PCRs for rapid SCCmec type assignments, which could differentiate SCCmec types I, II, III, IV, V, VI, and representative J regions to define subtypes in 2007 [34]. In 2007, Dr. Milheirico communicated multiplex PCR strategies for the assignment of SCCmec types I, II, III, IV and its major subtypes, and V [37,45].
The limitations of multiplex PCR methods should be considered when interpreting the results. For example, the partial deletion of the ccrB2 gene present in Japanese USA300, which was designated as ψUSA300, could cause failure in identifying the SCCmec type in some multiplex PCR methods [46]. In 2020, Dr. Yamaguchi published a substantially beneficial review covering the history and concept of SCCmec and detailed the methods of SCCmec typing using multiplex PCR formulated by Dr. Kondo [34,47]. The limitations of multiplex PCR methods, such as problems in identifying the SCCmec type if one isolate had both SCCmec and SCC (i.e., structure of SCCmec without the mec complex) components or the failure of PCR caused by the deletion/insertion of sequences of targeted sites were discussed. Approaches for solutions of the problems were also mentioned, mainly about the necessity of additional single PCRs to detect indefinite components in SCCmec with different primer sets and PCR conditions.
6.2. Whole-Genome Sequencing-Based Methods
SCCmecFinder is a web-based tool for SCCmec typing using whole-genome sequences and was released by Dr. Kaya et al. in 2018 (https://cge.cbs.dtu.dk/services/SCCmecFinder/, accessed on 14 December 2021). SCCmecFinder identifies SCCmec elements in sequenced S. aureus isolates. The read data from major platforms for whole-genome sequencing or preassembled genome/contigs can be submitted to the website of SCCmecFinder. Users can obtain information about the prediction of SCCmec types based on the genes in the ccr complex, mec complex, and J regions, including the homology to the entire cassette [48]. Recently, a rapid method to detect MRSA, covering multiple SCCmec types and subtypes, directly from clinical samples has been developed [49]. The rapid confirmation of MRSA followed by whole-genome sequencing using the same samples and implementation of SCCmecFinder may contribute to reveal the diversity of SCCmec in various settings. However, as mentioned later, the objective confirmation of the structure is necessary to report new SCCmec, even though whole-genome sequencing has the potential to discover new ones easily.
7. How to Report New SCCmec and SCCmec Subtypes in the Era of Whole-Genome Sequencing: Role of IWG-SCC
Molecular cloning and conventional sequencing of SCCmec have been adopted for several years to identify and verify the structures of new SCCmec. However, these methods have rarely appeared in recent publications regarding new SCCmec elements. Alternatively, whole-genome sequencing has been widely used. In the era of whole-genome sequencing, in addition to S. aureus, numerous mec homologs, ccr homologs, and other components of SCCmec can be identified from other staphylococci and numerous species other than staphylococci. Furthermore, new SCCmec elements can be identified more frequently in isolates from animals or the environment. To maintain the value of identification of the SCCmec elements as a tool for molecular epidemiology, investigations on the evolution of bacteria and the relationship with antimicrobial resistance, researchers should contact the curator and obtain approval from the IWG-SCC before reporting/publishing new SCCmec and/or SCCmec subtypes to avoid duplicated names/numbers and reporting structures that are not SCCmec.
As aforementioned, a complete SCCmec sequence is necessary. The IWG-SCC strongly recommends the use of long-read sequencing technology (e.g., nanopore systems, PacBio systems, etc.) to verify the sequence of a new cassette chromosome, because the assembly of the entire SCCmec sequence sometimes solely fails with the results of short-read sequencing technology, possibly due to multiple insertion sequences.
To maintain the appropriate nomenclature of SCCmec types and subtypes, researchers are requested to send the isolates containing candidates of new SCCmec elements to the Statens Serum Institut (SSI) in Denmark and/or the National Institute of Infectious Diseases (NIID) in Japan, both of which serve as reference laboratories and reference strain banks of SCCmec. Subsequently, SSI and NIID will conduct long-read whole-genome sequencing and annotation to verify the suggested nomenclature by researchers. After depositing the strains to SSI and NIID, SSI and NIID will share the strains when requested from outside to contribute to SCCmec research worldwide. The original researchers will be acknowledged and asked permission to share the strains from SSI and/or NIID whenever necessary.
To date, complete structures of SCCmec present in S. aureus have been designated as new SCCmec type names, regardless of the host. However, the IWG-SCC has decided not to annotate new SCCmec subtypes in species other than S. aureus owing to the high complexity of the elements present in isolates other than S. aureus. An alternative nomenclature is “SCCmec[NAME OF THE STRAIN]”, which has already been adopted for non-aureus staphylococci. Discussions should be continued on the appropriate way of differentiating and naming the SCCmec structures present in both S. aureus and other species using whole-genome sequencing data and/or in silico investigations from public genome databases.
8. Conclusions
The discovery of SCCmec was based on the findings from multiple researchers about MRSA in the 20th century, and its importance in the fields of molecular epidemiology, infection control and prevention, and investigation of the evolution of MRSA has been developed along with the evolution of molecular analysis technologies. The implementation of whole-genome sequencing will contribute to reveal the entire relatedness of the SCCmec elements found not only in MRSA but also in other Staphylococcus species and Gram-positive bacteria. Importance as a tool of molecular typing might be declined if whole-genome sequencing is more widely used in clinical settings, but PCR-based methods will continue to be used as easy and costless tools for molecular epidemiology, infection control, and prevention. Therefore, the nomenclature rule of SCCmec should be followed, and the IWG-SCC should continue to update the most appropriate policy to designate SCCmec elements, especially for isolates other than S. aureus, or MRSA isolated from animals or the environment.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
This review is dedicated to the late Teruyo Ito and Keiichi Hiramatsu, both of whom were crucial to the research on staphylococci and contributed immensely to the IWG-SCC, as well as were great mentors in this field for several years.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Jevons, M.P. “Celbenin”—Resistant Staphylococci. Br. Med. J. 1961, 1, 124–125. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention. Brief History of Resistance and Antibiotics. Available online: https://www.cdc.gov/drugresistance/about.html#anchor_1552062951754 (accessed on 14 December 2021).
- Johnson, A.P.; Pearson, A.; Duckworth, G. Surveillance and epidemiology of MRSA bacteraemia in the UK. J Antimicrob. Chemother. 2005, 56, 455–462. [Google Scholar] [CrossRef] [PubMed]
- Horner, C.; Mushtaq, S.; Allen, M.; Longshaw, C.; Reynolds, R.; Livermore, D.M. Are resistance rates among bloodstream isolates a good proxy for other infections? Analysis from the BSAC resistance surveillance programme. J. Antimicrob. Chemother. 2021, 76, 1822–1831. [Google Scholar] [CrossRef] [PubMed]
- Centers for Disease Control and Prevention (CDC). Antibiotic/ Antimicrobial Resistance (AR/AMR) U.S. Nation Action Plan. 2013. Available online: https://www.cdc.gov/drugresistance/us-activities/national-action-plan.html (accessed on 1 January 2022).
- The Japan Goverment. National Action Plan on Antimicrobial Resistance (AMR) 2016–2020. 2016. Available online: https://www.mhlw.go.jp/file/06-Seisakujouhou-10900000-Kenkoukyoku/0000138942.pdf (accessed on 1 January 2022).
- Ubukata, K.; Yamashita, N.; Konno, M. Occurrence of a beta-lactam-inducible penicillin-binding protein in methicillin-resistant staphylococci. Antimicrob. Agents Chemother. 1985, 27, 851–857. [Google Scholar] [CrossRef]
- Utsui, Y.; Yokota, T. Role of an altered penicillin-binding protein in methicillin- and cephem-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 1985, 28, 397–403. [Google Scholar] [CrossRef] [PubMed]
- Matsuhashi, M.; Song, M.D.; Ishino, F.; Wachi, M.; Doi, M.; Inoue, M.; Ubukata, K.; Yamashita, N.; Konno, M. Molecular cloning of the gene of a penicillin-binding protein supposed to cause high resistance to beta-lactam antibiotics in Staphylococcus aureus. J. Bacteriol. 1986, 167, 975–980. [Google Scholar] [CrossRef]
- Hiramatsu, K.; Asada, K.; Suzuki, E.; Okonogi, K.; Yokota, T. Molecular cloning and nucleotide sequence determination of the regulator region of mecA gene in methicillin-resistant Staphylococcus aureus (MRSA). FEBS Lett. 1992, 298, 133–136. [Google Scholar] [CrossRef]
- Ito, T.; Katayama, Y.; Hiramatsu, K. Cloning and nucleotide sequence determination of the entire mec DNA of pre-methicillin-resistant Staphylococcus aureus N315. Antimicrob. Agents Chemother. 1999, 43, 1449–1458. [Google Scholar] [CrossRef]
- Katayama, Y.; Ito, T.; Hiramatsu, K. A new class of genetic element, staphylococcus cassette chromosome mec, encodes methicillin resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 2000, 44, 1549–1555. [Google Scholar] [CrossRef]
- Ito, T.; Katayama, Y.; Asada, K.; Mori, N.; Tsutsumimoto, K.; Tiensasitorn, C.; Hiramatsu, K. Structural comparison of three types of staphylococcal cassette chromosome mec integrated in the chromosome in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 2001, 45, 1323–1336. [Google Scholar] [CrossRef]
- Kuroda, M.; Ohta, T.; Uchiyama, I.; Baba, T.; Yuzawa, H.; Kobayashi, I.; Cui, L.; Oguchi, A.; Aoki, K.; Nagai, Y.; et al. Whole genome sequencing of meticillin-resistant Staphylococcus aureus. Lancet 2001, 357, 1225–1240. [Google Scholar] [CrossRef]
- Ito, T.; Tsubakishita, S.; Kuwahara-Arai, K.; Han, X.; Hiramatsu, K. Staphylococcal cassette chromosome (SCC): A unique gene transfer system in staphylococci. In Bacterial Integrative Mobile Genetic Elements; Roberts, A., Mullany, P., Eds.; Landes Bioscience: Austin, TX, USA, 2013; pp. 1–15. [Google Scholar]
- Harada, S.; Aoki, K.; Okamoto, K.; Kinoshita, O.; Nawata, K.; Ishii, Y.; Tateda, K.; Sasaki, M.; Saga, T.; Doi, Y.; et al. Left ventricular assist device-associated endocarditis involving multiple clones of Staphylococcus aureus with distinct antimicrobial susceptibility patterns. Int. J. Infect. Dis. 2019, 84, 44–47. [Google Scholar] [CrossRef] [PubMed]
- International Working Group on the Classification of Staphylococcal Cassette Chromosome. Classification of staphylococcal cassette chromosome mec (SCCmec): Guidelines for reporting novel SCCmec elements. Antimicrob. Agents Chemother. 2009, 53, 4961–4967. [Google Scholar] [CrossRef] [PubMed]
- Ito, T.; Hiramatsu, K.; Tomasz, A.; de Lencastre, H.; Perreten, V.; Holden, M.T.; Coleman, D.C.; Goering, R.; Giffard, P.M.; Skov, R.L.; et al. Guidelines for reporting novel mecA gene homologues. Antimicrob. Agents Chemother. 2012, 56, 4997–4999. [Google Scholar] [CrossRef] [PubMed]
- García-Álvarez, L.; Holden, M.T.G.; Lindsay, H.; Webb, C.R.; Brown, D.F.J.; Curran, M.D.; Walpole, E.; Brooks, K.; Pickard, D.J.; Teale, C.; et al. Meticillin-resistant Staphylococcus aureus with a novel mecA homologue in human and bovine populations in the UK and Denmark: A descriptive study. Lancet Infect. Dis. 2011, 11, 595–603. [Google Scholar] [CrossRef]
- Tsubakishita, S.; Kuwahara-Arai, K.; Sasaki, T.; Hiramatsu, K. Origin and molecular evolution of the determinant of methicillin resistance in staphylococci. Antimicrob. Agents Chemother. 2010, 54, 4352–4359. [Google Scholar] [CrossRef]
- Ma, X.X.; Ito, T.; Tiensasitorn, C.; Jamklang, M.; Chongtrakool, P.; Boyle-Vavra, S.; Daum, R.S.; Hiramatsu, K. Novel type of staphylococcal cassette chromosome mec identified in community-acquired methicillin-resistant Staphylococcus aureus strains. Antimicrob. Agents Chemother. 2002, 46, 1147–1152. [Google Scholar] [CrossRef] [PubMed]
- Ito, T.; Ma, X.X.; Takeuchi, F.; Okuma, K.; Yuzawa, H.; Hiramatsu, K. Novel type V staphylococcal cassette chromosome mec driven by a novel cassette chromosome recombinase, ccrC. Antimicrob. Agents Chemother. 2004, 48, 2637–2651. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, D.C.; Milheirico, C.; de Lencastre, H. Redefining a structural variant of staphylococcal cassette chromosome mec, SCCmec type VI. Antimicrob. Agents Chemother. 2006, 50, 3457–3459. [Google Scholar] [CrossRef] [PubMed]
- Berglund, C.; Ito, T.; Ikeda, M.; Ma, X.X.; Soderquist, B.; Hiramatsu, K. Novel type of staphylococcal cassette chromosome mec in a methicillin-resistant Staphylococcus aureus strain isolated in Sweden. Antimicrob. Agents Chemother. 2008, 52, 3512–3516. [Google Scholar] [CrossRef]
- Zhang, K.; McClure, J.A.; Elsayed, S.; Conly, J.M. Novel staphylococcal cassette chromosome mec type, tentatively designated type VIII, harboring class A mec and type 4 ccr gene complexes in a Canadian epidemic strain of methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 2009, 53, 531–540. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Li, S.; Skov, R.L.; Han, X.; Larsen, A.R.; Larsen, J.; Sorum, M.; Wulf, M.; Voss, A.; Hiramatsu, K.; Ito, T. Novel types of staphylococcal cassette chromosome mec elements identified in clonal complex 398 methicillin-resistant Staphylococcus aureus strains. Antimicrob. Agents Chemother. 2011, 55, 3046–3050. [Google Scholar] [CrossRef]
- Wu, Z.; Li, F.; Liu, D.; Xue, H.; Zhao, X. Novel type XII staphylococcal cassette chromosome mec harboring a new cassette chromosome recombinase, CcrC2. Antimicrob. Agents Chemother. 2015, 59, 7597–7601. [Google Scholar] [CrossRef]
- Baig, S.; Johannesen, T.B.; Overballe-Petersen, S.; Larsen, J.; Larsen, A.R.; Stegger, M. Novel SCCmec type XIII (9A) identified in an ST152 methicillin-resistant Staphylococcus aureus. Infect. Genet. Evol. 2018, 61, 74–76. [Google Scholar] [CrossRef] [PubMed]
- Urushibara, N.; Aung, M.S.; Kawaguchiya, M.; Kobayashi, N. Novel staphylococcal cassette chromosome mec (SCCmec) type XIV (5A) and a truncated SCCmec element in SCC composite islands carrying speG in ST5 MRSA in Japan. J. Antimicrob. Chemother. 2020, 75, 46–50. [Google Scholar] [CrossRef]
- Shore, A.; Rossney, A.S.; Keane, C.T.; Enright, M.C.; Coleman, D.C. Seven novel variants of the staphylococcal chromosomal cassette mec in methicillin-resistant Staphylococcus aureus isolates from Ireland. Antimicrob. Agents Chemother. 2005, 49, 2070–2083. [Google Scholar] [CrossRef]
- Han, X.; Ito, T.; Takeuchi, F.; Ma, X.X.; Takasu, M.; Uehara, Y.; Oliveira, D.C.; de Lencastre, H.; Hiramatsu, K. Identification of a novel variant of staphylococcal cassette chromosome mec, type II.5, and its truncated form by insertion of putative conjugative transposon Tn6012. Antimicrob. Agents Chemother. 2009, 53, 2616–2619. [Google Scholar] [CrossRef]
- Oliveira, D.C.; Milheirico, C.; Vinga, S.; de Lencastre, H. Assessment of allelic variation in the ccrAB locus in methicillin-resistant Staphylococcus aureus clones. J. Antimicrob. Chemother. 2006, 58, 23–30. [Google Scholar] [CrossRef]
- Hisata, K.; Kuwahara-Arai, K.; Yamanoto, M.; Ito, T.; Nakatomi, Y.; Cui, L.; Baba, T.; Terasawa, M.; Sotozono, C.; Kinoshita, S.; et al. Dissemination of methicillin-resistant Staphylococci among healthy Japanese children. J. Clin. Microbiol. 2005, 43, 3364–3372. [Google Scholar] [CrossRef]
- Kondo, Y.; Ito, T.; Ma, X.X.; Watanabe, S.; Kreiswirth, B.N.; Etienne, J.; Hiramatsu, K. Combination of multiplex PCRs for staphylococcal cassette chromosome mec type assignment: Rapid identification system for mec, ccr, and major differences in junkyard regions. Antimicrob. Agents Chemother. 2007, 51, 264–274. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.X.; Ito, T.; Chongtrakool, P.; Hiramatsu, K. Predominance of clones carrying Panton-Valentine leukocidin genes among methicillin-resistant Staphylococcus aureus strains isolated in Japanese hospitals from 1979 to 1985. J. Clin. Microbiol. 2006, 44, 4515–4527. [Google Scholar] [CrossRef] [PubMed]
- Kwon, N.H.; Park, K.T.; Moon, J.S.; Jung, W.K.; Kim, S.H.; Kim, J.M.; Hong, S.K.; Koo, H.C.; Joo, Y.S.; Park, Y.H. Staphylococcal cassette chromosome mec (SCCmec) characterization and molecular analysis for methicillin-resistant Staphylococcus aureus and novel SCCmec subtype IVg isolated from bovine milk in Korea. J. Antimicrob. Chemother. 2005, 56, 624–632. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Milheirico, C.; Oliveira, D.C.; de Lencastre, H. Multiplex PCR strategy for subtyping the staphylococcal cassette chromosome mec type IV in methicillin-resistant Staphylococcus aureus: ‘SCCmec IV multiplex’. J. Antimicrob. Chemother. 2007, 60, 42–48. [Google Scholar] [CrossRef]
- Berglund, C.; Ito, T.; Ma, X.X.; Ikeda, M.; Watanabe, S.; Soderquist, B.; Hiramatsu, K. Genetic diversity of methicillin-resistant Staphylococcus aureus carrying type IV SCCmec in Orebro County and the western region of Sweden. J. Antimicrob. Chemother. 2009, 63, 32–41. [Google Scholar] [CrossRef] [PubMed]
- Iwao, Y.; Ishii, R.; Tomita, Y.; Shibuya, Y.; Takano, T.; Hung, W.C.; Higuchi, W.; Isobe, H.; Nishiyama, A.; Yano, M.; et al. The emerging ST8 methicillin-resistant Staphylococcus aureus clone in the community in Japan: Associated infections, genetic diversity, and comparative genomics. J. Infect. Chemother. 2012, 18, 228–240. [Google Scholar] [CrossRef]
- Hosoya, S.; Ito, T.; Misawa, S.; Yoshiike, T.; Oguri, T.; Hiramatsu, T. MRSA clones identified in outpatients dermatology clinic. J. Jpn. Assoc. Intectious Dis. 2014, 88, 840–848. (In Japanese) [Google Scholar]
- Harris, T.M.; Bowen, A.C.; Holt, D.C.; Sarovich, D.S.; Stevens, K.; Currie, B.J.; Howden, B.P.; Carapetis, J.R.; Giffard, P.M.; Tong, S.Y.C. Investigation of trimethoprim/sulfamethoxazole resistance in an emerging sequence type 5 methicillin-resistant Staphylococcus aureus clone reveals discrepant resistance reporting. Clin. Microbiol. Infect. 2018, 24, 1027–1029. [Google Scholar] [CrossRef]
- Hisata, K.; Ito, T.; Matsunaga, N.; Komatsu, M.; Jin, J.; Li, S.; Watanabe, S.; Shimizu, T.; Hiramatsu, K. Dissemination of multiple MRSA clones among community-associated methicillin-resistant Staphylococcus aureus infections from Japanese children with impetigo. J. Infect. Chemother. 2011, 17, 609–621. [Google Scholar] [CrossRef]
- Zhang, K.; McClure, J.A.; Conly, J.M. Enhanced multiplex PCR assay for typing of staphylococcal cassette chromosome mec types I to V in methicillin-resistant Staphylococcus aureus. Mol. Cell Probes 2012, 26, 218–221. [Google Scholar] [CrossRef]
- Zhang, K.; McClure, J.A.; Elsayed, S.; Louie, T.; Conly, J.M. Novel multiplex PCR assay for characterization and concomitant subtyping of staphylococcal cassette chromosome mec types I to V in methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 2005, 43, 5026–5033. [Google Scholar] [CrossRef]
- Milheirico, C.; Oliveira, D.C.; de Lencastre, H. Update to the multiplex PCR strategy for assignment of mec element types in Staphylococcus aureus. Antimicrob. Agents Chemother. 2007, 51, 3374–3377. [Google Scholar] [CrossRef] [PubMed]
- Takadama, S.; Nakaminami, H.; Takii, T.; Noguchi, N. Identification and detection of USA300 methicillin-resistant Staphylococcus aureus clones with a partial deletion in the ccrB2 gene on the type IV SCCmec element. Diagn. Microbiol. Infect. Dis. 2019, 94, 86–87. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, T.; Ono, D.; Sato, A. Staphylococcal cassette chromosome mec (SCCmec) analysis of MRSA. Methods Mol. Biol. 2020, 2069, 59–78. [Google Scholar] [CrossRef] [PubMed]
- Kaya, H.; Hasman, H.; Larsen, J.; Stegger, M.; Johannesen, T.B.; Allesoe, R.L.; Lemvigh, C.K.; Aarestrup, F.M.; Lund, O.; Larsen, A.R. SCCmecFinder, a web-based tool for typing of staphylococcal cassette chromosome mec in Staphylococcus aureus using whole-genome sequence data. mSphere 2018, 3, e00612-17. [Google Scholar] [CrossRef]
- McClure, J.A.; Conly, J.M.; Obasuyi, O.; Ward, L.; Ugarte-Torres, A.; Louie, T.; Zhang, K. A novel assay for detection of methicillin-resistant Staphylococcus aureus directly from clinical samples. Front. Microbiol. 2020, 11, 1295. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).