D-Mannoside FimH Inhibitors as Non-Antibiotic Alternatives for Uropathogenic Escherichia coli
Abstract
1. Introduction
2. Results and Discussion
2.1. Bacterial Strains, Genotypic Confirmation, and Phylogenetic Analysis
2.2. Detection of Virulence Factors and Biofilm-Forming Ability
Biofilm-Forming Abilities and Congo Red Agar Method
2.3. Antimicrobial Susceptibility of Isolated UPEC
2.4. In Silico Evaluation of the Previously Characterized UPEC Isolates
Molecular Sequences of the fimH Gene of Uropathogenic Strains
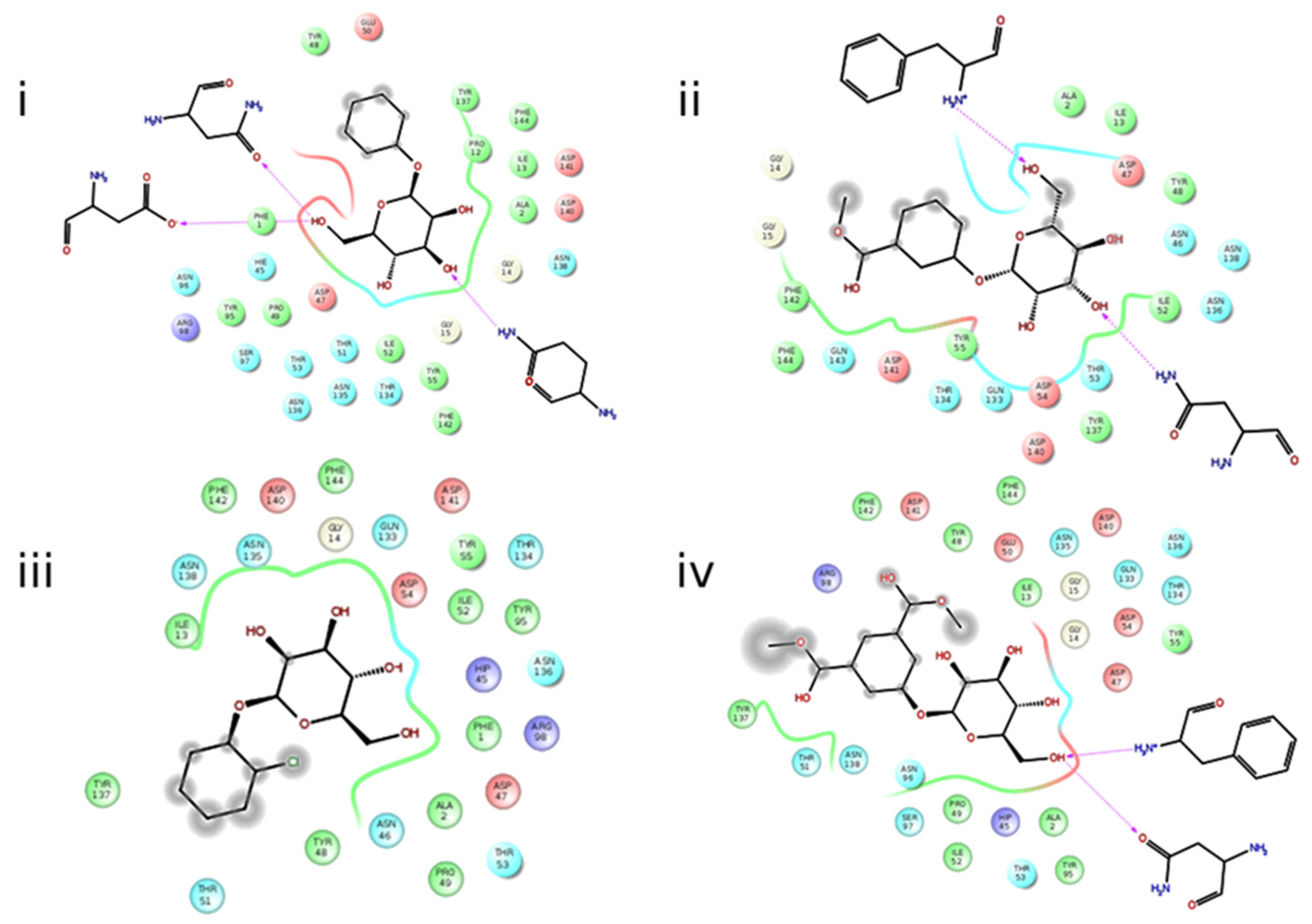
2.5. Molecular Docking
2.6. Molecular Dynamics Simulations
3. Conclusions
4. Materials and Methods
4.1. Bacterial Strains, Genotypic Confirmation, and Phylogenetic Analysis
4.2. Virulence Factors
4.2.1. Virulence Genes
4.2.2. Biofilm-Forming Ability
4.3. Antibiotic Susceptibility Testing and Detection of ESBL
4.3.1. Antibiotic Susceptibility Testing
4.3.2. Detection of ESBL
4.4. Statistical Analysis
4.5. In Silico Phase: Evaluation of the Previously Characterized UPEC Isolates
4.5.1. Molecular Sequences of the fimH Gene in the UPEC Strains
4.5.2. Obtaining Clean Sequences and Protein Analysis
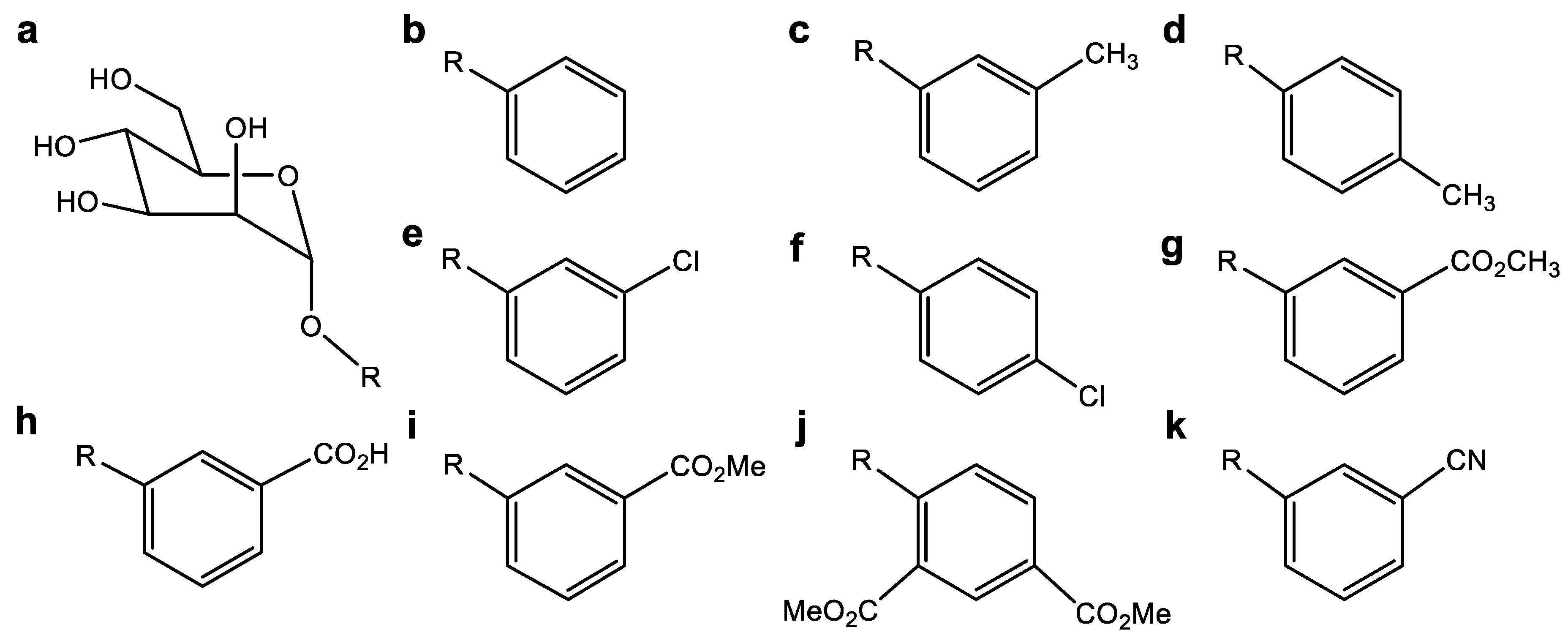
4.6. Inhibitor Molecule Design (Ligand Assay)
4.7. Molecular Docking
4.8. Molecular Dynamic Simulation
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Acknowledgments
Conflicts of Interest
References
- Foxman, B. Urinary tract infection syndromes: Occurrence, recurrence, bacteriology, risk factors, and disease burden. Infect. Dis. Clin. N. Am. 2014, 28, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Ren, Y.; Palusiak, A.; Wang, W.; Wang, Y.; Li, X.; Wei, H.; Wang, Q. A high-resolution typing assay for uropathogenic Escherichia coli based on fimbrial diversity. Front. Microbiol. 2016, 7, 623. [Google Scholar] [CrossRef] [PubMed]
- Wirth, T.; Falush, D.; Lan, R.; Colles, F.; Mensa, P.; Wieler, L.H.; Achtman, M. Sex and virulence in Escherichia coli: An evolutionary perspective. Mol. Microbiol. 2006, 60, 1136–1151. [Google Scholar] [CrossRef]
- Vagarali, M.A.; Karadesai, S.G.; Patil, C.S.; Metgud, S.C.; Mutnal, M.B. Haemagglutination and siderophore production as the urovirulence markers of uropathogenic Escherichia coli. Indian J. Med. Microbiol. 2008, 26, 68. [Google Scholar] [CrossRef]
- Hossain, M.; Tabassum, T.; Rahman, A.; Hossain, A.; Afroze, T.; Momen, A.M.I.; Khaleque, A. Genotype–phenotype correlation of β-lactamase-producing uropathogenic Escherichia coli (UPEC) strains from Bangladesh. Sci. Rep. 2020, 10, 14549. [Google Scholar] [CrossRef]
- Spaulding, C.; Hultgren, S. Adhesive pili in UTI pathogenesis and drug development. Pathogens 2010, 5, 30. [Google Scholar] [CrossRef] [PubMed]
- Tenaillon, O.; Skurnik, D.; Picard, B.; Denamur, E. The population genetics of commensal Escherichia coli. Nat. Rev. Microbiol. 2010, 8, 207. [Google Scholar] [CrossRef]
- Terlizzi, M.E.; Gribaudo, G.; Maffei, M.E. UroPathogenic Escherichia coli (UPEC) infections: Virulence factors, bladder responses, antibiotic, and non-antibiotic antimicrobial strategies. Front. Microbiol. 2007, 8, 1566. [Google Scholar] [CrossRef] [PubMed]
- Kline, K.A.; Fälker, S.; Dahlberg, S.; Normark, S.; Henriques-Normark, B. Bacterial adhesins in host-microbe interactions. Cell Host Microbe 2009, 5, 580–592. [Google Scholar] [CrossRef] [PubMed]
- Behzadi, P. Classical chaperone-usher (CU) adhesive fimbriome: Uropathogenic Escherichia coli (UPEC) and urinary tract infections (UTIs). Folia Microbiol. 2020, 65, 45–65. [Google Scholar] [CrossRef]
- Schwartz, D.J.; Chen, S.L.; Hultgren, S.J.; Seed, P. CPopulation dynamics and niche distribution of uropathogenic Escherichia coli during acute and chronic urinary tract infection. Infect. Immun. 2011, 79, 4250–4259. [Google Scholar] [CrossRef] [PubMed]
- Mydock-McGrane, L.K.; Hannan, T.J.; Janetka, J.W. Rational design strategies for FimH antagonists: New drugs on the horizon for urinary tract infection and Crohn’s disease. Expert Opin. Drug Discov. 2017, 12, 711–731. [Google Scholar] [CrossRef] [PubMed]
- Klein, T.; Abgottspon, D.; Wittwer, M.; Rabbani, S.; Herold, J.; Jiang, X.; Gubler, E. FimH antagonists for the oral treatment of urinary tract infections: From design and synthesis to in vitro and in vivo evaluation. J. Med. Chem. 2010, 53, 8627–8641. [Google Scholar] [CrossRef] [PubMed]
- Chalopin, T.; Brissonnet, Y.; Sivignon, A.; Deniaud, D.; Cremet, L.; Barnich, N.; Bouckaert, J.; Gouin, S.G. Inhibition profiles of mono- and polyvalent FimH antagonists against 10 different Escherichia coli strains. Org. Biomol. Chem. 2015, 13, 11369–11375. [Google Scholar] [CrossRef] [PubMed]
- Gouin, S.G.; Wellens, A.; Bouckaert, J.; Kovensky, J. Synthetic multimeric heptyl mannosides as potent antiadhesives of uropathogenic Escherichia coli. ChemMedChem Chem. Enabling Drug Discov. 2009, 4, 749–755. [Google Scholar] [CrossRef]
- Scharenberg, M.; Abgottspon, D.; Cicek, E.; Jiang, X.; Schwardt, O.; Rabbani, S.; Ernst, B. A flow cytometry-based assay for screening FimH antagonists. Assay Drug Dev. Technol. 2011, 9, 455–464. [Google Scholar] [CrossRef]
- Mian, M.F.; Lauzon, N.M.; Andrews, D.W.; Lichty, B.D.; Ashkar, A.A. FimH can directly activate human and murine natural killer cells via TLR4. Mol. Ther. 2010, 18, 1379–1388. [Google Scholar] [CrossRef]
- Eldridge, G.R.; Hughey, H.; Rosenberger, L.; Martin, S.M.; Shapiro, A.M.; D’Antonio, E.; Starks, C.M. Safety and immunogenicity of an adjuvanted Escherichia coli adhesin vaccine in healthy women with and without histories of recurrent urinary tract infections: Results from a first-in-human phase 1 study. Hum. Vaccines Immunother. 2021, 17, 1262–1270. [Google Scholar] [CrossRef]
- Starks, C.M.; Miller, M.M.; Broglie, P.M.; Cubbison, J.; Martin, S.M.; Eldridge, G.R. Optimization and qualification of an assay that demonstrates that a FimH vaccine induces functional antibody responses in women with histories of urinary tract infections. Hum. Vaccines Immunother. 2021, 17, 283–292. [Google Scholar] [CrossRef]
- Gordon, D.M.; O'Brien, C.L.; Pavli, P. Escherichia coli diversity in the lower intestinal tract of humans. Environ. Microbiol. Rep. 2015, 7, 642–648. [Google Scholar] [CrossRef]
- Johnson, J.R.; Johnston, B.D.; Porter, S.; Thuras, P.; Aziz, M.; Price, L.B. Accessory traits and phylogenetic background predict Escherichia coli extraintestinal virulence better than does ecological source. J. Infact. Dis. 2019, 219, 121–132. [Google Scholar] [CrossRef] [PubMed]
- Johnson, J.R.; Russo, T.A. Molecular epidemiology of extraintestinal pathogenic (Uropathogenic) Escherichia coli. Int. J. Med. Microbiol. 2005, 295, 383–404. [Google Scholar] [CrossRef] [PubMed]
- Dadi, B.R.; Abebe, T.; Zhang, L.; Mihret, A.; Abebe, W.; Amogne, W. Distribution of virulence genes and phylogenetics of uropathogenic Escherichia coli among urinary tract infection patients in Addis Ababa, Ethiopia. BMC. Infact. Dis. 2020, 20, 108. [Google Scholar] [CrossRef] [PubMed]
- Mojaz-Dalfardi, N.; Kalantar-Neyestanaki, D.; Hashemizadeh, Z.; Mansouri, S. Comparison of virulence genes and phylogenetic groups of Escherichia coli isolates from urinary tract infections and normal fecal flora. Gene Rep. 2020, 100709. [Google Scholar] [CrossRef]
- Baldiris-Avila, R.; Montes-Robledo, A.; Buelvas-Montes, Y. Phylogenetic Classification, Biofilm-Forming Capacity, Virulence Factors, and Antimicrobial Resistance in Uropathogenic Escherichia coli (UPEC). Curr. Microbiol. 2020, 3, 1–10. [Google Scholar] [CrossRef]
- Spurbeck, R.R.; Dinh, P.C.; Walk, S.T.; Stapleton, A.E.; Hooton, T.M.; Nolan, L.K.; Mobley, H.L. Escherichia coli that carry vat, fyuA, chuA, and yfcV efficiently colonize the urinary tract. Infect. Immun. 2012, 80, 4115–4122. [Google Scholar] [CrossRef]
- Watnick, P.; Kolter, R.; Bacteriol, J. Biofilm, city of microbes. J. Bacteriol. 2000, 182, 2675–2679. [Google Scholar] [CrossRef]
- Baldiris, R.; Teheran, V.; Vivas-Reyes, R.; Montes, A.; Arzuza, O. Anti-biofilm activity of ibuprofen and diclofenac against some biofilm producing Escherichia coli and Klebsiella pneumoniae uropathogens. Afr. J. Microbiol. Res. 2016, 10, 1675–1684. [Google Scholar] [CrossRef]
- Barnhart, M.M.; Lynem, J.; Chapman, M.R. GlcNAc-6P levels modulate the expression of Curli fibers by Escherichia coli. J. Bacteriol. 2006, 188, 5212–5219. [Google Scholar] [CrossRef] [PubMed]
- Machado-Alba, J.E.; Valladales-Restrepo, L.F.; Gaviria-Mendoza, A.; Machado-Duque, M.E.; Figueras, A. Patterns of antibiotic prescription in Colombia: Are there differences between capital cities and municipalities? Antibiotics 2020, 9, 389. [Google Scholar] [CrossRef] [PubMed]
- De La Cadena, E.; Mojica, M.F.; Castillo, N.; Correa, A.; Appel, T.M.; García-Betancur, J.C.; Villegas, M.V. Genomic analysis of ctx-m-group-1-producing extraintestinal pathogenic E. coli (ExPEC) from patients with urinary tract infections (UTI) from Colombia. Antibiotics 2020, 9, 899. [Google Scholar] [CrossRef] [PubMed]
- Toscano, Y.G.; Támara, M.F.; Urbina, M.C.; Rodriguez, L.G.; Martínez, A.B. Perfiles de los fenotipos de resistencia en Escherichia coli y Klebsiella pneumoniae en Barranquilla, Colombia. J. Biomed. Sci. 2020, 9, 15–24. [Google Scholar] [CrossRef]
- Mathers, A.J.; Peirano, G.; Pitout, J.D. Escherichia coli ST131: The quintessential example of an international multiresistant high-risk clone. Adv. Appl. Microbiol. 2015, 90, 109–154. [Google Scholar] [CrossRef]
- Mirkalantari, S.; Masjedian, F.; Irajian, G.; Siddig, E.E.; Fattahi, A. Determination of the frequency of β-lactamase genes (bla SHV, bla TEM, bla CTX-M) and phylogenetic groups among ESBL-producing uropathogenic Escherichia coli isolated from outpatients. J. Lab. Med. 2020, 44, 27–33. [Google Scholar] [CrossRef]
- Hasanzadeh, S.; Habibi, M.; Shokrgozar, M.A.; Cohan, R.A.; Ahmadi, K.; Karam, M.R.A.; Bouzari, S. In silico analysis and in vivo assessment of a novel epitope-based vaccine candidate against uropathogenic Escherichia coli. Sci. Rep. 2020, 10, 16258. [Google Scholar] [CrossRef]
- Yun, K.W.; Do Soo Kim, D.S.; Kim, W.Y.; Lim, I.S. Molecular typing of uropathogenic Escherichia coli isolated from Korean children with urinary tract infection. Korean J. Pediatr. 2015, 58, 20. [Google Scholar] [CrossRef]
- Bouckaert, J.; Berglund, J.; Schembri, M.; De Genst, E.; Cools, L.; Wuhrer, M.; Hung, C.S.; Pinkner, J.; Slättegård, R.; Zavialov, A.; et al. Receptor binding studies disclose a novel class of high-affinity inhibitors of the Escherichia coli FimH adhesin. Mol. Microbiol. 2005, 55, 441–455. [Google Scholar] [CrossRef] [PubMed]
- Warren, G.L.; Andrews, C.W.; Capelli, A.M.; Clarke, B.; LaLonde, J.; Lambert, M.H.; Tedesco, G. A critical assessment of docking programs and scoring functions. J. Med. Chem. 2006, 49, 5912–5931. [Google Scholar] [CrossRef]
- Alam, A.; Hosen, M.A.; Hosen, A.; Fujii, Y.; Ozeki, Y.; Abe Kawsar, S.M. Synthesis, Characterization, and Molecular Docking Against a Receptor Protein FimH of Escherichia coli (4XO8) of Thymidine Derivatives. J. Mex. Chem. Soc. 2021, 65, 256–276. [Google Scholar] [CrossRef]
- Mydock-McGrane, L.; Cusumano, Z.; Han, Z.; Binkley, J.; Kostakioti, M.; Hannan, T.; Rath, N.P. Antivirulence C-mannosides as antibiotic-sparing, oral therapeutics for urinary tract infections. J. Med. Chem. 2016, 59, 9390–9408. [Google Scholar] [CrossRef]
- Mashraqi, M.M.; Chaturvedi, N.; Alam, Q.; Alshamrani, S.; Bahnass, M.M.; Ahmad, K.; Rizvi, S.M.D. Biocomputational Prediction Approach Targeting FimH by Natural SGLT2 Inhibitors: A Possible Way to Overcome the Uropathogenic Effect of SGLT2 Inhibitor Drugs. Molecules 2021, 26, 582. [Google Scholar] [CrossRef]
- Singaravelu, M.; Selvan, A.; Anishetty, S. Molecular dynamics simulations of lectin domain of FimH and immunoinformatics for the design of potential vaccine candidates. Comput. Biol. Chem. 2014, 52, 18–24. [Google Scholar] [CrossRef] [PubMed]
- Tomašič, T.; Rabbani, S.; Jakob, R.P.; Reisner, A.; Jakopin, Ž.; Maier, T.; Anderluh, M. Does targeting Arg98 of FimH lead to high affinity antagonists? Eur. J. Med. Chem. 2021, 211, 113093. [Google Scholar] [CrossRef] [PubMed]
- Ardila-Leal, L.D.; Monterey-Gutiérrez, P.A.; Poutou-Piñales, R.A.; Quevedo-Hidalgo, B.E.; Galindo, J.F.; Pedroza-Rodríguez, A.M. Recombinant laccase rPOXA 1B real-time, accelerated and molecular dynamics stability study. BMC Biotechnol. 2021, 21, 1–18. [Google Scholar] [CrossRef]
- Dumych, T.; Bridot, C.; Gouin, S.G.; Lensink, M.F.; Paryzhak, S.; Szunerits, S.; Krammer, E.M. A novel integrated way for deciphering the glycan code for the FimH lectin. Molecules 2018, 23, 2794. [Google Scholar] [CrossRef]
- Bouckaert, J.; Mackenzie, J.; de Paz, J.L.; Chipwaza, B.; Choudhury, D.; Zavialov, A.; Mannerstedt, K.; Anderson, J.; Piérard, D.; Wyns, L.; et al. The affinity of the FimH fimbrial adhesin is receptor-driven and quasi-independent of Escherichia coli pathotypes. Mol. Microbiol. 2006, 61, 1556–1568. [Google Scholar] [CrossRef] [PubMed]
- Munera, D.; Hultgren, S.; Fernández, L.Á. Recognition of the N-terminal lectin domain of FimH adhesin by the usher FimD is required for type 1 pilus biogenesis. Mol. Microbiol. 2007, 64, 333–346. [Google Scholar] [CrossRef] [PubMed]
- Acosta-Tapia, N.; Galindo, J.F.; Baldiris, R. Insights into the Effect of Lowe Syndrome-Causing Mutation p. Asn591Lys of OCRL-1 through Protein–Protein Interaction Networks and Molecular Dynamics Simulations. J. Chem. Inf. Model. 2020, 60, 1019–1027. [Google Scholar] [CrossRef] [PubMed]
- Weinstein, M.P.; Patel, J.B.; Bobenchik, A.M. Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing, 27th ed.; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2017; p. 296. [Google Scholar]
- Gómez-Duarte, O.G.; Arzuza, O.; Urbina, D.; Bai, J.; Guerra, J.; Montes, O.; Castro, G.Y. Detection of Escherichia coli enteropathogens by multiplex polymerase chain reaction from children′s diarrheal stools in two Caribbean–Colombian cities. Foodborne Pathog. Dis. 2010, 7, 199–206. [Google Scholar] [CrossRef]
- Clermont, O.; Christenson, J.K.; Denamur, E.; Gordon, D.M. The C lermont Escherichia coli phylo-typing method revisited: Improvement of specificity and detection of new phylo-groups. Environ. Microbiol. Rep. 2013, 5, 58–65. [Google Scholar] [CrossRef]
- Johnson, J.R.; Stell, A.L. Extended virulence genotypes of Escherichia coli strains from patients with urosepsis in relation to phylogeny and host compromise. J. Infact. Dis. 2000, 181, 261–272. [Google Scholar] [CrossRef] [PubMed]
- Nakano, M.; Yamamoto, S.; Terai, A.; Ogawa, O.; Makino, S.I.; Hayashi, H.; Kurazono, H. Structural and sequence diversity of the pathogenicity island of uropathogenic Escherichia coli which encodes the USP protein. FEMS Microbiol. Lett. 2001, 205, 71–76. [Google Scholar] [CrossRef]
- Freeman, D.J.; Falkiner, F.R.; Keane, C.T. New method for detecting slime production by coagulase negative staphylococci. J. Clin. Pathog. 1989, 42, 872–874. [Google Scholar] [CrossRef] [PubMed]
- Bokranz, W.; Wang, X.; Tschäpe, H.; Römling, U. Expression of cellulose and curli fimbriae by Escherichia coli isolated from the gastrointestinal tract. J. Med. Microbiol. 2005, 54, 1171–1182. [Google Scholar] [CrossRef]
- O'Toole, G.A.; Kolter, R. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: A genetic analysis. Mol. Microbiol. 1998, 28, 449–461. [Google Scholar] [CrossRef]
- Stepanović, S.; Vuković, D.; Hola, V.; Bonaventura, G.D.; Djukić, S.; Ćirković, I.; Ruzicka, F. Quantification of biofilm in microtiter plates: Overview of testing conditions and practical recommendations for assessment of biofilm production by staphylococci. J. Appl. Microbiol. 2007, 115, 891–899. [Google Scholar] [CrossRef] [PubMed]
- Krumperman, P.H. Multiple antibiotic resistance indexing of Escherichia coli to identify high-risk sources of fecal contamination of foods. Appl. Environ. Microbiol. 1983, 46, 165–170. [Google Scholar] [CrossRef]
- Lezameta, L.; Gonzáles-Escalante, E.; Tamariz, J.H. Comparación de cuatro métodos fenotípicos para la detección de beta-lactamasas de espectro extendido. Revista Peruana de Medicina Experimental Y Salud Publica 2010, 27, 345–351. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Oliver, A.; Weigel, L.M.; Rasheed, J.K.; McGowan, J.E.; Raney, P.; Tenover, F.C. Mechanisms of decreased susceptibility to cefpodoxime in Escherichia coli. Antimicrob. Agents Chemother. 2002, 46, 3829–3836. [Google Scholar] [CrossRef]
- Conceicao, T.; Brizio, A.; Duarte, A.; Lito, L.M.; Cristino, J.M.; Salgado, M.J. First description of CTX-M-15-producing Klebsiella pneumoniae in Portugal. Antimicrob. Agents Chemother. 2005, 49, 477–478. [Google Scholar] [CrossRef] [PubMed]
- Sokurenko, E.V.; Feldgarden, M.; Trintchina, E.; Weissman, S.J.; Avagyan, S.; Chattopadhyay, S.; Dykhuizen, D.E. Selection footprint in the FimH adhesin shows pathoadaptive niche differentiation in Escherichia coli. Mol. Biol. Evol. 2004, 21, 1373–1383. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Mydock-McGrane, L.K.; Cusumano, Z.T.; Janetka, J.W. Mannose-derived FimH antagonists: A promising anti-virulence therapeutic strategy for urinary tract infections and Crohn’s disease. Expert Opin. Ther. Pat. 2016, 26, 175–197. [Google Scholar] [CrossRef] [PubMed]
- Han, Z.; Pinkner, J.S.; Ford, B.; Obermann, R.; Nolan, W.; Wildman, S.A.; Janetka, J.W. Structure-based drug design and optimization of mannoside bacterial FimH antagonists. J. Med. Chem. 2010, 53, 4779–4792. [Google Scholar] [CrossRef] [PubMed]
- Sanner, M.F.; Olson, A.J.; Spehner, J.C. Reduced surface: An efficient way to compute molecular surfaces. Biopolymers 1996, 38, 305–320. [Google Scholar] [CrossRef]
- Menon, V.V.; Mary, Y.S.; Mary, Y.S.; Panicker, C.Y.; Bielenica, A.; Armaković, S.; Van Alsenoy, C. Combined spectroscopic, DFT, TD-DFT and MD study of newly synthesized thiourea derivative. J. Mol. Struct. 2018, 1155, 184–195. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef]
- Morris, G.M.; Goodsell, D.S.; Halliday, R.S.; Huey, R.; Hart, W.E.; Belew, R.K.; Olson, A.J. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J. Comput. Chem. 1998, 19, 1639–1662. [Google Scholar] [CrossRef]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef]
- Kameshwar, A.K.S.; Barber, R.; Qin, W. Comparative modeling and molecular docking analysis of white, brown and soft rot fungal laccases using lignin model compounds for understanding the structural and functional properties of laccases. J. Mol. Grap. Mod. 2018, 79, 15–26. [Google Scholar] [CrossRef] [PubMed]
- Mu, Y.; Kosov, D.S.; Stock, G. Conformational dynamics of trialanine in water. 2. Comparison of AMBER, CHARMM, GROMOS, and OPLS force fields to NMR and infrared experiments. J. Phys. Chem. B 2003, 107, 5064–5073. [Google Scholar] [CrossRef]
- Lee, T.S.; Cerutti, D.S.; Mermelstein, D.; Lin, C.; LeGrand, S.; Giese, T.J.; York, D.M. GPU-accelerated molecular dynamics and free energy methods in Amber18: Performance enhancements and new features. J. Chem. Inf. Model. 2018, 58, 2043–2050. [Google Scholar] [CrossRef]
- Wang, J.; Wang, W.; Kollman, P.A.; Case, D.A. Antechamber: An accessory software package for molecular mechanical calculations. J. Am. Chem. Soc. 2001, 222, U403. [Google Scholar]
- Pearlman, D.A.; Case, D.A.; Caldwell, J.W.; Ross, W.S.; Cheatham, T.E.; DeBolt, S.; Kollman, P. AMBER, a package of computer programs for applying molecular mechanics, normal mode analysis, molecular dynamics and free energy calculations to simulate the structural and energetic properties of molecules. Comput. Phys. Commun. 1995, 91, 1–41. [Google Scholar] [CrossRef]
- Lai, Z.; Zhang, K.; Wang, J. Exploring multi-dimensional coordinate-dependent diffusion dynamics on the energy landscape of protein conformation change. Comput. Phys. Commun. 2014, 16, 6486–6495. [Google Scholar] [CrossRef] [PubMed]

| Phylogroup | A (14.7%) n = 15 | B1 (5%) n = 6 | B2 (23.5%) n = 24 | C (9.8%) n = 10 | D (26.4%) n = 27 | E (0.98%) n = 1 | F (3.9%) n = 4 | Clade I (7.8%) n = 8 | N/D (6.8%) n = 7 |
|---|---|---|---|---|---|---|---|---|---|
| Characteristics | |||||||||
| Biofilm | |||||||||
| Non-adherent | 0 (0%) | 1 (16.6%) | 0 (0%) | 0 (0%) | 1 (3%) | 1 (100%) | 0 (0%) | 1 (12.5%) | 1 (14.2%) |
| Weak | 9 (60%) | 3 (50%%) | 7 (29.1%) | 5 (50%) | 10 (37%) | 0 (0%) | 1 (25%) | 2 (25%) | 3 (42.8%) |
| Moderate | 6 (40%) | 2 (33.3%) | 10 (41.6%) | 5 (50%) | 13 (48.1%) | 0 (0%) | 2 (50%) | 5 (62.5%) | 3 (42.8%) |
| Strong | 0 (0%) | 0 (0%) | 7 (29.1%) | 0 (0%) | 3 (11.1%) | 0 (0%) | 1 (25%) | 0 (0%) | 0 (0%) |
| Pathogenicity | |||||||||
| pdar | 5 (33.3%) | 3 (50%) | 0 (0%) | 4 (40%) | 16 (59.2%) | 0 (0%) | 0 (0%) | 2 (25%) | 0 (0%) |
| rdar | 6 (40%) | 0 (0%) | 4 (16.6%) | 3 (30%) | 1 (3.7%) | 0 (0%) | 1 (25%) | 2 (25%) | 5 (71.4%) |
| bdar | 4 (26.6%) | 3 (50%) | 20 (83.3%) | 3 (30%) | 10 (37%) | 1 (100%) | 3 (75%) | 4 (50%) | 2 (28.5%) |
| Virulence Factors | |||||||||
| fimH | 14 (93.3%) | 6 (100%) | 24 (100%) | 10 (100%) | 26 (96.2%) | 0 (0%) | 4 (100%) | 8 (100%) | 5 (71.4%) |
| fyuA | 11 (73.3%) | 4 (66.6%) | 22 (91.6%) | 7 (70%) | 24 (88.8%) | 1 (100%) | 4 (100%) | 7 (87.5%) | 6 (85.7%) |
| PAI | 2 (13.3%) | 1 (16.6%) | 11 (45.8%) | 6 (60%) | 7 (25.9%) | 0 (0%) | 1 (25%) | 2 (25%) | 2 (28.5%) |
| Usp | 0 (0%) | 0 (0%) | 10 (41.6%) | 0 (0%) | 2 (7.4%) | 0 (0%) | 1 (25%) | 0 (0%) | 2 (28.5%) |
| papAH | 2 (13.3%) | 0 (0%) | 9 (37.5%) | 5 (50%) | 9 (33.3%) | 0 (0%) | 2 (50%) | 2 (25%) | 2 (28.5%) |
| kpsMTII | 5 (33.3%) | 1 (16.6%) | 23 (95.8%) | 6 (60%) | 23 (85.1) | 0 (0%) | 4 (100%) | 3 (37.5%) | 4 (57.1%) |
| Antibiotics (Disk Concentration) | % R | % I | % S |
|---|---|---|---|
| Aminoglycosides | |||
| Amikacin (AMK) (30 µg) | 8.4 | - | 91.6 |
| Gentamicin (GEN) (10 µg) | 27.4 | - | 72.6 |
| Tobramycin (TOB) (10 µg) | 24.2 | 13.2 | 62.6 |
| Cephalosporins | |||
| Cefazolin (CFZ) (30 µg) | 45.8 | 5.8 | 48.4 |
| Cefepime (FEP) (30 µg) | 54.2 | - | 45.8 |
| Cefotaxime (CTX) (30 µg) | 44.2 | - | 55.8 |
| Cefoxitin (FOX) (30 µg) | 6.3 | 4.2 | 89.5 |
| Ceftazidime (CAZ) (30 µg) | 11.1 | - | 88.9 |
| Ceftriaxone (CAX) (30 µg) | 5.3 | 3.7 | 91 |
| Quinolones | |||
| Ciprofloxacin (CIP) (5 µg) | 49.5 | - | 50.5 |
| Carbapenems | |||
| Doripenem (DOR) (8 µg) | 4.2 | - | 95.8 |
| Ertapenem (ETP) (4 µg) | 4.2 | - | 95.8 |
| Aztreonam (ATM) (30 µg) | 33 | 13 | 54 |
| Meropenem (MEM) (10 µg) | 9.5 | - | 90.5 |
| Nitrofurans | |||
| Nitrofurantoin (NIT) (300 µg) | 11.1 | - | 88.9 |
| Penicillins | |||
| Ampicillin (AMP) (10 µg) | 88.4 | - | 12.6 |
| Pip/tazo (TZP) (100/10 µg) | 5.8 | 7.9 | 86.3 |
| Piperacillin (PIP) (100 µg) | 45.2 | - | 51.1 |
| Amp/Sulbactam (SAM) (10 µg) | 42.6 | 19 | 38.4 |
| Sulfonamides | |||
| Trimet/sulfa (SXT) (0.25/23.7 µg) | 50.5 | - | 49.5 |
| Strains | Phylogenetic Group | Virulence Factors | Biofilm Forming Ability | Resistance to Antibiotics | |
|---|---|---|---|---|---|
| Morphotypes/ Type of Biofilm | Antibiotic Susceptibility | MDR/ ESBL Genes | |||
| AMR1638 | A | fyuA, fimH | rdar/Moderate | AMP | -/- |
| AMR1676 | B1 | fimH | bdar/weak | - | -/- |
| AMR4129 | B1 | fyuA, kpsMTII, fimH | bdar/non-producer | AMK; GEN; TOB; SXT; CIP; CEZ; FEP; CAZ; FOX; CRO; AMP; PIP; SAM; AZM. | +/- |
| AMR1201 | B2 | fyuA, kpsMTII, usp, fimH | bdar/Moderate | GEN; TOB; NIT; SXT; CIP; CEZ; CTX; FEP; CAZ; FOX; CRO; AMP; PIP; * SAM. | +/SHV |
| AMR4620 | B2 | fyuA, kpsMTII, usp, PAI, fimH | bdar/strong | GEN; TOB; SXT; CIP; CTX; FEP; CAZ; FOX; CRO; AMP; PIP; SAM; TZP; AZM. | +/OXA |
| AMR1898 | B2 | fyuA, kpsMTII, usp, papAH, fimH | bdar/Moderate | - | -/SHV |
| AMR1740 | B2 | fyuA, kpsMTII, papAH, fimH | bdar/strong | GEN; TOB; * NIT; SXT; CTX; FEP; CAZ; FOX; CRO; AMP; PIP; SAM; AZM. | +/- |
| AMR2919 | B2 | fyuA, kpsMTII, usp, papAH, fimH | bdar/non-producer | GEN; * TOB; NIT; SXT; CIP; CEZ; CTX; FEP; CAZ; CRO; AMP; PIP; * SAM | +/- |
| AMR3633 | C | fimH | bdar/weak | - | -/CTX-M-1 |
| AMR1598 | Clade I | fyuA, kpsMTII, papAH, fimH | rdar/Moderate | GEN; TOB; * NIT; SXT; CIP; CTX; FEP; CAZ; FOX; CRO; AMP; PIP; SAM; * TZP; * AZM | +/TEM |
| AMR4671 | D | fyuA, papAH, fimH | pdar/Moderate | - | -/- |
| AMR0843 | D | fimH | rdar/Moderate | - | -/TEM |
| AMR1642 | D | fuyA, fimH | bdar/Moderate | GEN; TOB; SXT; CIP; CEZ; CTX; FEP; CAZ; CRO; AMP; PIP; SAM. | +/OXA |
| AMR0864 | F | fyuA, kpsMTII, fimH | rdar/strong | GEN; * TOB; CEZ; CTX; FEP; CAZ; CRO; AMP; PIP; SAM. | +/- |
| AMR3525 | F | fyuA, kpsMTII, fimH | bdar/weak | AMK; GEN; TOB; NIT; SXT; CIP; CEZ; CTX; FEP; CAZ; FOX; CRO; AMP; PIP; SAM; TZP; AZM. | +/- |
| Protein 4X5P | Protein 4XO9 | |||||
|---|---|---|---|---|---|---|
| Ligand | Affinity (kcal/mol) | Distance RMSD (Å) | Mode RMSD (Å) | Affinity (kcal/mol) | Distance RMSD (Å) | Mode RMSD (Å) |
| Native | −6.6 | 0.097 | 1.899 | −6.6 | 0.080 | 2.122 |
| b | −6.7 | 0.462 | 1.215 | −6.6 | 0.050 | 1.144 |
| c | −6.8 | 0.371 | 1.900 | −6.9 | 0.570 | 1.554 |
| d | −7.0 | 0.096 | 1.111 | −6.9 | 1.826 | 2.232 |
| e | −6.5 | 0.080 | 1.117 | −6.7 | 0.063 | 1.109 |
| f | −6.8 | 1.437 | 1.750 | −6.6 | 0.159 | 1.125 |
| g | −6.5 | 0.094 | 1.625 | −6.8 | 0.527 | 1.137 |
| h | −6.7 | 0.166 | 1.111 | −6.9 | 2.228 | 3.856 |
| i | −6.6 | 0.525 | 1.343 | −6.8 | 0.257 | 1.064 |
| j | −6.5 | 0.366 | 3.804 | −6.7 | 0.293 | 3.726 |
| k | −6.5 | 0.370 | 1.483 | −7.0 | 0.649 | 1.133 |
| MM/GBSA | MM/PBSA | |||||
|---|---|---|---|---|---|---|
| Ligand | Energy Average | Standard Deviation | Standard Error | Energy Average | Standard Deviation | Standard Error |
| Native | −20.5170 | 3.1661 | 0.1837 | −2.2483 | 4.0371 | 0.3291 |
| b | −40.0576 | 5.6342 | 0.1783 | −22.0351 | 5.4900 | 0.1737 |
| c | −19.8246 | 7.2451 | 1.7077 | 6.4182 | 9.509 | 2.2413 |
| d | −9.0984 | 5.9672 | 0.1888 | −0.2142 | 4.6101 | 0.1484 |
| e | −9.6600 | 6.1696 | 0.1952 | 0.1840 | 4.4867 | 0.1420 |
| f | −20.4026 | 6.3241 | 0.2001 | −2.3836 | 6.4959 | 0.2055 |
| g | −27.5406 | 7.8822 | 0.2494 | −8.2752 | 11.2063 | 0.3546 |
| h | −18.8009 | 7.5942 | 0.2403 | −2.7163 | 6.9245 | 0.2191 |
| i | −18.9992 | 3.5661 | 0.1128 | −2.3183 | 5.6019 | 0.1772 |
| j | −7.8151 | 5.9003 | 0.1867 | 0.3845 | 3.2391 | 0.1022 |
| k | −10.7836 | 6.7607 | 0.2139 | −0.9951 | 6.0892 | 0.1927 |
| MM/GBSA | MM/PBSA | |||||
|---|---|---|---|---|---|---|
| Ligand | Energy Average | Standard Deviation | Standard Error | Energy Average | Standard Deviation | Standard Error |
| Native | −42.3971 | 4.1549 | 0.1171 | −21.6338 | 5.1121 | 0.1732 |
| b | −41.1313 | 4.7338 | 0.1498 | −24.6143 | 4.5592 | 0.1442 |
| c | −43.0973 | 3.8352 | 0.1213 | −23.7164 | 4.8470 | 0.1534 |
| d | −40.2022 | 5.1058 | 0.1615 | −22.4473 | 5.5330 | 0.1750 |
| e | −45.9053 | 3.6061 | 0.1141 | −28.1108 | 3.5312 | 0.1170 |
| f | −44.3710 | 4.5614 | 0.1443 | −26.7404 | 3.9121 | 0.1238 |
| g | −42.7031 | 4.8568 | 0.1537 | −22.9272 | 5.1634 | 0.1634 |
| h | −42.5463 | 4.6572 | 0.1473 | −25.2265 | 4.4345 | 0.1403 |
| i | −45.0402 | 3.8545 | 0.1220 | −27.0876 | 3.7791 | 0.1196 |
| j | −46.5859 | 4.0885 | 0.1294 | −24.6417 | 4.7458 | 0.1502 |
| k | −44.4012 | 4.1211 | 0.1304 | −24.3189 | 5.0090 | 0.1585 |
| Protein 4XO9 | ||
|---|---|---|
| Ligand | GLN 133 | ASP 54 |
| b | 71.30% | 68.20% |
| f | - | 54.30% |
| g | 98.90% | 99.90% |
| Protein 4X5P | ||
| e | 99.90% | 100% |
| i | 99.90% | 99.80% |
| j | - | 99.90% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Montes-Robledo, A.; Baldiris-Avila, R.; Galindo, J.F. D-Mannoside FimH Inhibitors as Non-Antibiotic Alternatives for Uropathogenic Escherichia coli. Antibiotics 2021, 10, 1072. https://doi.org/10.3390/antibiotics10091072
Montes-Robledo A, Baldiris-Avila R, Galindo JF. D-Mannoside FimH Inhibitors as Non-Antibiotic Alternatives for Uropathogenic Escherichia coli. Antibiotics. 2021; 10(9):1072. https://doi.org/10.3390/antibiotics10091072
Chicago/Turabian StyleMontes-Robledo, Alfredo, Rosa Baldiris-Avila, and Johan Fabian Galindo. 2021. "D-Mannoside FimH Inhibitors as Non-Antibiotic Alternatives for Uropathogenic Escherichia coli" Antibiotics 10, no. 9: 1072. https://doi.org/10.3390/antibiotics10091072
APA StyleMontes-Robledo, A., Baldiris-Avila, R., & Galindo, J. F. (2021). D-Mannoside FimH Inhibitors as Non-Antibiotic Alternatives for Uropathogenic Escherichia coli. Antibiotics, 10(9), 1072. https://doi.org/10.3390/antibiotics10091072






