ESBL-Producing Escherichia coli Carrying CTX-M Genes Circulating among Livestock, Dogs, and Wild Mammals in Small-Scale Farms of Central Chile
Abstract
1. Introduction
2. Materials and Methods
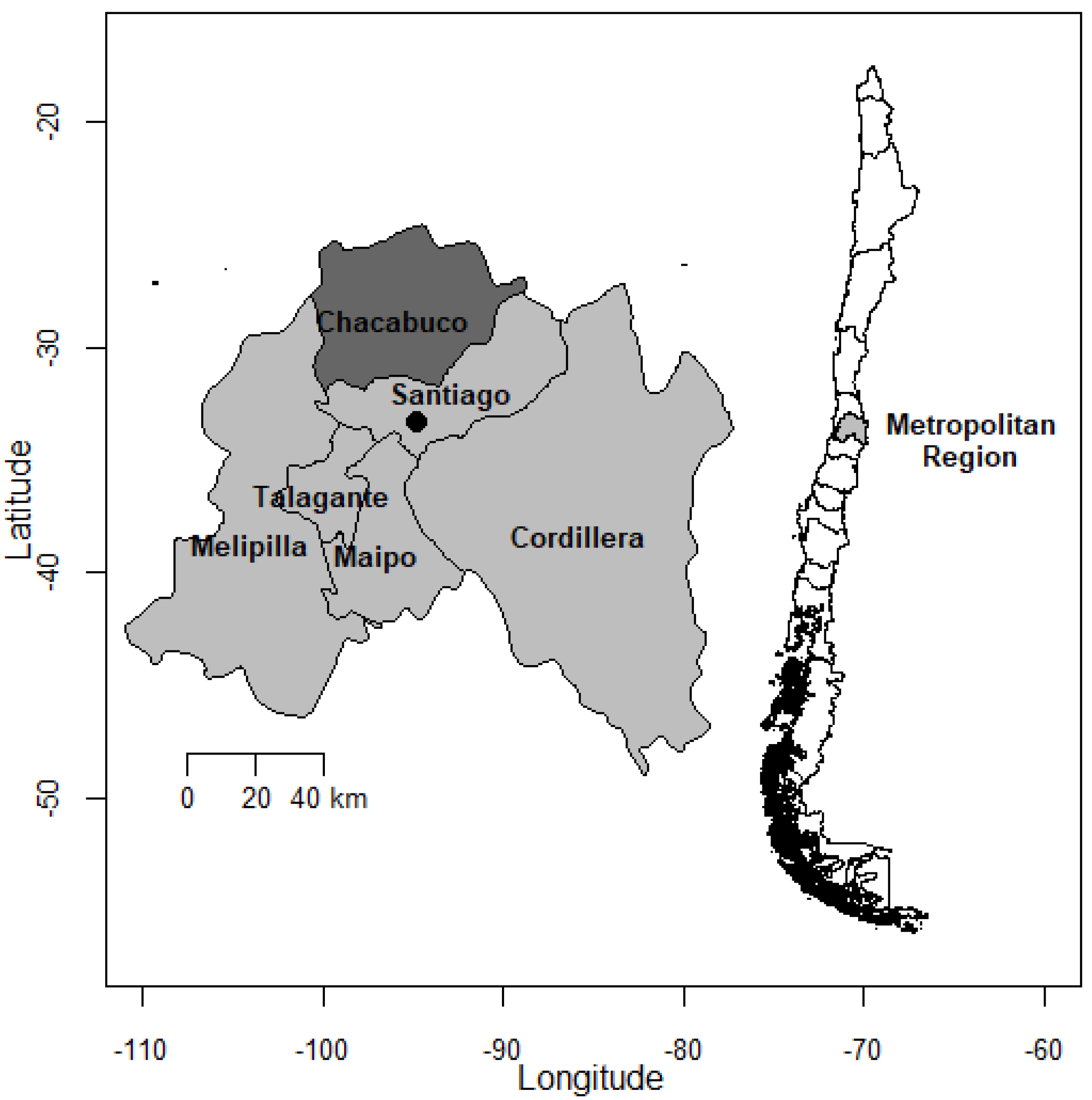
2.1. Sample Collection
2.2. Sample Size and Prevalence Estimation
2.3. Microbiology Analyses
2.4. Statistical Analyses
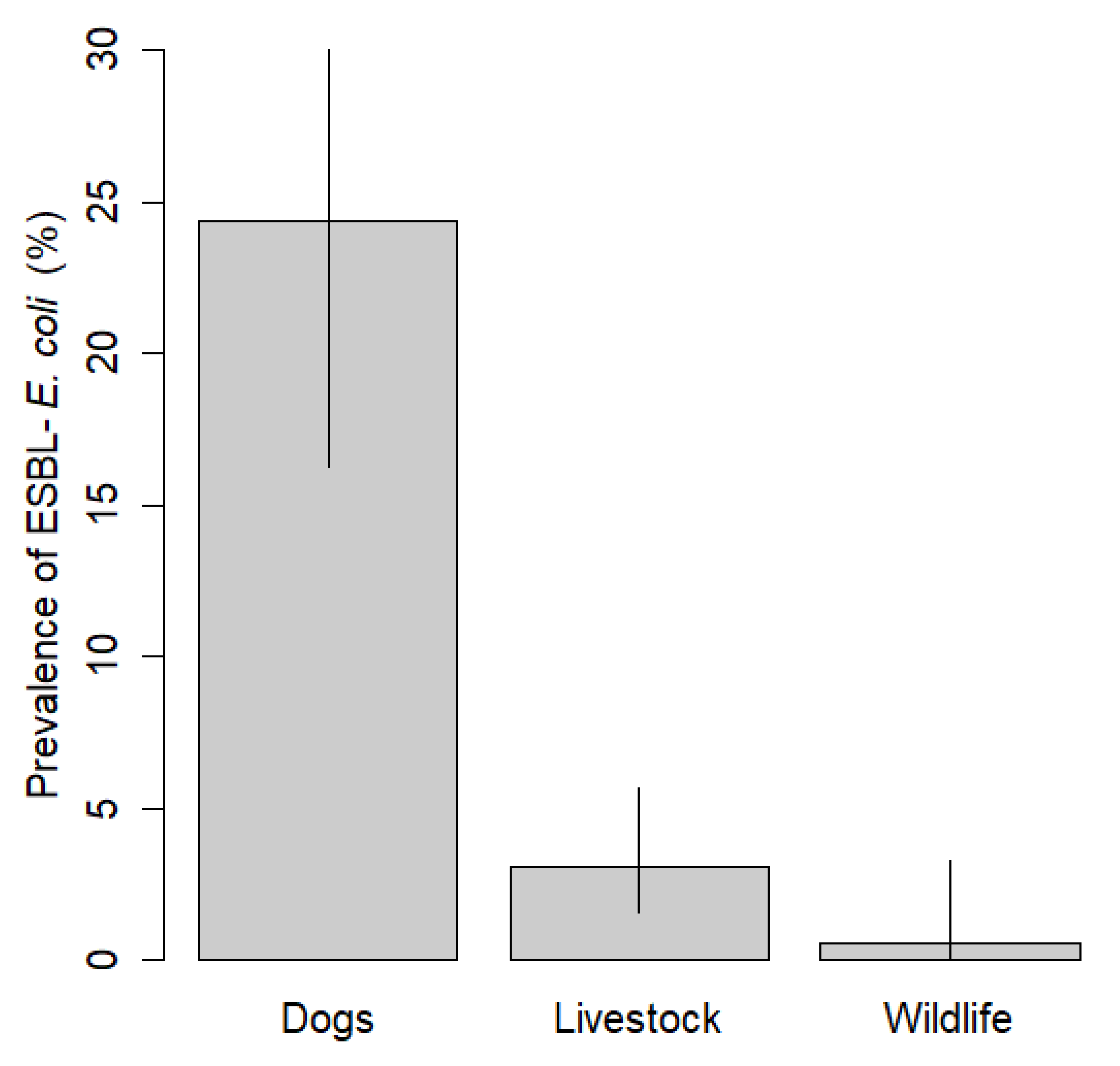
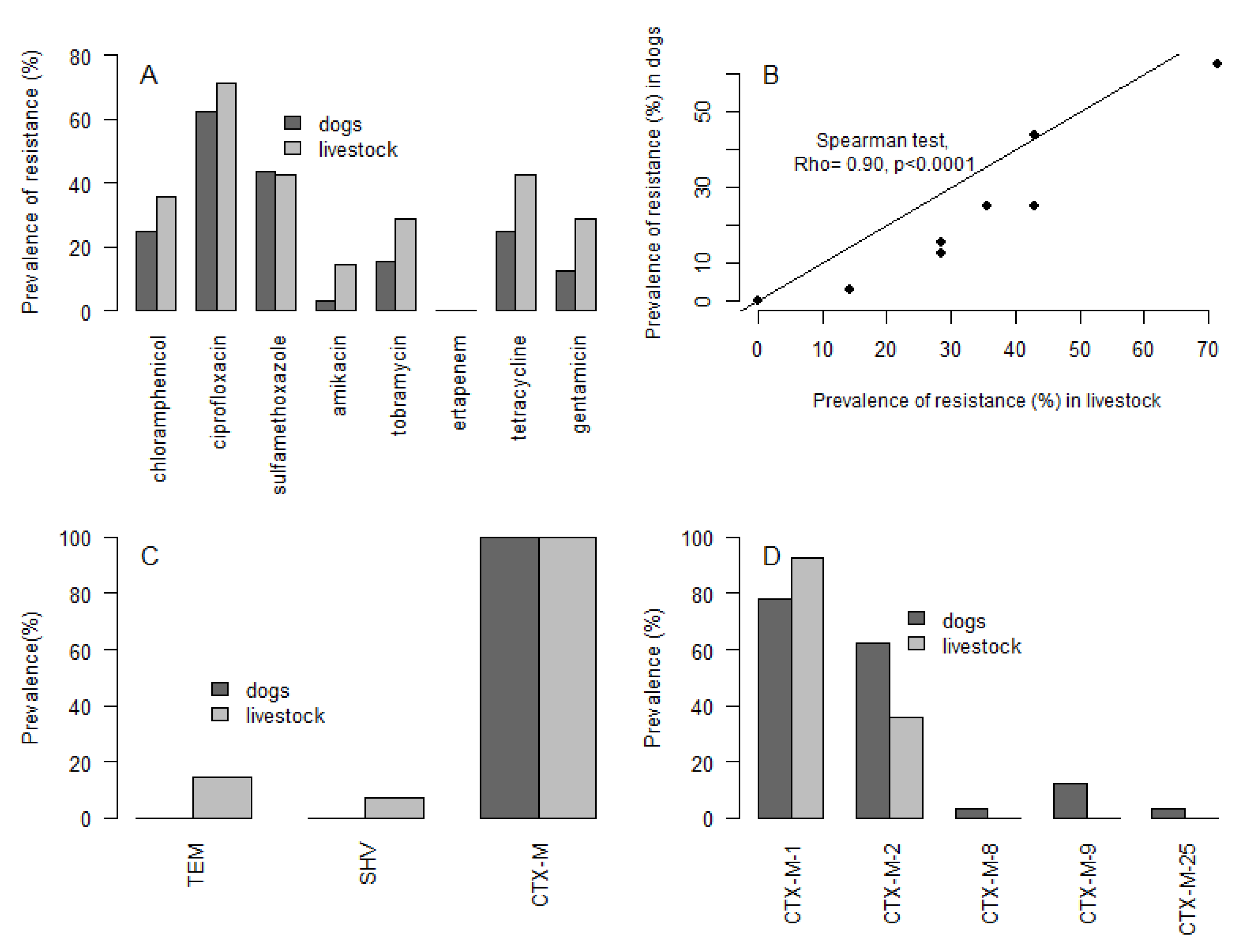
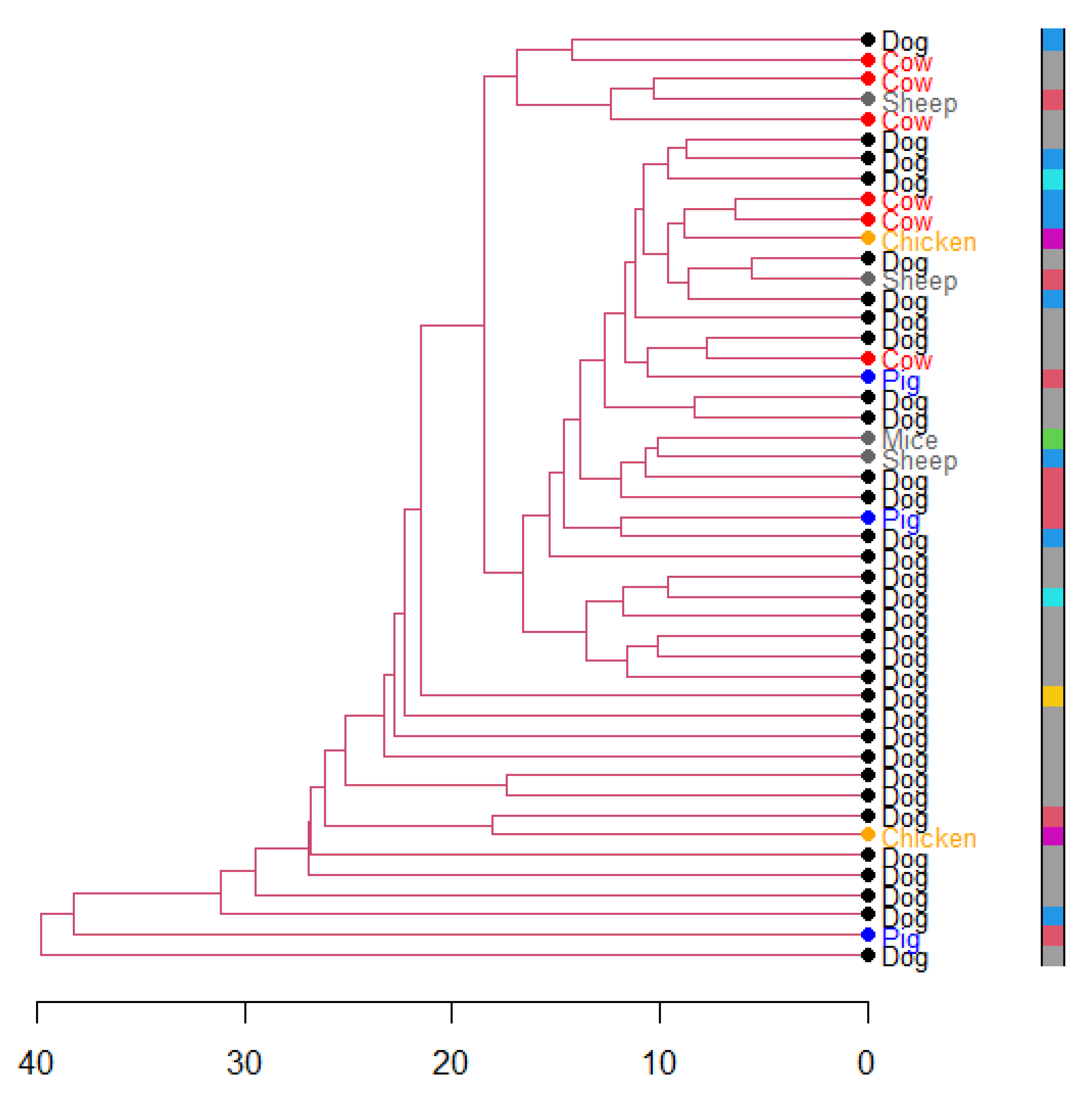
3. Results
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Health Organization. 2019 Antibacterial Agents in Clinical Development: An Analysis of the Antibacterial Clinical Development Pipeline; WHO: Geneva, Switzerland, 2019; Available online: https://www.who.int/medicines/areas/rational_use/antibacterial_agents_clinical_development/en/ (accessed on 28 December 2020).
- Wall, B.A.; Mateus, A.; Marshall, L.; Pfeiffer, D.; Lubroth, J.; Ormel, H.J.; Otto, P.; Patriarchi, A.; Food and Agriculture Organization of the United Nations. Drivers, Dynamics and Epidemiology of Antimicrobial Resistance in Animal Production; Food and Agriculture Organization (FAO): Rome, Italy, 2016; ISBN 978-92-5-109441-9. [Google Scholar]
- IACG. No Time to Wait: Securing the Future from Drug-Resistant Infections. Available online: http://www.who.int/antimicrobial-resistance/interagency-coordination-group/final-report/en/ (accessed on 23 July 2020).
- World Bank. Drug-Resistant Infections: A Threat to Our Economic Future; World Bank: Washington, DC, USA, 2017. [Google Scholar]
- Antimicrobial Resistance: Global Report on Surveillance; World Health Organization: Geneva, Switzerland, 2014; ISBN 978-92-4-156474-8.
- O’Neill, J. The Review on Antimicrobial Resistance—Tackling Drug-Resistant Infections Globally: Final Report and Recommendations. Available online: https://amr-review.org/ (accessed on 29 July 2020).
- Van Boeckel, T.P.; Brower, C.C.; Gilbert, M.; Grenfell, B.T.; Levin, S.A.S.; Robinson, T.P.; Teillant, A.; Laxminarayan, R.R. Global trends in antimicrobial use in food animals. Proc. Natl. Acad. Sci. USA 2015, 112, 5649–5654. [Google Scholar] [CrossRef]
- Doi, Y.; Iovleva, A.; Bonomo, R.A. The ecology of extended-spectrum β-lactamases (ESBLs) in the developed world. J. Travel Med. 2017, 24, S44–S51. [Google Scholar] [CrossRef]
- Smet, A.; Martel, A.; Persoons, D.; Dewulf, J.; Heyndrickx, M.; Herman, L.; Haesebrouck, F.; Butaye, P. Broad-spectrum β-lactamases among Enterobacteriaceaeof animal origin: Molecular aspects, mobility and impact on public health. FEMS Microbiol. Rev. 2010, 34, 295–316. [Google Scholar] [CrossRef]
- Eguenther, S.; Eewers, C.; Wieler, L.H. Extended-Spectrum Beta-Lactamases Producing E. coli in Wildlife, yet Another Form of Environmental Pollution? Front. Microbiol. 2011, 2, 246. [Google Scholar] [CrossRef]
- Loayza, F.; Graham, J.P.; Trueba, G. Factors Obscuring the Role of E. coli from Domestic Animals in the Global Antimicrobial Resistance Crisis: An Evidence-Based Review. Int. J. Environ. Res. Public Health 2020, 17, 3061. [Google Scholar] [CrossRef]
- Benavides, J.A.; Shiva, C.; Virhuez, M.; Tello, C.; Appelgren, A.; Vendrell, J.; Solassol, J.; Godreuil, S.; Streicker, D.G. Extended-spectrum beta-lactamase-producing Escherichia coli in common vampire bats Desmodus rotundus and livestock in Peru. Zoonoses Public Health 2018, 65, 454–458. [Google Scholar] [CrossRef]
- Snow, L.; Warner, R.; Cheney, T.; Wearing, H.; Stokes, M.; Harris, K.; Teale, C.; Coldham, N. Risk factors associated with extended spectrum beta-lactamase Escherichia coli (CTX-M) on dairy farms in North West England and North Wales. Prev. Vet. Med. 2012, 106, 225–234. [Google Scholar] [CrossRef]
- Atterby, C.; Börjesson, S.; Ny, S.; Järhult, J.D.; Byfors, S.; Bonnedahl, J. ESBL-producing Escherichia coli in Swedish gulls—A case of environmental pollution from humans? PLoS ONE 2017, 12, e0190380. [Google Scholar] [CrossRef]
- Jamborova, I.; Johnston, B.D.; Papousek, I.; Kachlikova, K.; Micenkova, L.; Clabots, C.; Skalova, A.; Chudejova, K.; Dolejska, M.; Literak, I.; et al. Extensive Genetic Commonality among Wildlife, Wastewater, Community, and Nosocomial Isolates of Escherichia coli Sequence Type 131 (H30R1 and H30Rx Subclones) That Carry blaCTX-M-27 or blaCTX-M-15. Antimicrob. Agents Chemother. 2018, 62, 00519-18. [Google Scholar] [CrossRef]
- Joosten, P.; Ceccarelli, D.; Odent, E.; Sarrazin, S.; Graveland, H.; Van Gompel, L.; Battisti, A.; Caprioli, A.; Franco, A.; Wagenaar, J.A.; et al. Antimicrobial Usage and Resistance in Companion Animals: A Cross-Sectional Study in Three European Countries. Antibiotics 2020, 9, 87. [Google Scholar] [CrossRef]
- Van Boeckel, T.P.; Pires, J.; Silvester, R.; Zhao, C.; Song, J.; Criscuolo, N.G.; Gilbert, M.; Bonhoeffer, S.; Laxminarayan, R. Global trends in antimicrobial resistance in animals in low- and middle-income countries. Science 2019, 365, eaaw1944. [Google Scholar] [CrossRef]
- Chantziaras, I.; Boyen, F.; Callens, B.; Dewulf, J. Correlation between veterinary antimicrobial use and antimicrobial resistance in food-producing animals: A report on seven countries. J. Antimicrob. Chemother. 2014, 69, 827–834. [Google Scholar] [CrossRef]
- de Jong, A.; Thomas, V.; Klein, U.; Marion, H.; Moyaert, H.; Simjee, S.; Vallé, M. Pan-European resistance monitoring programmes encompassing food-borne bacteria and target pathogens of food-producing and companion animals. Int. J. Antimicrob. Agents 2013, 41, 403–409. [Google Scholar] [CrossRef]
- Hille, K.; Felski, M.; Ruddat, I.; Woydt, J.; Schmid, A.; Friese, A.; Fischer, J.; Sharp, H.; Valentin, L.; Michael, G.B.; et al. Association of farm-related factors with characteristics profiles of extended-spectrum β-lactamase-/plasmid-mediated AmpC β-lactamase-producing Escherichia coli isolates from German livestock farms. Vet. Microbiol. 2018, 223, 93–99. [Google Scholar] [CrossRef]
- Gay, N.; LeClaire, A.; Laval, M.; Miltgen, G.; Jégo, M.; Stéphane, R.; Jaubert, J.; Belmonte, O.; Cardinale, E. Risk Factors of Extended-Spectrum β-Lactamase Producing Enterobacteriaceae Occurrence in Farms in Reunion, Madagascar and Mayotte Islands, 2016–2017. Vet. Sci. 2018, 5, 22. [Google Scholar] [CrossRef]
- Dahms, C.; Hübner, N.-O.; Kossow, A.; Mellmann, A.; Dittmann, K.; Kramer, A. Occurrence of ESBL-Producing Escherichia coli in Livestock and Farm Workers in Mecklenburg-Western Pomerania, Germany. PLoS ONE 2015, 10, e0143326. [Google Scholar] [CrossRef]
- Wang, J.; Ma, Z.B.; Zeng, Z.L.; Yang, X.W.; Huang, Y.; Liu, J.H. The role of wildlife (wild birds) in the global transmission of antimicrobial resistance genes. Zool. Res. 2017, 38, 55. [Google Scholar] [CrossRef]
- Poeta, P.; Radhouani, H.; Pinto, L.; Martinho, A.; Rego, V.; Rodrigues, R.; Gonçalves, A.; Rodrigues, J.; Estepa, V.; Torres, C.; et al. Wild boars as reservoirs of extended-spectrum beta-lactamase (ESBL) producing Escherichia coli of different phylogenetic groups. J. Basic Microbiol. 2009, 49, 584–588. [Google Scholar] [CrossRef]
- Alonso, C.; González-Barrio, D.; Tenorio, C.; Ruiz-Fons, F.; Torres, C. Antimicrobial resistance in faecal Escherichia coli isolates from farmed red deer and wild small mammals. Detection of a multiresistant E. coli producing extended-spectrum beta-lactamase. Comp. Immunol. Microbiol. Infect. Dis. 2016, 45, 34–39. [Google Scholar] [CrossRef]
- Alonso, C.A.; Alcalá, L.; Simón, C.; Torres, C. Novel sequence types of extended-spectrum and acquired AmpC beta-lactamase producing Escherichia coli and Escherichia clade V isolated from wild mammals. FEMS Microbiol. Ecol. 2017, 93, fiy066. [Google Scholar] [CrossRef]
- Seni, J.; Falgenhauer, L.; Simeo, N.; Mirambo, M.M.; Imirzalioglu, C.; Matee, M.; Rweyemamu, M.; Chakraborty, T.; Mshana, S.E. Multiple ESBL-Producing Escherichia coli Sequence Types Carrying Quinolone and Aminoglycoside Resistance Genes Circulating in Companion and Domestic Farm Animals in Mwanza, Tanzania, Harbor Commonly Occurring Plasmids. Front. Microbiol. 2016, 7, 142. [Google Scholar] [CrossRef] [PubMed]
- Dupouy, V.; Abdelli, M.; Moyano, G.; Arpaillange, N.; Bibbal, D.; Cadiergues, M.-C.; Lopez-Pulin, D.; Sayah-Jeanne, S.; De Gunzburg, J.; Saint-Lu, N.; et al. Prevalence of Beta-Lactam and Quinolone/Fluoroquinolone Resistance in Enterobacteriaceae From Dogs in France and Spain—Characterization of ESBL/pAmpC Isolates, Genes, and Conjugative Plasmids. Front. Vet. Sci. 2019, 6, 279. [Google Scholar] [CrossRef]
- Bunt, G.V.D.; Fluit, A.C.; Spaninks, M.P.; Timmerman, A.J.; Geurts, Y.; Kant, A.; Scharringa, J.; Mevius, D.; Wagenaar, J.A.; Bonten, M.J.M.; et al. Faecal carriage, risk factors, acquisition and persistence of ESBL-producing Enterobacteriaceae in dogs and cats and co-carriage with humans belonging to the same household. J. Antimicrob. Chemother. 2020, 75, 342–350. [Google Scholar] [CrossRef] [PubMed]
- Salgado-Caxito, M.; Benavides, J.A.; Munita, J.M.; Rivas, L.; García, P.; Listoni, F.J.; Moreno-Switt, A.I.; Paes, A.C. Risk factors associated with faecal carriage of extended-spectrum cephalosporin-resistant Escherichia coli among dogs in Southeast Brazil. Prev. Vet. Med. 2021, 190, 105316. [Google Scholar] [CrossRef]
- Barth, S.A.; Blome, S.; Cornelis, D.; Pietschmann, J.; Laval, M.; Maestrini, O.; Geue, L.; Charrier, F.; Etter, E.; Menge, C.; et al. FaecalEscherichia colias biological indicator of spatial interaction between domestic pigs and wild boar (Sus scrofa) in Corsica. Transbound. Emerg. Dis. 2018, 65, 746–757. [Google Scholar] [CrossRef] [PubMed]
- Mercat, M.; Clermont, O.; Massot, M.; Ruppe, E.; De Garine-Wichatitsky, M.; Miguel, E.; Fox, H.V.; Cornelis, D.; Andremont, A.; Denamur, E.; et al. Escherichia coli Population Structure and Antibiotic Resistance at a Buffalo/Cattle Interface in Southern Africa. Appl. Environ. Microbiol. 2015, 82, 1459–1467. [Google Scholar] [CrossRef]
- Founou, L.L.; Founou, R.C.; Essack, S.Y. Antibiotic Resistance in the Food Chain: A Developing Country-Perspective. Front. Microbiol. 2016, 7, 1881. [Google Scholar] [CrossRef]
- Ho, P.L.; Chow, K.H.; Lai, E.L.; Lo, W.U.; Yeung, M.K.; Chan, J.; Chan, P.Y.; Yuen, K.Y. Extensive dissemination of CTX-M-producing Escherichia coli with multidrug resistance to ‘critically important’ antibiotics among food animals in Hong Kong, 2008–2010. J. Antimicrob. Chemother. 2011, 66, 765–768. [Google Scholar] [CrossRef]
- Hasan, B.; Laurell, K.; Rakib, M.M.; Ahlstedt, E.; Hernandez, J.; Caceres, M.; Järhult, J.D. Fecal Carriage of Extended-Spectrum β-Lactamases in Healthy Humans, Poultry, and Wild Birds in León, Nicaragua—A Shared Pool of blaCTX-M Genes and Possible Interspecies Clonal Spread of Extended-Spectrum β-Lactamases-Producing Escherichia coli. Microb. Drug Resist. 2016, 22, 682–687. [Google Scholar] [CrossRef]
- Hernandez, J.; Johansson, A.; Stedt, J.; Bengtsson, S.; Porczak, A.; Granholm, S.; González-Acuña, D.; Olsen, B.; Bonnedahl, J.; Drobni, M. Characterization and Comparison of Extended-Spectrum β-Lactamase (ESBL) Resistance Genotypes and Population Structure of Escherichia coli Isolated from Franklin’s Gulls (Leucophaeus pipixcan) and Humans in Chile. PLoS ONE 2013, 8, e76150. [Google Scholar] [CrossRef]
- Nweneka, C.V.; Tapha-Sosseh, N.; Sosa, A. Curbing the menace of antimicrobial resistance in developing countries. Harm Reduct. J. 2009, 6, 31. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, M.; Khan, A.U. Global economic impact of antibiotic resistance: A review. J. Glob. Antimicrob. Resist. 2019, 19, 313–316. [Google Scholar] [CrossRef] [PubMed]
- González, C.M.A. Susceptibilidad Microbiana: Un Test Rápido Para el Análisis de Resistencia Bacteriana en Cepas Aisladas de Mastitis Clínica. Bachelor’s Thesis, Universidad de Chile, Santiago, Chile, 2006. [Google Scholar]
- Ewers, C.; Bethe, A.; Semmler, T.; Guenther, S.; Wieler, L. Extended-spectrum β-lactamase-producing and AmpC-producing Escherichia coli from livestock and companion animals, and their putative impact on public health: A global perspective. Clin. Microbiol. Infect. 2012, 18, 646–655. [Google Scholar] [CrossRef]
- Moreno, A.; Bello, H.; Guggiana, D.; Domínguez, M.; González, G. Extended-spectrum β-lactamases belonging to CTX-M group produced by Escherichia coli strains isolated from companion animals treated with enrofloxacin. Vet. Microbiol. 2008, 129, 203–208. [Google Scholar] [CrossRef]
- Fuentes-Castillo, D.; Farfán-López, M.; Esposito, F.; Moura, Q.; Fernandes, M.R.; Lopes, R.; Cardoso, B.; Muñoz, M.E.; Cerdeira, L.; Najle, I.; et al. Wild owls colonized by international clones of extended-spectrum β-lactamase (CTX-M)-producing Escherichia coli and Salmonella Infantis in the Southern Cone of America. Sci. Total Environ. 2019, 674, 554–562. [Google Scholar] [CrossRef]
- Fuentes-Castillo, D.; Esposito, F.; Cardoso, B.; Dalazen, G.; Moura, Q.; Fuga, B.; Fontana, H.; Cerdeira, L.; Dropa, M.; Rottmann, J.; et al. Genomic data reveal international lineages of critical priorityEscherichia coliharbouring wide resistome in Andean condors (Vultur gryphus Linnaeus, 1758). Mol. Ecol. 2020, 29, 1919–1935. [Google Scholar] [CrossRef]
- Simonetti, J.A. Diversity and Conservation of Terrestrial Vertebrates in Mediterranean Chile. Rev. Chil. Hist. Nat. 1999, 72, 493–500. [Google Scholar]
- Cofre, H.; A Marquet, P. Conservation status, rarity, and geographic priorities for conservation of Chilean mammals: An assessment. Biol. Conserv. 1999, 88, 53–68. [Google Scholar] [CrossRef]
- Iriarte, J.A.; Lobos, G.A.; Jaksic, F.M. Invasive vertebrate species in Chile and their control and monitoring by governmental agencies. Rev. Chil. Hist. Nat. 2005, 78, 143–151. [Google Scholar] [CrossRef]
- Silva, C.; Saavedra, B. Knowing for controlling: Ecological effects of invasive vertebrates in Tierra del Fuego. Rev. Chil. Hist. Nat. 2008, 81, 123–136. [Google Scholar] [CrossRef]
- Sanguinetti, J.; Kitzberger, T. Factors controlling seed predation by rodents and non-native Sus scrofa in Araucaria araucana forests: Potential effects on seedling establishment. Biol. Invasions 2010, 12, 689–706. [Google Scholar] [CrossRef]
- Castro, S.; Bozinovic, F.; Jaksic, F. Ecological efficiency and legitimacy in seed dispersal of an endemic shrub (Lithrea caustica) by the European rabbit (Oryctolagus cuniculus) in central Chile. J. Arid. Environ. 2008, 72, 1164–1173. [Google Scholar] [CrossRef]
- Muñoz-Zanzi, C.; Mason, M.; Encina, C.; Gonzalez, M.; Berg, S. Household Characteristics Associated with Rodent Presence and Leptospira Infection in Rural and Urban Communities from Southern Chile. Am. J. Trop. Med. Hyg. 2014, 90, 497–506. [Google Scholar] [CrossRef]
- Salvatori, V.; Vaglio-Laurin, G.; Meserve, P.L.; Boitani, L.; Campanella, A. Spatial Organization, Activity, and Social Interactions of Culpeo Foxes (Pseudalopex culpaeus) in North-Central Chile. J. Mammal. 1999, 80, 980–985. [Google Scholar] [CrossRef]
- Cevidanes, A.; Esperón, F.; Di Cataldo, S.; Neves, E.; Sallaberry-Pincheira, N.; Millán, J. Antimicrobial resistance genes in Andean foxes inhabiting anthropized landscapes in central Chile. Sci. Total Environ. 2020, 724, 138247. [Google Scholar] [CrossRef]
- INE. Instituto Nacional de Estadísticas—Censo Agropecuario. Available online: http://www.ine.cl/estadisticas/economia/agricultura-agroindustria-y-pesca/censos-agropecuarios (accessed on 24 March 2021).
- Milstead, W.B.; Meserve, P.L.; Campanella, A.; Previtali, M.A.; Kelt, D.A.; Gutiérrez, J.R. Spatial Ecology of Small Mammals in North-central Chile: Role of Precipitation and Refuges. J. Mammal. 2007, 88, 1532–1538. [Google Scholar] [CrossRef]
- Jaksic, F.M.; Soriguer, R.C. Predation Upon the European Rabbit (Oryctolagus cuniculus) in Mediterranean Habitats of Chile and Spain: A Comparative Analysis. J. Anim. Ecol. 1981, 50, 269. [Google Scholar] [CrossRef]
- Cevidanes, A.; Ulloa-Contreras, C.; Di Cataldo, S.; Latrofa, M.S.; Gonzalez-Acuña, D.; Otranto, D.; Millán, J. Marked host association and molecular evidence of limited transmission of ticks and fleas between sympatric wild foxes and rural dogs. Med. Vet. Èntomol. 2021. [Google Scholar] [CrossRef]
- CDC. Downloads|Support|Epi InfoTM|CDC. Available online: https://www.cdc.gov/epiinfo/support/downloads.html (accessed on 18 June 2020).
- Ho, P.-L.; Liu, M.C.-J.; Lo, W.-U.; Lai, E.L.-Y.; Lau, T.C.-K.; Law, O.-K.; Chow, K.-H. Prevalence and characterization of hybrid blaCTX-M among Escherichia coli isolates from livestock and other animals. Diagn. Microbiol. Infect. Dis. 2015, 82, 148–153. [Google Scholar] [CrossRef]
- Silva, N.; Igrejas, G.; Figueiredo, N.; Gonçalves, A.; Radhouani, H.; Rodrigues, J.; Poeta, P. Molecular characterization of antimicrobial resistance in enterococci and Escherichia coli isolates from European wild rabbit (Oryctolagus cuniculus). Sci. Total Environ. 2010, 408, 4871–4876. [Google Scholar] [CrossRef]
- Radhouani, H.; Igrejas, G.; Gonçalves, A.; Estepa, V.; Sargo, R.; Torres, C.; Poeta, P. Molecular characterization of extended-spectrum-beta-lactamase-producing Escherichia coli isolates from red foxes in Portugal. Arch. Microbiol. 2012, 195, 141–144. [Google Scholar] [CrossRef]
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing, 28th ed.; CLSI Supplement M100; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2018; ISBN 978-1-56238-838-6. [Google Scholar]
- Magiorakos, A.-P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef]
- Dallenne, C.; Da Costa, A.; Decré, D.; Favier, C.; Arlet, G. Development of a set of multiplex PCR assays for the detection of genes encoding important β-lactamases in Enterobacteriaceae. J. Antimicrob. Chemother. 2010, 65, 490–495. [Google Scholar] [CrossRef]
- Woodford, N.; Fagan, E.J.; Ellington, M.J. Multiplex PCR for rapid detection of genes encoding CTX-M extended-spectrum β-lactamases. J. Antimicrob. Chemother. 2005, 57, 154–155. [Google Scholar] [CrossRef] [PubMed]
- Bilung, L.M.; Pui, C.F.; Su’Ut, L.; Apun, K. Evaluation of BOX-PCR and ERIC-PCR as Molecular Typing Tools for PathogenicLeptospira. Dis. Markers 2018, 2018, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Brasil. Decreto—Lei n° 227, de 28 de Fevereiro de 1967. Dá nova Redação ao Decreto-Lei n° 1.985, de 29 de Janeiro de 1940 (Código de Minas) Brasília. 1967. Available online: http://www.planalto.gov.br/ccivil_03/Decreto-Lei/Del0227.htm (accessed on 19 October 2020).
- Palmeira, J.D.; Haenni, M.; Metayer, V.; Madec, J.-Y.; Ferreira, H.M.N. Epidemic spread of IncI1/pST113 plasmid carrying the Extended-Spectrum Beta-Lactamase (ESBL) blaCTX-M-8 gene in Escherichia coli of Brazilian cattle. Vet. Microbiol. 2020, 243, 108629. [Google Scholar] [CrossRef] [PubMed]
- Résapath, B. Réseau D’épidémiosurveillance de L’antibiorésistance des Bactéries Pathogènes Animales; Ploufragan-Plouzané-Niort: Lyon, France, 2020; p. 155. [Google Scholar]
- Levy, S. Reduced Antibiotic Use in Livestock: How Denmark TackledResistance. Environ. Health Perspect. 2014, 122, A160-5. [Google Scholar] [CrossRef]
- Elgorriaga-Islas, E.; Guggiana-Nilo, P.; Domínguez-Yévenes, M.; González-Rocha, G.; Mella-Montecinos, S.; Labarca-Labarca, J.; García-Cañete, P.; Bello-Toledo, H. Prevalencia del determinante de resistencia plasmídica a quinolonas aac(6’)-Ib-cr en cepas de Escherichia coli y Klebsiella pneumoniae productoras de BLEE aisladas en diez hospitales de Chile. Enferm. Infecc. Microbiol. Clín. 2012, 30, 466–468. [Google Scholar] [CrossRef]
- Pavez, M.; Troncoso, C.; Osses, I.; Salazar, R.; Illesca, V.; Reydet, P.; Rodríguez, C.; Chahin, C.; Concha, C.; Barrientos, L. High prevalence of CTX-M-1 group in ESBL-producing enterobacteriaceae infection in intensive care units in southern Chile. Braz. J. Infect. Dis. 2019, 23, 102–110. [Google Scholar] [CrossRef]
- Benavides, J.A.; Streicker, D.G.; Gonzales, M.S.; Rojas-Paniagua, E.; Shiva, C. Knowledge and use of antibiotics among low-income small-scale farmers of Peru. Prev. Vet. Med. 2021, 189, 105287. [Google Scholar] [CrossRef]
- Albrechtova, K.; Papousek, I.; De Nys, H.M.; Pauly, M.; Anoh, E.; Mossoun, A.; Dolejska, M.; Masarikova, M.; Metzger, S.; Couacy-Hymann, E.; et al. Low Rates of Antimicrobial-Resistant Enterobacteriaceae in Wildlife in Taï National Park, Côte d’Ivoire, Surrounded by Villages with High Prevalence of Multiresistant ESBL-Producing Escherichia coli in People and Domestic Animals. PLoS ONE 2014, 9, e113548. [Google Scholar] [CrossRef]
- Sacristán, I.; Esperón, F.; Acuña, F.; Aguilar, E.; García, S.; López, M.J.; Cevidanes, A.; Neves, E.; Cabello, J.; Hidalgo-Hermoso, E.; et al. Antibiotic resistance genes as landscape anthropization indicators: Using a wild felid as sentinel in Chile. Sci. Total Environ. 2020, 703, 134900. [Google Scholar] [CrossRef]
- Ortega-Paredes, D.; Haro, M.; Leoro-Garzón, P.; Barba, P.; Loaiza, K.; Mora, F.; Fors, M.; Vinueza-Burgos, C.; Fernández-Moreira, E. Multidrug-resistant Escherichia coli isolated from canine faeces in a public park in Quito, Ecuador. J. Glob. Antimicrob. Resist. 2019, 18, 263–268. [Google Scholar] [CrossRef] [PubMed]
- Melo, L.C.; Oresco, C.; Leigue, L.; Netto, H.M.; Melville, P.A.; Benites, N.R.; Saras, E.; Haenni, M.; Lincopan, N.; Madec, J.-Y. Prevalence and molecular features of ESBL/pAmpC-producing Enterobacteriaceae in healthy and diseased companion animals in Brazil. Vet. Microbiol. 2018, 221, 59–66. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, A.; Barbosa, A.; Arais, L.; Ribeiro, P.; Carneiro, V.; Cerqueira, A. Resistance patterns, ESBL genes, and genetic relatedness of Escherichia coli from dogs and owners. Braz. J. Microbiol. 2016, 47, 150–158. [Google Scholar] [CrossRef] [PubMed]
- Rocha-Gracia, R.; Cortés-Cortés, G.; Lozano-Zarain, P.; Bello, F.; Martínez-Laguna, Y.; Torres, C. Faecal Escherichia coli isolates from healthy dogs harbour CTX-M-15 and CMY-2 β-lactamases. Vet. J. 2015, 203, 315–319. [Google Scholar] [CrossRef] [PubMed]
- Salgado-Caxito, M.; Benavides, J.A.; Adell, A.D.; Paes, A.C.; Moreno-Switt, A.I. Global prevalence and molecular characterization of extended-spectrum β-lactamase producing- in dogs and cats—A scoping review and meta-analysis. One Health 2021, 100236, 100236. [Google Scholar] [CrossRef] [PubMed]
- Wedley, A.L.; Dawson, S.; Maddox, T.W.; Coyne, K.P.; Pinchbeck, G.L.; Clegg, P.; Nuttall, T.; Kirchner, M.; Williams, N.J. Carriage of antimicrobial resistant Escherichia coli in dogs: Prevalence, associated risk factors and molecular characteristics. Vet. Microbiol. 2017, 199, 23–30. [Google Scholar] [CrossRef]
- Ljungquist, O.; Ljungquist, D.; Myrenås, M.; Rydén, C.; Finn, M.; Bengtsson, B. Evidence of household transfer of ESBL-/pAmpC-producing Enterobacteriaceae between humans and dogs—A pilot study. Infect. Ecol. Epidemiol. 2016, 6, 31514. [Google Scholar] [CrossRef] [PubMed]
- Sag Resolución Exenta No: 4579/2018; Servicio Agricola Ganadero: Santiago, Chile, 2018.
- Cornejo, J.; Pokrant, E.; Figueroa, F.; Riquelme, R.; Galdames, P.; Di Pillo, F.; Jimenez-Bluhm, P.; Hamilton-West, C. Assessing Antibiotic Residues in Poultry Eggs from Backyard Production Systems in Chile, First Approach to a Non-Addressed Issue in Farm Animals. Animals 2020, 10, 1056. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Benavides, J.A.; Salgado-Caxito, M.; Opazo-Capurro, A.; González Muñoz, P.; Piñeiro, A.; Otto Medina, M.; Rivas, L.; Munita, J.; Millán, J. ESBL-Producing Escherichia coli Carrying CTX-M Genes Circulating among Livestock, Dogs, and Wild Mammals in Small-Scale Farms of Central Chile. Antibiotics 2021, 10, 510. https://doi.org/10.3390/antibiotics10050510
Benavides JA, Salgado-Caxito M, Opazo-Capurro A, González Muñoz P, Piñeiro A, Otto Medina M, Rivas L, Munita J, Millán J. ESBL-Producing Escherichia coli Carrying CTX-M Genes Circulating among Livestock, Dogs, and Wild Mammals in Small-Scale Farms of Central Chile. Antibiotics. 2021; 10(5):510. https://doi.org/10.3390/antibiotics10050510
Chicago/Turabian StyleBenavides, Julio A., Marília Salgado-Caxito, Andrés Opazo-Capurro, Paulina González Muñoz, Ana Piñeiro, Macarena Otto Medina, Lina Rivas, Jose Munita, and Javier Millán. 2021. "ESBL-Producing Escherichia coli Carrying CTX-M Genes Circulating among Livestock, Dogs, and Wild Mammals in Small-Scale Farms of Central Chile" Antibiotics 10, no. 5: 510. https://doi.org/10.3390/antibiotics10050510
APA StyleBenavides, J. A., Salgado-Caxito, M., Opazo-Capurro, A., González Muñoz, P., Piñeiro, A., Otto Medina, M., Rivas, L., Munita, J., & Millán, J. (2021). ESBL-Producing Escherichia coli Carrying CTX-M Genes Circulating among Livestock, Dogs, and Wild Mammals in Small-Scale Farms of Central Chile. Antibiotics, 10(5), 510. https://doi.org/10.3390/antibiotics10050510






