Microscopic Analysis of Bacterial Inoculum Effect Using Micropatterned Biochip
Abstract
1. Introduction
2. Results
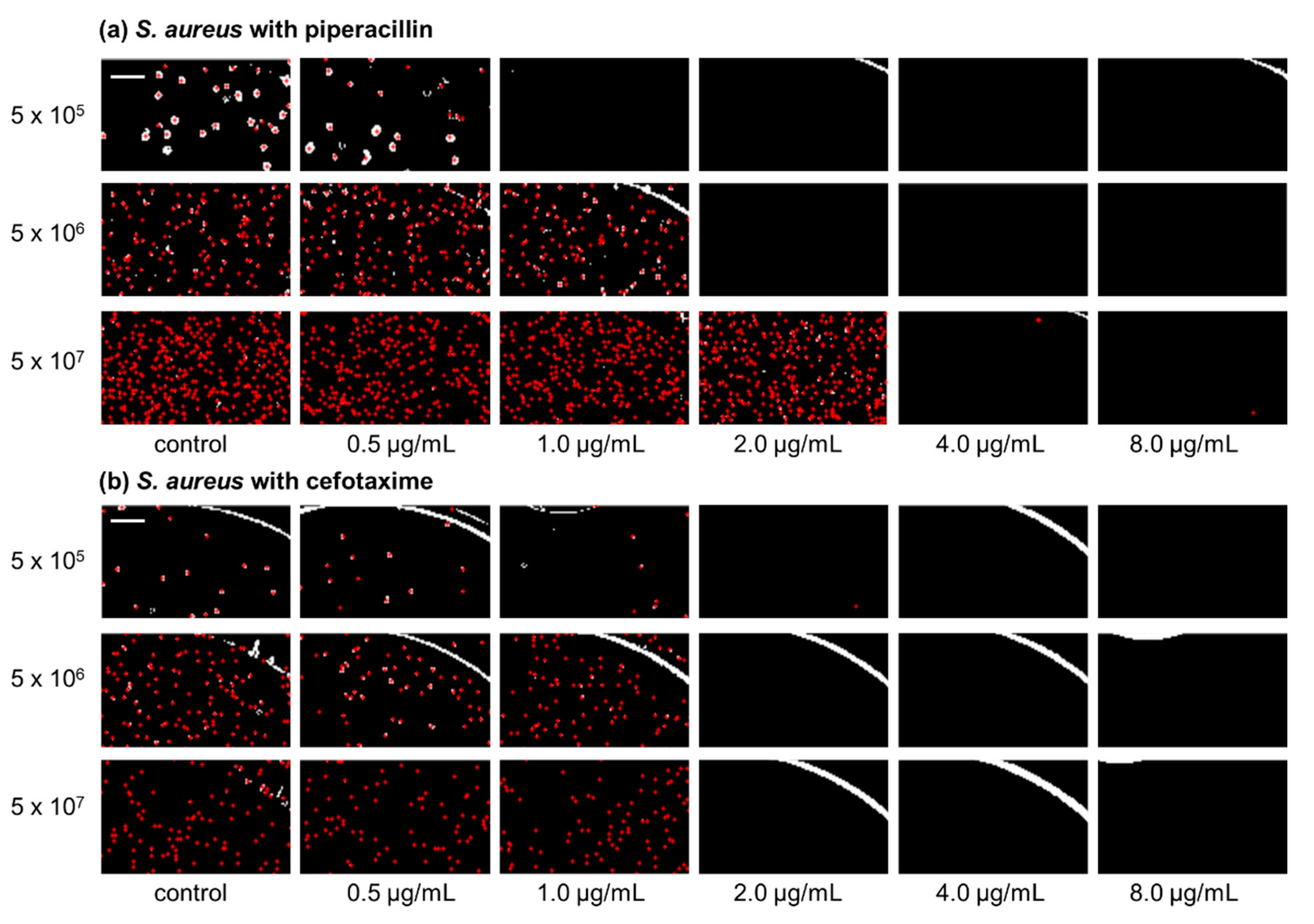
2.1. Inoculum Effect of Staphylococcus aureus ATCC 29213
2.2. Inoculum Effect of Pseudomonas aeruginosa ATCC 27853
2.3. Inoculum Effect of Escherichia coli ATCC 25922
2.4. Inoculum Effect of Enterococcus faecalis ATCC 29212
3. Discussion
4. Materials and Methods
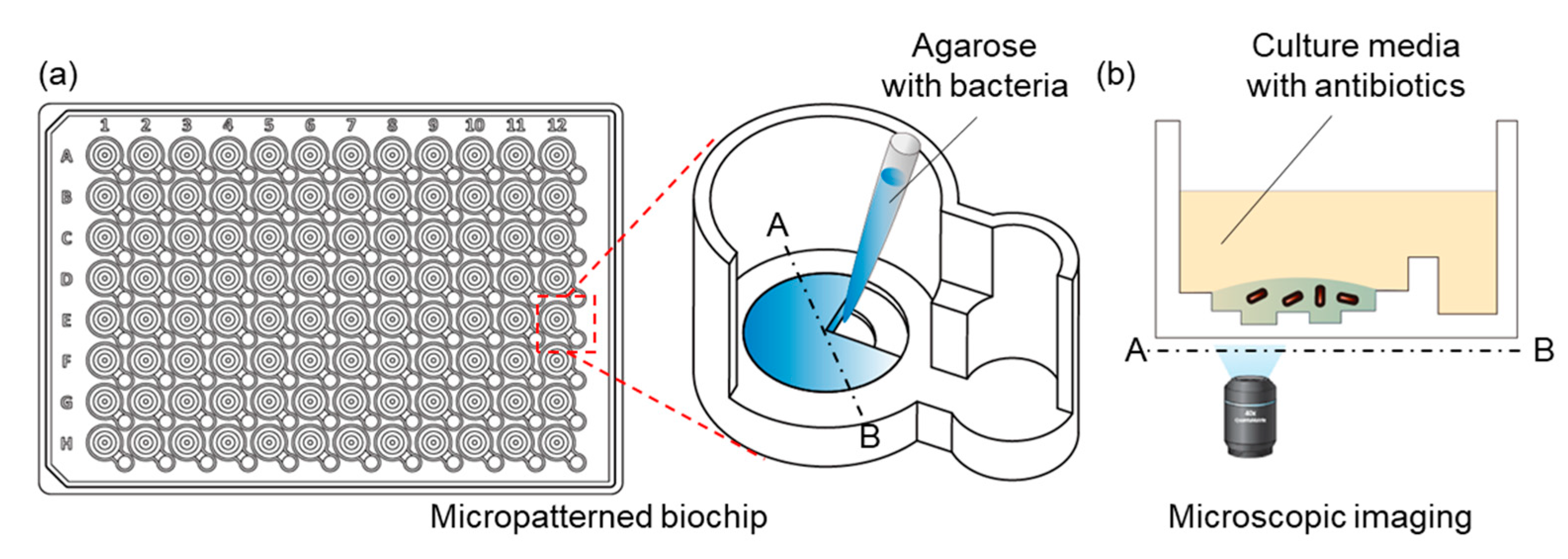
4.1. Fabrication of the IBAST Chip
4.2. Stock Preparation and Subculture of Bacteria
4.3. Antibiotic Preparation
4.4. Inoculum Sizes of Bacteria
4.5. Determination of Growth of Bacteria
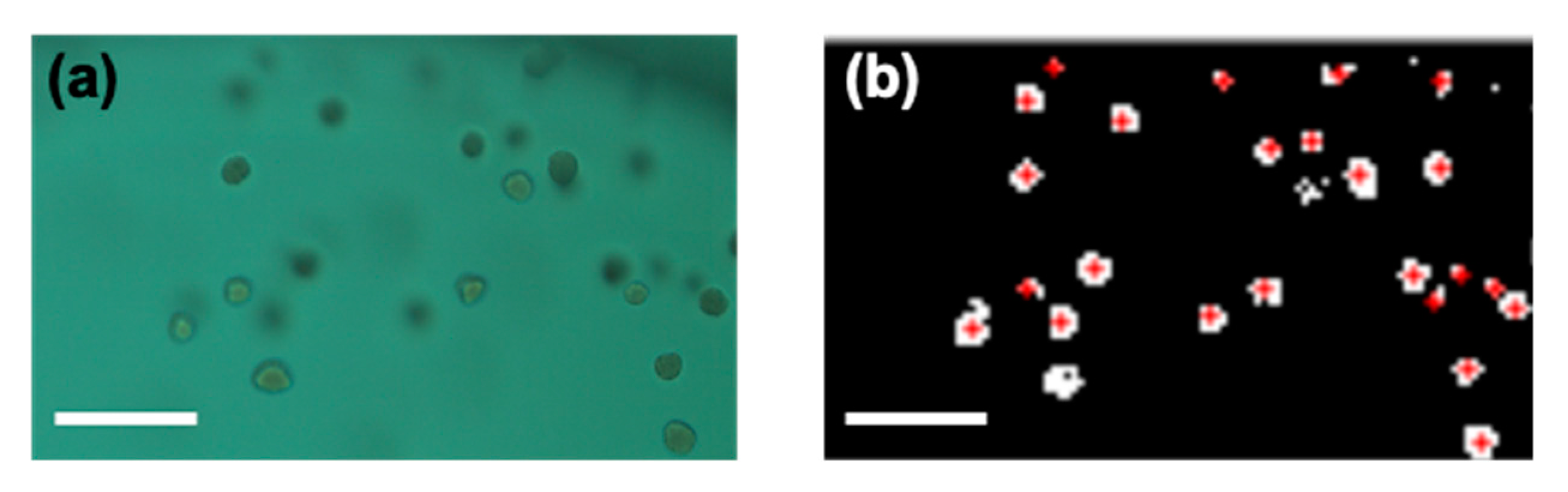
4.6. Image Processing
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Blair, J.M.A.; Webber, M.A.; Baylay, A.J.; Ogbolu, D.O.; Piddock, L.J.V. Molecular mechanisms of antibiotic resistance. Nat. Rev. Microbiol. 2015, 13, 42–51. [Google Scholar] [CrossRef]
- Coll, F.; Harrison, E.M.; Toleman, M.S.; Reuter, S.; Raven, K.E.; Blane, B.; Palmer, B.; Kappeler, A.R.M.; Brown, N.M.; Torok, M.E.; et al. Longitudinal genomic surveillance of MRSA in the UK reveals transmission patterns in hospitals and the community. Sci. Transl. Med. 2017, 9. [Google Scholar] [CrossRef] [PubMed]
- WHO. Antimicrobial Resistance; Global report on surveillance; World Health Organization: Geneva, Switzerland, 2014. [Google Scholar] [CrossRef]
- CDC. Antibiotic Resistance Threats in the United States, 2013; CDC: Atlanta, GA, USA, 2013. [Google Scholar] [CrossRef]
- Balouiri, M.; Sadiki, M.; Ibnsouda, S.K. Methods for in vitro evaluating antimicrobial activity: A review. J. Pharm. Anal. 2016, 6, 71–79. [Google Scholar] [CrossRef]
- Wiegand, I.; Hilpert, K.; Hancock, R.E.W. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat. Protoc. 2008. [Google Scholar] [CrossRef]
- Jorgensen, J.H.; Turnidge, J.D. Susceptibility Test Methods: Dilution and Disk Diffusion Methods. Man. Clin. Microbiol. 2015. [Google Scholar] [CrossRef]
- Kara, V.; Duan, C.H.; Gupta, K.; Kurosawa, S.; Stearns-Kurosawa, D.J.; Ekinci, K.L. Microfluidic detection of movements of Escherichia coli for rapid antibiotic susceptibility testing. Lab Chip 2018, 18, 743–753. [Google Scholar] [CrossRef]
- Li, Y.Y.; Yang, X.; Zhao, W.A. Emerging Microtechnologies and Automated Systems for Rapid Bacterial Identification and Antibiotic Susceptibility Testing. Slas. Technol. 2017, 22, 585–608. [Google Scholar] [CrossRef]
- Rossello, G.A.M.; Perez, M.A.B. Rapid antibiotic susceptibility test in Clinical Microbiology. Enferm. Infec. Micr. Clin. 2016, 34, 61–68. [Google Scholar] [CrossRef]
- Wistrand-Yuen, P.; Malmberg, C.; Fatsis-Kavalopoulos, N.; Lubke, M.; Tangden, T.; Kreuger, J. A Multiplex Fluidic Chip for Rapid Phenotypic Antibiotic Susceptibility Testing. Mbio 2020, 11. [Google Scholar] [CrossRef]
- Choi, J.; Yoo, J.; Lee, M.; Kim, E.G.; Lee, J.S.; Lee, S.; Joo, S.; Song, S.H.; Kim, E.C.; Lee, J.C.; et al. A rapid antimicrobial susceptibility test based on single-cell morphological analysis. Sci. Transl. Med. 2014, 6. [Google Scholar] [CrossRef]
- Lukačišinová, M.; Bollenbach, T. Toward a quantitative understanding of antibiotic resistance evolution. Curr. Opin. Biotechnol. 2017. [Google Scholar] [CrossRef]
- Nicoloff, H.; Andersson, D.I. Indirect resistance to several classes of antibiotics in cocultures with resistant bacteria expressing antibiotic-modifying or -degrading enzymes. J. Antimicrob. Chemother. 2016, 71, 100–110. [Google Scholar] [CrossRef]
- Soriano, F.; Ponte, C.; Santamaria, M.; Jimenez-Arriero, M. Relevance of the inoculum effect of antibiotics in the outcome of experimental infections caused by Escherichia coli. J. Antimicrob. Chemother. 1990, 25, 621–627. [Google Scholar] [CrossRef]
- Martinez, J.L.; Baquero, F. Mutation frequencies and antibiotic resistance. Antimicrob. Agents Chemother. 2000. [Google Scholar] [CrossRef]
- Choi, J.; Jung, Y.G.; Kim, J.; Kim, S.; Jung, Y.; Na, H.; Kwon, S. Rapid antibiotic susceptibility testing by tracking single cell growth in a microfluidic agarose channel system. Lab Chip 2013, 13, 280–287. [Google Scholar] [CrossRef]
- Georgopapadakou, N.H. Penicillin-binding proteins and bacterial resistance to β-lactams. Antimicrob. Agents Chemother. 1993. [Google Scholar] [CrossRef]
- Kohanski, M.A.; Dwyer, D.J.; Collins, J.J. How antibiotics kill bacteria: From targets to networks. Nat. Rev. Microbiol. 2010. [Google Scholar] [CrossRef]
- Konig, C.; Simmen, H.P.; Blaser, J. Bacterial concentrations in pus and infected peritoneal fluid--implications for bactericidal activity of antibiotics. J. Antimicrob. Chemother. 1998, 42, 227–232. [Google Scholar] [CrossRef]
- Song, K.H.; Jung, S.I.; Lee, S.; Park, S.; Kim, E.S.; Park, K.H.; Park, W.B.; Choe, P.G.; Kim, Y.K.; Kwak, Y.G.; et al. Inoculum effect of methicillin-susceptible Staphylococcus aureus against broad-spectrum beta-lactam antibiotics. Eur. J. Clin. Microbiol. Infect. Dis. 2019, 38, 67–74. [Google Scholar] [CrossRef]
- Palmer, S.M.; Kang, S.L.; Cappelletty, D.M.; Rybak, M.J. Bactericidal killing activities of cefepime, ceftazidime, cefotaxime, and ceftriaxone against Staphylococcus aureus and beta-lactamase-producing strains of Enterobacter aerogenes and Klebsiella pneumoniae in an in vitro infection model. Antimicrob. Agents. Chemother. 1995, 39, 1764–1771. [Google Scholar] [CrossRef]
- Steckelberg, J.M.; Rouse, M.S.; Tallan, B.M.; Osmon, D.R.; Henry, N.K.; Wilson, W.R. Relative efficacies of broad-spectrum cephalosporins for treatment of methicillin-susceptible Staphylococcus aureus experimental infective endocarditis. Antimicrob. Agents Chemother. 1993, 37, 554–558. [Google Scholar] [CrossRef][Green Version]
- Mizuguchi, Y.; Ogawa, M.; Udou, T. Morphological changes induced by beta-lactam antibiotics in Mycobacterium avium-intracellulare complex. Antimicrob. Agents Chemother. 1985, 27, 541–547. [Google Scholar] [CrossRef] [PubMed]
- Li, R.C.; Ma, H.H. Parameterization of inoculum effect via mathematical modeling: Aminoglycosides against Staphylococcus aureus and Escherichia coli. J. Chemother. 1998, 10, 203–207. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Xie, S.; Ahmed, S.; Wang, F.; Gu, Y.; Zhang, C.; Chai, X.; Wu, Y.; Cai, J.; Cheng, G. Antimicrobial activity and resistance: Influencing factors. Front. Pharmacol. 2017. [Google Scholar] [CrossRef] [PubMed]
- Mizunaga, S.; Kamiyama, T.; Fukuda, Y.; Takahata, M.; Mitsuyama, J. Influence of inoculum size of Staphylococcus aureus and Pseudomonas aeruginosa on in vitro activities and in vivo efficacy of fluoroquinolones and carbapenems. J. Antimicrob. Chemother. 2005, 56, 91–96. [Google Scholar] [CrossRef]
- Chan, E.; Zhou, S.; Srikumar, S.; Duan, W. Use of in vitro critical inhibitory concentration, a novel approach to predict in vivo synergistic bactericidal effect of combined amikacin and piperacillin against Pseudomonas aeruginosa in a systemic rat infection model. Pharm. Res. 2006, 23, 729–741. [Google Scholar] [CrossRef]
- Eng, R.H.; Smith, S.M.; Cherubin, C. Inoculum effect of new beta-lactam antibiotics on Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 1984, 26, 42–47. [Google Scholar] [CrossRef]
- Brook, I. Inoculum effect. Rev. Infect. Dis. 1989, 11, 361–368. [Google Scholar] [CrossRef]
- Corrado, M.L.; Landesman, S.H.; Cherubin, C.E. Influence of inoculum size on activity of cefoperazone, cefotaxime, moxalactam, piperacillin, and N-formimidoyl thienamycin (MK0787) against Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 1980, 18, 893–896. [Google Scholar] [CrossRef]
- Masuyoshi, S.; Arai, S.; Miyamoto, M.; Mitsuhashi, S. In vitro antimicrobial activity of cefotaxime, a new cephalosporin. Antimicrob. Agents Chemother. 1980, 18, 1–8. [Google Scholar] [CrossRef]
- Kresken, M.; Korber-Irrgang, B.; Lauffer, J.; Decker-Burgard, S.; Davies, T. In vitro activities of ceftobiprole combined with amikacin or levofloxacin against Pseudomonas aeruginosa: Evidence of a synergistic effect using time-kill methodology. Int. J. Antimicrob. Agents 2011, 38, 70–75. [Google Scholar] [CrossRef]
- Gombert, M.E.; Aulicino, T.M. Comparison of agar dilution, microtitre broth dilution and tube macrodilution susceptibility testing of ciprofloxacin against several pathogens at two different inocula. J. Antimicrob. Chemother. 1985, 16, 709–712. [Google Scholar] [CrossRef]
- Odenholt, I.; Lowdin, E.; Cars, O. Bactericidal effects of levofloxacin in comparison with those of ciprofloxacin and sparfloxacin. Clin. Microbiol. Infect. 1998, 4, 264–270. [Google Scholar] [CrossRef] [PubMed]
- Firsov, A.A.; Vostrov, S.N.; Kononenko, O.V.; Zinner, S.H.; Portnoy, Y.A. Prediction of the effects of inoculum size on the antimicrobial action of trovafloxacin and ciprofloxacin against Staphylococcus aureus and Escherichia coli in an in vitro dynamic model. Antimicrob. Agents Chemother. 1999, 43, 498–502. [Google Scholar] [CrossRef] [PubMed]
- Fass, R.J. In vitro activity of ciprofloxacin (Bay o 9867). Antimicrob. Agents Chemother. 1983, 24, 568–574. [Google Scholar] [CrossRef] [PubMed]
- Queenan, A.M.; Foleno, B.; Gownley, C.; Wira, E.; Bush, K. Effects of inoculum and beta-lactamase activity in AmpC-and extended-spectrum beta-lactamase (ESBL)-producing Escherichia coli and Klebsiella pneumoniae clinical isolates tested by using NCCLS ESBL methodology. J. Clin. Microbiol. 2004, 42, 269–275. [Google Scholar] [CrossRef] [PubMed]
- Soriano, F.; Garcia-Corbeira, P.; Ponte, C.; Fernandez-Roblas, R.; Gadea, I. Correlation of pharmacodynamic parameters of five beta-lactam antibiotics with therapeutic efficacies in an animal model. Antimicrob. Agents Chemother. 1996, 40, 2686–2690. [Google Scholar] [CrossRef] [PubMed]
- Thomson, K.S.; Moland, E.S. Cefepime, piperacillin-tazobactam, and the inoculum effect in tests with extended-spectrum beta-lactamase-producing Enterobacteriaceae. Antimicrob. Agents Chemother. 2001, 45, 3548–3554. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, Y.; Nakano, R.; Kasahara, K.; Mizuno, T.; Hirai, N.; Nakano, A.; Suzuki, Y.; Kakuta, N.; Masui, T.; Yano, H.; et al. Comparison of the inoculum size effects of antibiotics on IMP-6 beta-lactamase-producing Enterobacteriaceae co-harboring plasmid-mediated quinolone resistance genes. PLoS ONE 2019, 14, e0225210. [Google Scholar] [CrossRef]
- Goldstein, E.J.; Citron, D.M.; Cherubin, C.E. Comparison of the inoculum effects of members of the family Enterobacteriaceae on cefoxitin and other cephalosporins, beta-lactamase inhibitor combinations, and the penicillin-derived components of these combinations. Antimicrob. Agents Chemother. 1991, 35, 560–566. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Patterson, J.E.; Zervos, M.J. Susceptibility and bactericidal activity studies of four beta-lactamase-producing enterococci. Antimicrob. Agents Chemother. 1989, 33, 251–253. [Google Scholar] [CrossRef] [PubMed]
- Murray, B.E.; Mederski-Samoraj, M.; Foster, S.K.; Brunton, J.L.; Harford, P. In vitro studies of plasmid-mediated penicillinase from Streptococcus faecalis suggest a staphylococcal origin. J. Clin. Investig. 1986. [Google Scholar] [CrossRef]
- Markowitz, S.M.; Wells, V.D.; Williams, D.S.; Stuart, C.G.; Coudron, P.E.; Wong, E.S. Antimicrobial susceptibility and molecular epidemiology of β-lactamase-producing, aminoglycoside-resistant isolates of Enterococcus faecalis. Antimicrob. Agents Chemother. 1991. [Google Scholar] [CrossRef]
| Antibiotics | Ampicillin | Cefotaxime | Levofloxacin | Amikacin | Piperacillin | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Inoculum size (CFU/mL) | 5 × 105 | 5 × 106 | 5 × 107 | 5 × 105 | 5 × 106 | 5 × 107 | 5 × 105 | 5 × 106 | 5 × 107 | 5 × 105 | 5 × 106 | 5 × 107 | 5 × 105 | 5 × 106 | 5 × 107 | |
| Volume of culture media w/antibiotics (μL) | 45 | 0.25 | 1 | 4 | − | − | − | 0.25 | 0.25 | 0.25 | 1 | 1 | 2 | 1 | 2 | 8 |
| 90 | 0.25 | 0.5 | 2 | 2 | 2 | 2 | 0.12 | 0.25 | 0.25 | 1 | 1 | 2 | 1 | 2 | 4 | |
| 135 | 0.25 | 0.5 | 2 | − | − | − | 0.12 | 0.25 | 0.25 | 1 | 1 | 2 | 1 | 2 | 2 | |
| Antibiotics | Cefotaxime | Levofloxacin | Amikacin | Piperacillin | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Inoculum size (CFU/mL) | 5 × 105 | 5 × 106 | 5 × 107 | 5 × 105 | 5 × 106 | 5 × 107 | 5 × 105 | 5 × 106 | 5 × 107 | 5 × 105 | 5 × 106 | 5 × 107 | |
| Volume of culture media w/antibiotics (μL) | 45 | 8 | 32 | 32 | 0.5 | 1 | 1 | 0.5 | 1 | 2 | 2 | 4 | >16 |
| 90 | 8 | 16 | 16 | 0.5 | 1 | 1 | 0.5 | 1 | 2 | 2 | 4 | >16 | |
| 135 | 8 | 16 | >64 | 0.5 | 1 | 1 | 0.5 | 1 | 2 | 2 | 4 | >16 | |
| Antibiotics | Ampicillin | Cefotaxime | Levofloxacin | Amikacin | Piperacillin | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Inoculum size (CFU/mL) | 5 × 105 | 5 × 106 | 5 × 107 | 5 × 105 | 5 × 106 | 5 × 107 | 5 × 105 | 5 × 106 | 5 × 107 | 5 × 105 | 5 × 106 | 5 × 107 | 5 × 105 | 5 × 106 | 5 × 107 | |
| Volume of culture media w/antibiotics (μL) | 45 | − | − | − | 0.12 | 0.12 | >0.25 | − | − | − | 0.5 | 0.5 | 1 | − | − | − |
| 90 | 8 | 8 | 8 | 0.06 | 0.12 | 0.12 | 0.015 | 0.015 | 0.015 | 0.5 | 0.5 | 1 | 4 | 4 | 4 | |
| 135 | − | − | − | 0.06 | 0.12 | >0.25 | − | − | − | 0.5 | 0.5 | 1 | − | − | − | |
| Antibiotics | Ampicillin | Levofloxacin | Piperacillin | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Inoculum size (CFU/mL) | 5 × 105 | 5 × 106 | 5 × 107 | 5 × 105 | 5 × 106 | 5 × 107 | 5 × 105 | 5 × 106 | 5 × 107 | |
| Volume of culture media w/antibiotics (μL) | 45 | − | − | − | 0.5 | 1 | 2 | − | − | − |
| 90 | 1 | 1 | 1 | 0.5 | 1 | 1 | 2 | 2 | 2 | |
| 135 | − | − | − | 0.5 | 1 | 1 | − | − | − | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hwang, J.H.; Lee, S.Y.; Choi, J. Microscopic Analysis of Bacterial Inoculum Effect Using Micropatterned Biochip. Antibiotics 2021, 10, 300. https://doi.org/10.3390/antibiotics10030300
Hwang JH, Lee SY, Choi J. Microscopic Analysis of Bacterial Inoculum Effect Using Micropatterned Biochip. Antibiotics. 2021; 10(3):300. https://doi.org/10.3390/antibiotics10030300
Chicago/Turabian StyleHwang, Jung Ho, Sang Young Lee, and Jungil Choi. 2021. "Microscopic Analysis of Bacterial Inoculum Effect Using Micropatterned Biochip" Antibiotics 10, no. 3: 300. https://doi.org/10.3390/antibiotics10030300
APA StyleHwang, J. H., Lee, S. Y., & Choi, J. (2021). Microscopic Analysis of Bacterial Inoculum Effect Using Micropatterned Biochip. Antibiotics, 10(3), 300. https://doi.org/10.3390/antibiotics10030300







