Identification of Risk Factors Associated with Resistant Escherichia coli Isolates from Poultry Farms in the East Coast of Peninsular Malaysia: A Cross Sectional Study
Abstract
1. Introduction
2. Results
3. Discussion
4. Materials and Methods
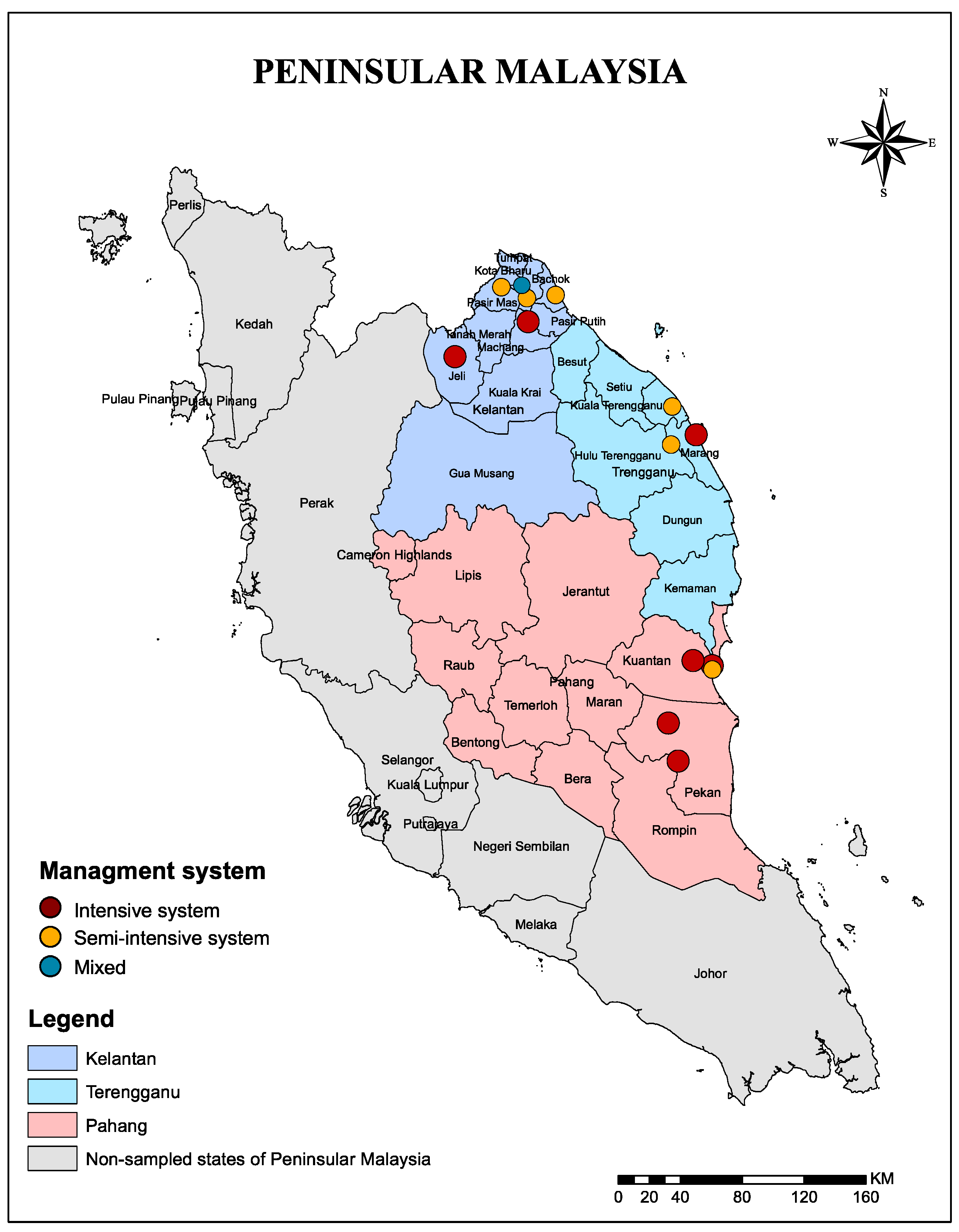
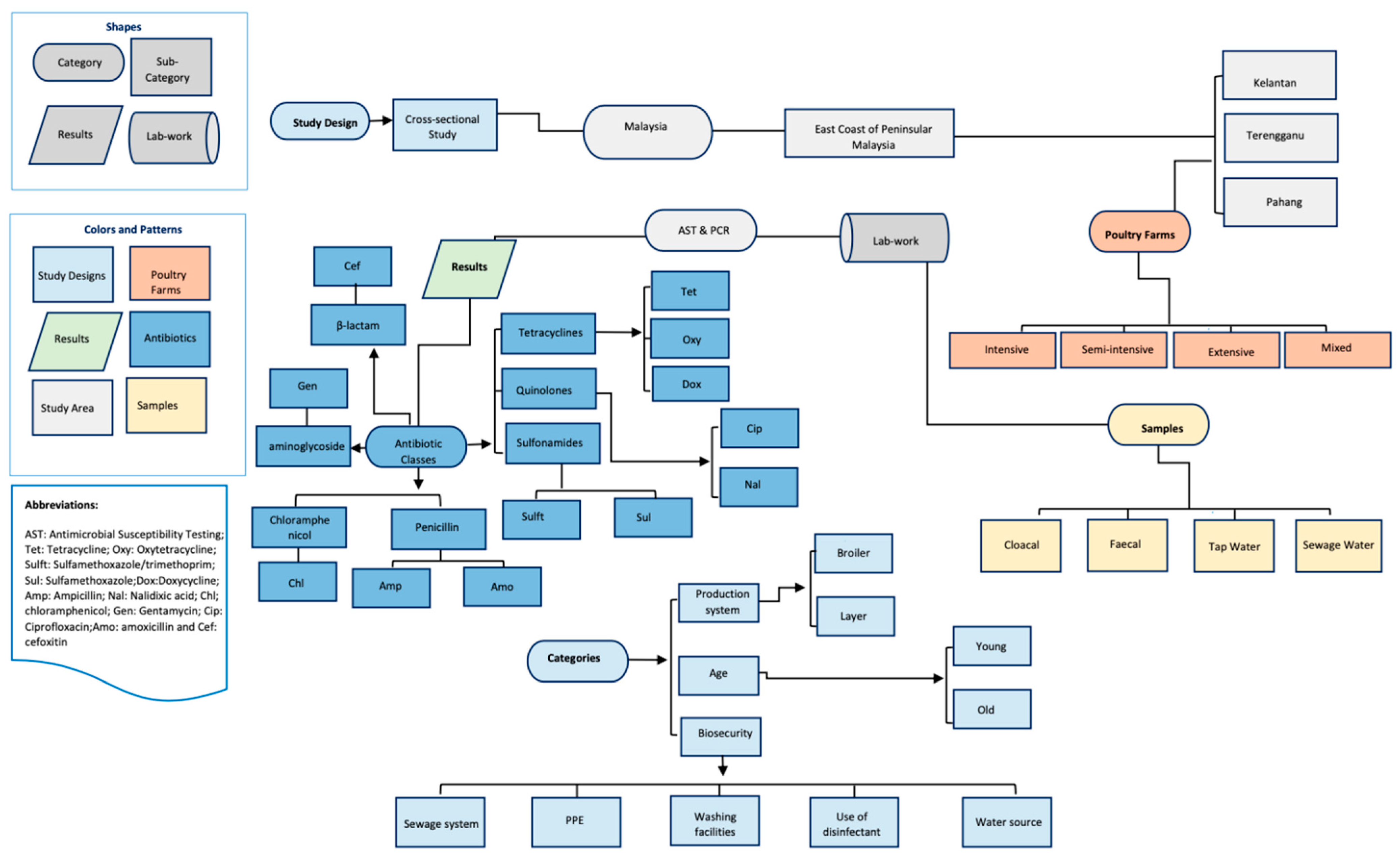
4.1. Study Area
4.2. Study Design, Definitions, and Data Sources
- Intensive management system is defined as mainly concentrated and often mechanized operations that use controlled-environment systems to provide the ideal thermal environment for the poultry.
- Semi-intensive system is that which relies on natural airflow though the shed for ventilation.
- Extensive system is mainly pasture-based and land-based where birds in the household flock are typically housed overnight in the shelter and are let out in the morning to forage during the day.
- The criteria of the farm size included large-scale commercial farms that has more than ≥10,000 birds, medium-scale commercial farms that has more 5000–10,000, and small-scale farms where birds are often kept in single-age groups of >1000.
- A poor sewage system is defined as that which retains high volumes of wastewater with low flow rate, blackish appearance, and sewage smell odour as a result of composing agricultural waste—probably as leakage from nearby irrigated effluent which is used for agricultural land application along with the presence of food waste, green waste, plastic, and heavy materials.
- A good sewage system is that which has good drainage with no agricultural waste and relatively low heavy materials.
- Excellent swage system is that which has significant drainage, no agriculture, and heavy materials.
4.3. Microbiological Testing
4.4. Antimicrobial Susceptibility Testing
4.5. PCR
4.5.1. DNA Extraction of Escherichia Coli Isolates
4.5.2. Primers and PCR Assay for Specific Genes
4.6. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Prestinaci, F.; Pezzotti, P.; Pantosti, A. Antimicrobial resistance: A global multifaceted phenomenon. Pathog. Glob. Health 2015, 109, 309–318. [Google Scholar] [CrossRef] [PubMed]
- Holmes, A.H.; Moore, L.S.; Sundsfjord, A.; Steinbakk, M.; Regmi, S.; Karkey, A.; Guerin, P.J.; Piddock, L.J. Understanding the mechanisms and drivers of antimicrobial resistance. Lancet 2016, 387, 176–187. [Google Scholar] [CrossRef]
- Winckler, C.; Grafe, A. Use of veterinary drugs in intensive animal production. J. Soils Sediments 2001, 1, 66. [Google Scholar] [CrossRef]
- Sun, P.; Barmaz, D.; Cabrera, M.L.; Pavlostathis, S.G.; Huang, C.-H. Detection and quantification of ionophore antibiotics in runoff, soil and poultry litter. J. Chromatogr. A 2013, 1312, 10–17. [Google Scholar] [CrossRef] [PubMed]
- Kay, P.; Blackwell, P.A.; Boxall, A.B. Transport of veterinary antibiotics in overland flow following the application of slurry to arable land. Chemosphere 2005, 59, 951–959. [Google Scholar] [CrossRef] [PubMed]
- Phillips, I. Withdrawal of growth-promoting antibiotics in Europe and its effects in relation to human health. Int. J. Antimicrob. Agents 2007, 30, 101–107. [Google Scholar] [CrossRef]
- Akbar, A.; Sitara, U.; Ali, I.; Khan, M.I.; Phadungchob, T.; Anal, A.K. Presence of Escherichia coli in poultry meat: A potential food safety threat. Int. Food Res. J. 2014, 21, 941. [Google Scholar]
- Van, T.T.H.; Chin, J.; Chapman, T.; Tran, L.T.; Coloe, P.J. Safety of raw meat and shellfish in Vietnam: An analysis of Escherichia coli isolations for antibiotic resistance and virulence genes. Int. J. Food Microbiol. 2008, 124, 217–223. [Google Scholar] [CrossRef]
- Acar, J.; Moulin, G. Antimicrobial resistance at farm level. Rev. Sci. Tech. 2006, 25, 775–792. [Google Scholar] [CrossRef]
- Sarkar, P.; Gould, I.M. Antimicrobial agents are societal drugs. Drugs 2006, 66, 893–901. [Google Scholar] [CrossRef]
- Hammerum, A.M.; Heuer, O.E.; Emborg, H.-D.; Bagger-Skjøt, L.; Jensen, V.F.; Rogues, A.-M.; Skov, R.L.; Agersø, Y.; Brandt, C.T.; Seyfarth, A.M. Danish integrated antimicrobial resistance monitoring and research program. Emerg. Infect. Dis. 2007, 13, 1633. [Google Scholar] [CrossRef] [PubMed]
- Mathew, A.G.; Cissell, R.; Liamthong, S. Antibiotic resistance in bacteria associated with food animals: A United States perspective of livestock production. Foodborne Pathog. Dis. 2007, 4, 115–133. [Google Scholar] [CrossRef] [PubMed]
- Prescott, J.F. Antimicrobial use in food and companion animals. Anim. Health Res. Rev. 2008, 9, 127. [Google Scholar] [CrossRef] [PubMed]
- Donado-Godoy, P.; Gardner, I.; Byrne, B.A.; Leon, M.; Perez-Gutierrez, E.; Ovalle, M.; Tafur, M.; Miller, W. Prevalence, risk factors, and antimicrobial resistance profiles of Salmonella from commercial broiler farms in two important poultry-producing regions of Colombia. J. Food Prot. 2012, 75, 874–883. [Google Scholar] [CrossRef]
- Planta, M.B. The role of poverty in antimicrobial resistance. J. Am. Board Fam. Med. 2007, 20, 533–539. [Google Scholar] [CrossRef]
- Abo-Amer, A.E.; Shobrak, M.Y.; Altalhi, A.D. Isolation and antimicrobial resistance of Escherichia coli isolated from farm chickens in Taif, Saudi Arabia. J. Glob. Antimicrob. Resist. 2018, 15, 65–68. [Google Scholar] [CrossRef]
- Messele, Y.E.; Abdi, R.D.; Yalew, S.T.; Tegegne, D.T.; Emeru, B.A.; Werid, G.M. Molecular determination of antimicrobial resistance in Escherichia coli isolated from raw meat in Addis Ababa and Bishoftu, Ethiopia. Ann. Clin. Microbiol. Antimicrob. 2017, 16, 55. [Google Scholar] [CrossRef]
- Torkan, S.; Khamesipour, F.; Anyanwu, M. Detection of virulence and antibacterial resistance genes in Salmonella isolates from diarrhoeic dogs in Iran. Rev. Méd. Vét. 2015, 166, 221–228. [Google Scholar]
- Aliyu, A.; Saleha, A.; Jalila, A.; Zunita, Z. Risk factors and spatial distribution of extended spectrum β-lactamase-producing-Escherichia coli at retail poultry meat markets in Malaysia: A cross-sectional study. BMC Public Health 2016, 16, 699. [Google Scholar] [CrossRef]
- Imtiaz, J.; Hashmi, I.; Saeed, A.; Qazi, I.; Arshad, M. Development of PCR Protocol for Detection of Escherichia Coli in Drinking Water. In Water Resources Management VII; Brebbia, C.A., Ed.; WIT PRESS: Southhampton, UK, 2013; pp. 225–232. [Google Scholar]
- Tang, K.L.; Caffrey, N.P.; Nóbrega, D.B.; Cork, S.C.; Ronksley, P.E.; Barkema, H.W.; Polachek, A.J.; Ganshorn, H.; Sharma, N.; Kellner, J.D. Restricting the use of antibiotics in food-producing animals and its associations with antibiotic resistance in food-producing animals and human beings: A systematic review and meta-analysis. Lancet Planet. Health 2017, 1, e316–e327. [Google Scholar] [CrossRef]
- Usui, M.; Ozawa, S.; Onozato, H.; Kuge, R.; Obata, Y.; Uemae, T.; Ngoc, P.T.; Heriyanto, A.; Chalemchaikit, T.; Makita, K. Antimicrobial susceptibility of indicator bacteria isolated from chickens in Southeast Asian countries (Vietnam, Indonesia and Thailand). J. Vet. Med. Sci. 2014, 76. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, V.T.; Carrique-Mas, J.J.; Ngo, T.H.; Ho, H.M.; Ha, T.T.; Campbell, J.I.; Nguyen, T.N.; Hoang, N.N.; Pham, V.M.; Wagenaar, J.A. Prevalence and risk factors for carriage of antimicrobial-resistant Escherichia coli on household and small-scale chicken farms in the Mekong Delta of Vietnam. J. Antimicrob. Chemother. 2015, 70, 2144–2152. [Google Scholar] [PubMed]
- Shahaza, O.; Zakirah, S.; Muhammad, A.; Syamsyul, A.; Maswati, M. Antimicrobial resistance in veterinary clinical isolates of Escherichia coli from northern region of Peninsular Malaysia. Malays. J. Vet. Res. 2017, 8, 1–7. [Google Scholar]
- Cheong, Y.; Lim, V.; Jegathesan, M.; Suleiman, A. Antimicrobial resistance in 6 Malaysian general hospitals. Med. J. Malays. 1994, 49, 317–326. [Google Scholar]
- Aarestrup, F.M.; Woolhouse, M.E. Using sewage for surveillance of antimicrobial resistance. Science 2020, 367, 630–632. [Google Scholar] [CrossRef] [PubMed]
- Dangour, A.D.; Watson, L.; Cumming, O.; Boisson, S.; Che, Y.; Velleman, Y.; Cavill, S.; Allen, E.; Uauy, R. Interventions to improve water quality and supply, sanitation and hygiene practices, and their effects on the nutritional status of children. Cochrane Database Syst. Rev. 2013, 1. [Google Scholar] [CrossRef]
- Chuah, L.-O.; He, X.B.; Effarizah, M.E.; Syahariza, Z.A.; Shamila-Syuhada, A.K.; Rusul, G. Mislabelling of beef and poultry products sold in Malaysia. Food Control 2016, 62, 157–164. [Google Scholar] [CrossRef]
- Bong, C.W.; Chai, S.K.; Chai, L.C.; Wang, A.J.; Lee, C.W. Prevalence and characterization of Escherichia coli in the Kelantan River and its adjacent coastal waters. Water Supply 2020, 20, 930–942. [Google Scholar] [CrossRef]
- Alhaj, N.; Mariana, N.; Raha, A.; Ishak, Z. Prevalence of antibiotic resistance among Escherichia coli from different sources in Malaysia. Int. J. Poult. Sci. 2007, 6, 293–297. [Google Scholar] [CrossRef]
- Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing, 26th ed; CLSI Supplement M100; CLSI: Wayne, PA, USA, 2016. [Google Scholar]
- Murphy, C.P.; Carson, C.; Smith, B.A.; Chapman, B.; Marrotte, J.; McCann, M.; Primeau, C.; Sharma, P.; Parmley, E.J. Factors potentially linked with the occurrence of antimicrobial resistance in selected bacteria from cattle, chickens and pigs: A scoping review of publications for use in modelling of antimicrobial resistance (IAM. AMR Project). Zoonoses Public Health 2018, 65, 957–971. [Google Scholar] [CrossRef]
- Ibrahim, R.A.; Cryer, T.L.; Lafi, S.Q.; Basha, E.-A.; Good, L.; Tarazi, Y.H. Identification of Escherichia coli from broiler chickens in Jordan, their antimicrobial resistance, gene characterization and the associated risk factors. BMC Vet. Res. 2019, 15, 159. [Google Scholar] [CrossRef] [PubMed]
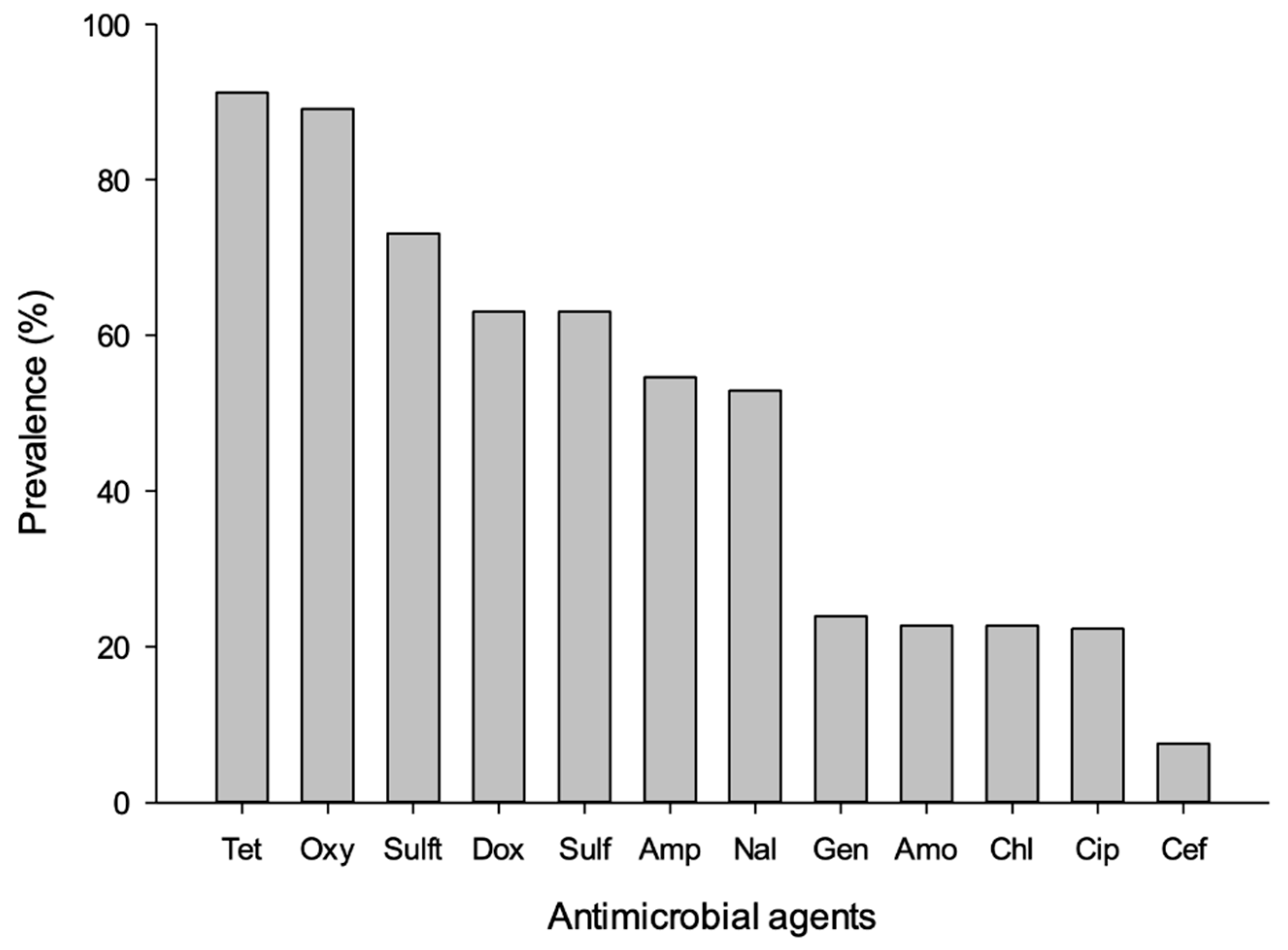
| Risk Factors | Samples Tested | Affected (%) | p-Value |
|---|---|---|---|
| Age | 0.511 | ||
| Young | 187 | 123 (65.8%) | |
| Adult | 184 | 115 (62.5%) | |
| Management system | 0.541 | ||
| Intensive | 187 | 115 (61.5%) | |
| Semi-intensive | 158 | 105 (66.5%) | |
| Mixed | 26 | 18 (69.2%) | |
| Production system | 0.278 | ||
| Broiler | 212 | 129 (60.8%) | |
| Layer | 53 | 35 (66%) | |
| Mixed | 106 | 74 (69.8%) | |
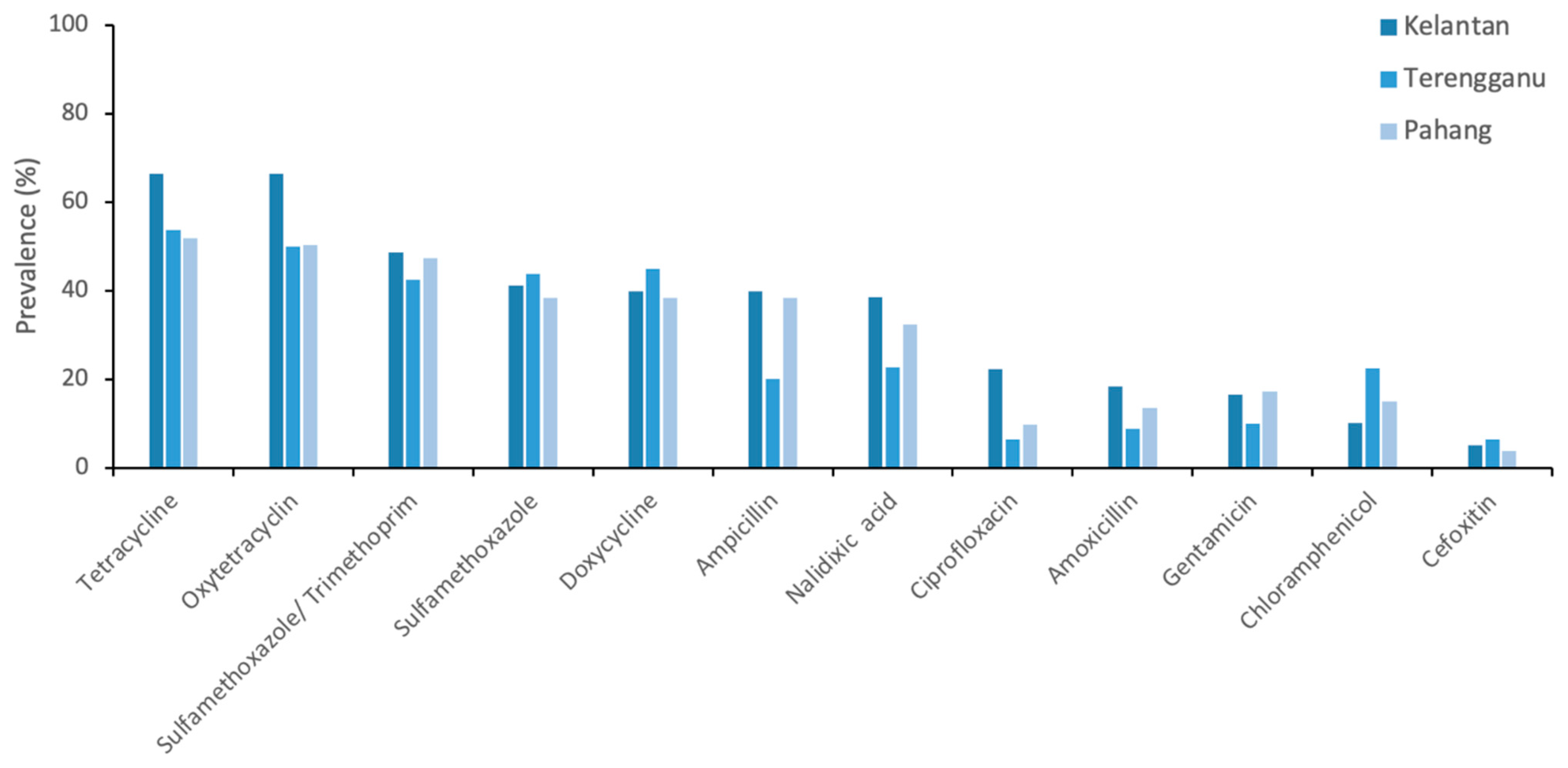
| State | 0.012 | ||
| Kelantan | 158 | 115 (72.8%) | |
| Terengganu | 80 | 46 (57.5%) | |
| Pahang | 133 | 77 (57.9%) | |
| Districts | 0.001 | ||
| Bachok | 52 | 36 (69.2%) | |
| Kota Bharu | 26 | 18 (69.2%) | |
| Machang | 28 | 24 (85.7%) | |
| Pasir Mas | 26 | 14 (53.8%) | |
| Jeli | 26 | 23 (88.5%) | |
| Kuantan | 79 | 49 (62%) | |
| Pekan | 54 | 28 (51.9%) | |
| kuala terengganu | 26 | 20 (76.9%) | |
| Marang | 54 | 26 (48.1%) | |
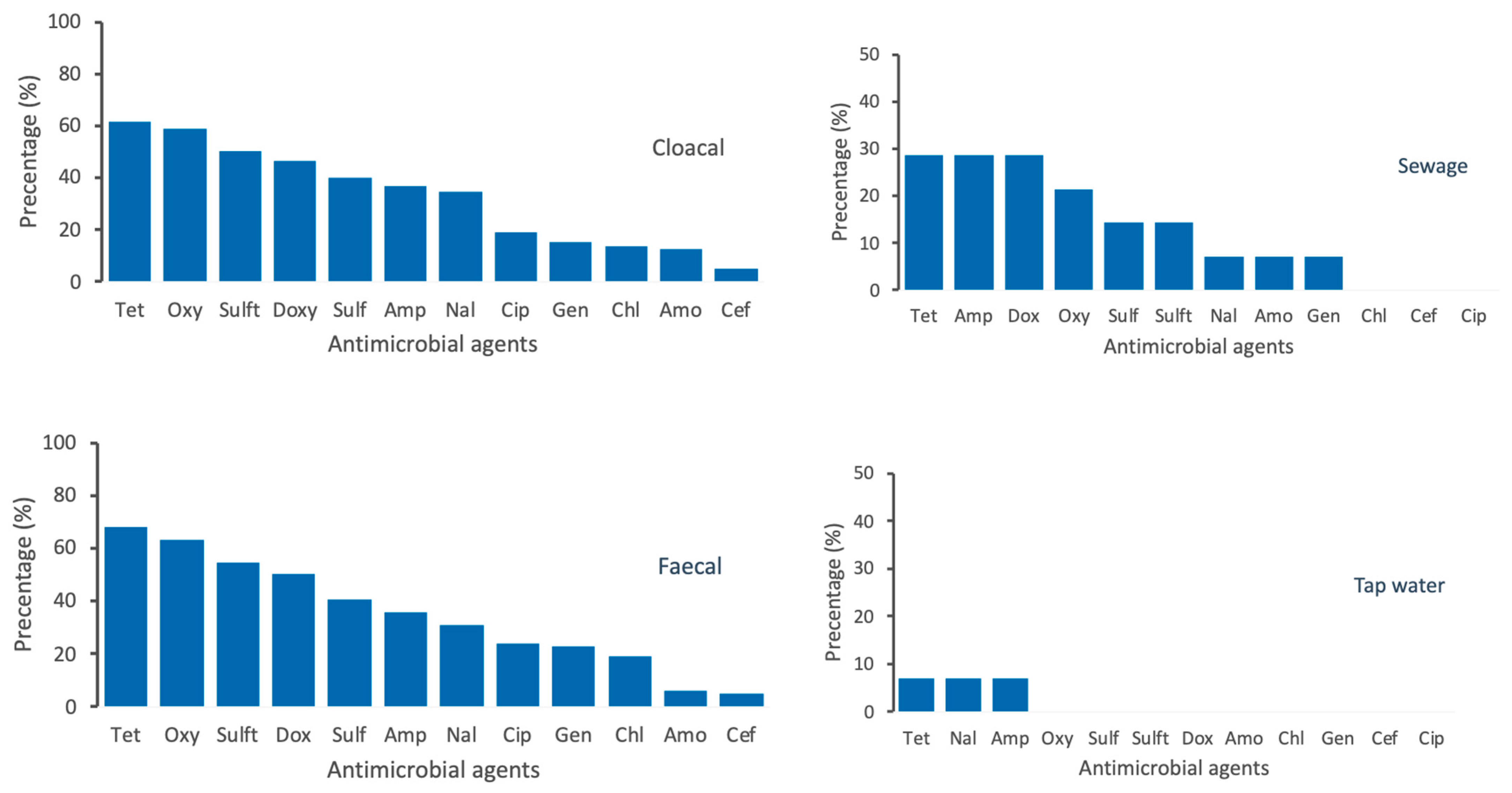
| Sample source | 0.001 | ||
| Cloaca swab | 259 | 172 (66.4%) | |
| Faecal Sample | 84 | 58 (69%) | |
| Sewage | 14 | 5 (35.7%) | |
| Tape Water | 14 | 3 (21.4%) | |
| Farm size | 0.013 | ||
| Small | 104 | 77 (74%) | |
| Medium | 188 | 119 (63.2%) | |
| Large | 79 | 42 (53.2%) | |
| Origin of the poultry | 0.005 | ||
| Local | 26 | 18 (69.2%) | |
| Imported | 133 | 71 (53.4%) | |
| Both | 212 | 149 (70.3%) |
| Risk Factors | No Antimicrobial Resistance n = 137 | Resistance to at least One Antimicrobial n = 234 | p-Value |
|---|---|---|---|
| Age | 0.44 | ||
| Young | 65 (47.4%) | 122 (52.1%) | |
| Adult | 72 (52.6%) | 112 (47.9%) | |
| Origin of the poultry | 0.01 | ||
| Local | 10 (7.3%) | 16 (6.8%) | |
| Imported | 63 (46%) | 70 (29.9%) | |
| Both | 64 (46.7%) | 148 (63.2%) | |
| Management system | 0.18 | ||
| Intensive | 80 (58.4%) | 115 (49.1%) | |
| Semi-intensive | 47 (34.3%) | 103 (44%) | |
| Mixed | 10 (7.3%) | 16 (6.8%) | |
| Production system | 0.21 | ||
| Broiler | 86 (62.8%) | 125 (53.4%) | |
| Layer | 18 (13.1%) | 37 (15.8%) | |
| Mixed | 33 (24.1%) | 72 (30.8%) | |
| Farm size | 0.02 | ||
| Small | 29 (21.2%) | 75 (32.1%) | |
| Medium | 70 (51.1%) | 117 (50%) | |
| Large | 38 (27.7%) | 42 (17.9%) | |
| Source of sample | <0.001 | ||
| Cloacal swab | 89 (65%) | 170 (72.6%) | |
| Faecal sample | 26 (19%) | 58 (24.8%) | |
| sewage | 9 (6.6%) | 5 (2.1%) | |
| Tap water | 13 (9.5%) | 1 (0.4%) | |
| Water source | 0.02 | ||
| Surface water | 37 (27%) | 69 (29.5%) | |
| Bond water | 61 (44.5%) | 72 (30.8%) | |
| Pump water | 39 (28.5%) | 93 (39.7%) | |
| Sewage system | 0.60 | ||
| Excellent | 38 (27.7%) | 71 (30.3%) | |
| Good | 82 (59.9%) | 128 (54.7%) | |
| Poor | 17 (12.4%) | 35 (15%) | |
| Feed source | 0.53 | ||
| Endogenous | 50 (36.5%) | 82 (35%) | |
| Exogenous | 75 (54.7%) | 138 (59%) | |
| Other | 12 (8.8%) | 14 (6%) |
| Antimicrobials | Cloacal n = 259 | Faecal n = 84 | Sewage n = 14 | Tape Water n = 14 | p-Value |
|---|---|---|---|---|---|
| No identified resistance | <0.001 | ||||
| No antimicrobial resistance | 89 (34.4%) | 26 (31%) | 9 (64.3%) | 13 (92.9%) | |
| Resistance to at least one antimicrobial | 170 (65.6%) | 58 (69%) | 5 (35.7%) | 1 (7.1%) | |
| Antimicrobial class resistance | 0.003 | ||||
| No antimicrobial resistance | 89 (34.4%) | 26 (31%) | 9 (64.3%) | 13 (92.9%) | |
| Resistant to 1 class | 4 (1.5%) | 1 (1.2%) | 0 (0%) | 0 (0%) | |
| Resistant to 2 classes | 13 (5%) | 3 (3.6%) | 2 (14.3%) | 0 (0%) | |
| Resistant to 3 classes | 46 (17.8%) | 19 (22.6%) | 2 (14.3%) | 1 (7.1%) | |
| Resistant to 4 classes | 74 (28.6%) | 19 (22.6%) | 1 (7.1%) | 0 (0%) | |
| Resistant to 5 or more classes | 33 (12.7%) | 16 (19%) | 0 (0%) | 0 (0%) | |
| Source of antimicrobials | 1 | ||||
| Drug supplier | 112 (43.2%) | 36 (42.9%) | 6 (42.9%) | 6 (42.9%) | |
| Feed store | 147 (56.8%) | 48 (57.1%) | 8 (57.1%) | 8 (57.1%) | |
| Tetracyclines | <0.001 | ||||
| Not resistant | 93 (35.9%) | 26 (31%) | 9 (64.3%) | 13 (92.9%) | |
| Resistant | 166 (64.1%) | 58 (69%) | 5 (35.7%) | 1 (7.1%) | |
| Penicillins | 0.048 | ||||
| Not resistant | 151 (58.3%) | 47 (56%) | 10 (71.4%) | 13 (92.9%) | |
| Resistant | 108 (41.7%) | 37 (44%) | 4 (28.6%) | 1 (7.1%) | |
| Aminoglycosides | 0.246 | ||||
| Not resistant | 219 (84.6%) | 68 (81%) | 13 (92.9%) | 14 (100%) | |
| Resistant | 40 (15.4%) | 16 (19%) | 1 (7.1%) | 0 (0%) | |
| Quinolones | 0.002 | ||||
| Not resistant | 142 (54.8%) | 49 (58.3%) | 13 (92.9%) | 13 (92.9%) | |
| Resistant | 117 (45.2%) | 35 (41.7%) | 1 (7.1%) | 1 (7.1%) | |
| Sulfonamides | <0.001 | ||||
| Not resistant | 104 (40.2%) | 29 (34.5%) | 11 (78.6%) | 14 (100%) | |
| Resistant | 155 (59.8%) | 55 (65.5%) | 3 (21.4%) | 0 (0%) | |
| Cephelosporins | 0.645 | ||||
| Not resistant | 246 (95%) | 79 (94%) | 14 (100%) | 14 (100%) | |
| Resistant | 13 (5%) | 5 (6%) | 0 (0%) | 0 (0%) | |
| Other classes | 0.025 | ||||
| Not resistant | 224 (86.5%) | 65 (77.4%) | 14 (100%) | 14 (100%) | |
| Resistant | 35 (13.5%) | 19 (22.6%) | 0 (0%) | 0 (0%) |
| Variables | OR | 2.5% | 97.5% | Pr (>|z|) | |
|---|---|---|---|---|---|
| Farms | |||||
| Farm 1 | 10.20 | 2.95 | 42.89 | <0.001 | *** |
| Farm 2 | 2.72 | 0.91 | 8.53 | 0.07 | . |
| Farm 3 | 13.03 | 3.46 | 65.56 | <0.001 | *** |
| Farm 4 | 1.98 | 0.66 | 6.09 | 0.22 | |
| Farm 5 | 2.31 | 0.78 | 7.18 | 0.134 | |
| Farm 6 | 7.14 | 2.16 | 27.16 | 0.002 | ** |
| Farm 7 | 2.89 | 0.97 | 9.02 | 0.05 | . |
| Farm 8 | Ref | Ref | Ref | Ref | |
| Farm 9 | 4.61 | 1.48 | 15.63 | 0.01 | * |
| Farm 10 | 3.82 | 1.25 | 12.52 | 0.02 | * |
| Farm 11 | 1.16 | 0.38 | 3.53 | 0.78 | |
| Farm 12 | 1.06 | 0.34 | 3.25 | 0.91 | |
| Farm 13 | 5.66 | 1.78 | 20.13 | 0.004 | ** |
| Farm 14 | 2.26 | 0.77 | 6.87 | 0.13 | |
| Sample source | |||||
| Cloaca swab | 24.83 | 4.82 | 454.74 | 0.002 | ** |
| Faecal sample | 29.0 | 5.35 | 540.73 | 0.001 | ** |
| Sewage | 7.22 | 0.95 | 151.21 | 0.09 | . |
| Tap water | Ref | Ref | Ref | Ref | |
| Age | |||||
| Young | 1.21 | 0.79 | 1.84 | 0.38 | |
| Adult | Ref | Ref | Ref | Ref | |
| Poultry origin | |||||
| Local | 1.44 | 0.61 | 3.50 | 0.41 | |
| Both | 2.08 | 1.32 | 3.26 | 0.001 | ** |
| Imported | Ref | Ref | Ref | Ref | |
| Management system | |||||
| Semi-intensive | 1.52 | 0.97 | 2.39 | 0.06 | . |
| Mixed | 1.11 | 0.48 | 2.65 | 0.80 | |
| Intensive | Ref | Ref | Ref | Ref | |
| Production system | |||||
| Layer | 1.41 | 0.76 | 2.69 | 0.27 | |
| Broiler | Ref | Ref | Ref | 0.11 | |
| Mixed | 1.50 | 0.91 | 2.48 | Ref | |
| Farm size | |||||
| Small | 2.33 | 1.27 | 4.35 | 0.001 | ** |
| Medium | 1.51 | 0.88 | 2.57 | 0.125 | |
| Large | Ref | Ref | Ref | Ref | |
| Water source | |||||
| Surface water | 1.57 | 0.93 | 2.68 | 0.08 | . |
| Pump water | 2.02 | 1.22 | 3.36 | 0.01 | ** |
| Bond water | |||||
| Sewage system | |||||
| Excellent | 0.91 | 0.44 | 1.81 | 0.786 | |
| Good | 0.75 | 0.39 | 1.42 | 0.398 | |
| Poor | Ref | Ref | Ref | Ref |
| OR | 2.5% | 97.5% | Pr (>|z|) | ||
|---|---|---|---|---|---|
| Cloaca swab | 26.50 | 5.08 | 487.69 | 0.001 | ** |
| Feacal sample | 30.92 | 5.63 | 579.63 | 0.001 | ** |
| Sewage | 7.43 | 0.96 | 156.87 | 0.09 | . |
| Farm size Small | 2.50 | 1.33 | 4.77 | 0.004 | ** |
| Farm size medium | 1.55 | 0.89 | 2.67 | 0.114 |
| Antimicrobial Class/Agent | Resistance Gene | % Isolates | Total # Tested |
|---|---|---|---|
| Gentamicin | aac(3)-IV | 12 (85.7%) | 14 |
| Tetracyclines | tet(A), tet(B) | 9 (64.2%) | 14 |
| Chloramphenicol | catA1 | 2 (14.2%) | 14 |
| Sulfonamides | sul1 | 14 (100%) | 14 |
| β-Lactams | blaSHV | 6 (42.8%) | 14 |
| Trimethoprim | dhfrI | 4 (28.5%) | 14 |
| Genes | Primer Sequence(5′ to 3′) | PCR Condition | Product Size | References |
|---|---|---|---|---|
| β-Lactams | F- CTATCGCCAGCAGGATCTGG R- ATTTGCTGATTTCGCTCGGC | 3 min at 95 °C; 35 cycles of 1 min at 94 °C, 90 s at 55 °C and 1 min at 72 °C; 10 min at 72 °C | 543 | [16] |
| Gentamicin aac(3)-IV | F-CTTCAGGATGGCAAGTTGGT R-TCATCTCGTTCTCCGCTCAT | 3 min at 95 °C; 35 cycles of 1 min at 94 °C, 90 s at 55 °C and 1 min at 72 °C; 10 min at 72 °C | 286 | [17] |
| Sulfonamide sul1 | F- ACTGCAGGCTGGTGGTTATG R- ACCGAGACCAATAGCGGAAG | 3 min at 95 C; 35 cycles of 1 min at 94 C, 90 s at 55 °C and 1 min at 72 °C; 10 min at 72 °C | 271 | [8] |
| Tetracycline tet(A) | F-CCTCAATTTCCTGACGGGCT R-GGCAGAGCAGGGAAAGGAAT | 3 min at 95 °C; 35 cycles of 1 min at 94 C, 90 s at 55 °C and 1 min at 72 °C; 10 min at 72 °C | 712 | [18] |
| Tetracycline tet(B) | F-ACCACCTCAGCTTCTCAACG R-GTAAAGCGATCCCACCACCA | 3 min at 95 °C; 35 cycles of 1 min at 94 C, 90 s at 55 °C and 1 min at 72 °C; 10 min at 72 °C | 586 | [18] |
| Chloramphenicol catA1 | F- GAAAGACGGTGAGCTGGTGA R- TAGCACCAGGCGTTTAAGGG | 3 min at 95 °C; 35 cycles of 1 min at 94 °C, 90 s at 55 °C and 1 min at 72 °C; 10 min at 72 °C | 473 | [8] |
| Trimethoprim dhfrI | F-AAGAATGGAGTTATCGGGAATG R-GGGTAAAAACTGGCCTAAAATTG | 15 min at 95 °C; 30 cycles of 30 s at 94 °C; 30 s at 58 °C; 1 min at 72 °C; 10 min 72 °C. | 391 | [8] |
| Ampicillin CITM | F-TGGCCAGAACTGACAGGCAAA R-TTTCTCCTGAACGTGGCTGGC | 15 min at 95 °C; 30 cycles of 30 s at 94 °C; 30 s at 58 °C; 1 min at 72 °C; 10 min 72 °C. | 462 | [8] |
| E.coli | F-TGACGTTACCCGCAGAAGAA R- CTCCAATCCGGACTACGACG | 3 min at 95 °C; 35 cycles of 15s at 95 °C, 90 s at 55 °C and 15s at 72 °C; 10 min at 72 °C | 832 | [19] |
| O157 | F-GTGTCCATTTATACGGACATCCATG R-CCTATAACGTCATGCCAATATTGCC | 2 min at 94 °C; 35 cycles of 30s at 94 °C, 30 s at 55 °C and 30s at 72 °C; 5 min at 72 °C | 292 | [20] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Elmi, S.A.; Simons, D.; Elton, L.; Haider, N.; Abdel Hamid, M.M.; Shuaib, Y.A.; Khan, M.A.; Othman, I.; Kock, R.; Osman, A.Y. Identification of Risk Factors Associated with Resistant Escherichia coli Isolates from Poultry Farms in the East Coast of Peninsular Malaysia: A Cross Sectional Study. Antibiotics 2021, 10, 117. https://doi.org/10.3390/antibiotics10020117
Elmi SA, Simons D, Elton L, Haider N, Abdel Hamid MM, Shuaib YA, Khan MA, Othman I, Kock R, Osman AY. Identification of Risk Factors Associated with Resistant Escherichia coli Isolates from Poultry Farms in the East Coast of Peninsular Malaysia: A Cross Sectional Study. Antibiotics. 2021; 10(2):117. https://doi.org/10.3390/antibiotics10020117
Chicago/Turabian StyleElmi, Sharifo Ali, David Simons, Linzy Elton, Najmul Haider, Muzamil Mahdi Abdel Hamid, Yassir Adam Shuaib, Mohd Azam Khan, Iekhsan Othman, Richard Kock, and Abdinasir Yusuf Osman. 2021. "Identification of Risk Factors Associated with Resistant Escherichia coli Isolates from Poultry Farms in the East Coast of Peninsular Malaysia: A Cross Sectional Study" Antibiotics 10, no. 2: 117. https://doi.org/10.3390/antibiotics10020117
APA StyleElmi, S. A., Simons, D., Elton, L., Haider, N., Abdel Hamid, M. M., Shuaib, Y. A., Khan, M. A., Othman, I., Kock, R., & Osman, A. Y. (2021). Identification of Risk Factors Associated with Resistant Escherichia coli Isolates from Poultry Farms in the East Coast of Peninsular Malaysia: A Cross Sectional Study. Antibiotics, 10(2), 117. https://doi.org/10.3390/antibiotics10020117









