Third-Generation Cephalosporin Resistance in Intrinsic Colistin-Resistant Enterobacterales Isolated from Retail Meat
Abstract
1. Introduction
2. Results
2.1. Isolated ICR Enterobacterales
2.2. Antimicrobial Resistance Profiles of the Isolated ICR Enterobacterales
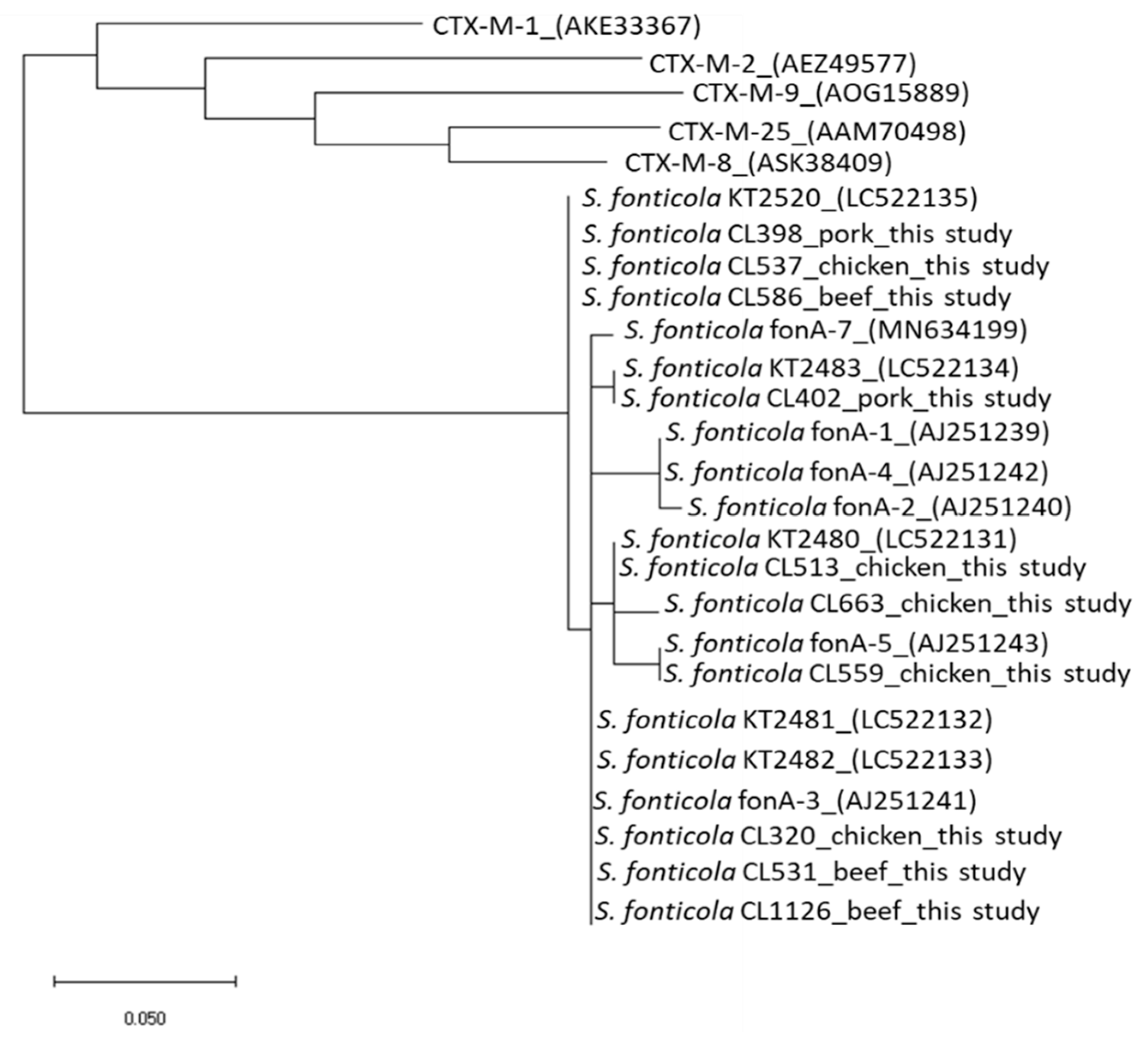
3. Discussion
4. Materials and Methods
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Read, A.F.; Woods, R.J. Antibiotic Resistance Management. Evol. Med. Public Health 2014, 2014, 147. [Google Scholar] [CrossRef]
- Ventola, C.L. The Antibiotic Resistance Crisis: Part 1: Causes and Threats. Pharm. Ther. 2015, 40, 277–283. [Google Scholar]
- Aghapour, Z.; Gholizadeh, P.; Ganbarov, K.; Bialvaei, A.Z.; Mahmood, S.S.; Tanomand, A.; Yousefi, M.; Asgharzadeh, M.; Yousefi, B.; Kafil, H.S. Molecular Mechanisms Related to Colistin Resistance in Enterobacteriaceae. Infect. Drug Resist. 2019, 12, 965–975. [Google Scholar] [CrossRef] [PubMed]
- Jayol, A.; Saly, M.; Nordmann, P.; Ménard, A.; Poirel, L.; Dubois, V. Hafnia, an Enterobacterial Genus Naturally Resistant to Colistin Revealed by Three Susceptibility Testing Methods. J. Antimicrob. Chemother. 2017, 72, 2507–2511. [Google Scholar] [CrossRef] [PubMed][Green Version]
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing, 29th ed.; M100; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2019; pp. 33–40. [Google Scholar]
- Olivares, J.; Bernardini, A.; Garcia-Leon, G.; Corona, F.; B Sanchez, M.; Martinez, J.L. The intrinsic resistome of bacterial pathogens. Front. Microbiol. 2013, 4, 103. [Google Scholar] [CrossRef] [PubMed]
- Economou, V.; Gousia, P. Agriculture and food animals as a source of antimicrobial-resistant bacteria. Infect. Drug Resist. 2015, 8, 49–61. [Google Scholar] [CrossRef]
- Nippon, A.M.R. One Health Report (NAOR). 2019. Available online: https://www.mhlw.go.jp/content/10900000/000714628.pdf (accessed on 25 December 2020).
- Hao, H.; Cheng, G.; Iqbal, Z.; Ai, X.; Hussain, H.I.; Huang, L.; Dai, M.; Wang, Y.; Liu, Z.; Yuan, Z. Benefits and risks of antimicrobial use in food-producing animals. Front. Microbiol. 2014, 5, 288. [Google Scholar] [CrossRef]
- Gwida, M.; Hotzel, H.; Geue, L.; Tomaso, H. Occurrence of Enterobacteriaceae in Raw Meat and in Human Samples from Egyptian Retail Sellers. Int. Sch. Res. Not. 2014, 2014, 565671. [Google Scholar] [CrossRef]
- Marshall, B.M.; Levy, S.B. Food animals and antimicrobials: Impacts on human health. Clin. Microbiol. Rev. 2011, 24, 718–733. [Google Scholar] [CrossRef]
- Tanimoto, K.; Nomura, T.; Hashimoto, Y.; Hirakawa, H.; Watanabe, H.; Tomita, H. Isolation of Serratia fonticola Producing FONA, a Minor Extended-Spectrum β-Lactamase (ESBL), from Imported Chicken Meat in Japan. Jpn. J. Infect. Dis. 2021, 74, 79–81. [Google Scholar] [CrossRef]
- Furuhata, K.; Ishizaki, N.; Sugi, Y.; Fukuyama, M. Isolation and Identification of Enterobacteriaceae from Raw Horsemeat Intended for Human Consumption (Basashi). Biocontrol. Sci. 2014, 19, 181–188. [Google Scholar] [CrossRef]
- Kilonzo-Nthenge, A.; Rotich, E.; Nahashon, S.N. Evaluation of Drug-Resistant Enterobacteriaceae in Retail Poultry and Beef. Poult. Sci. 2013, 92, 1098–1107. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.H.; Wei, C.I.; An, H. Molecular Characterization of Multidrug-Resistant Proteus mirabilis Isolates from Retail Meat Products. J. Food Prot. 2005, 68, 1408–1413. [Google Scholar] [CrossRef]
- Birk, T.; Fuentes, M.A.F.; Aabo, S.; Jensen, L.B. Horizontal Transmission of Antimicrobial Resistance Genes from E. coli to Serratia spp. in Minced Meat Using a Gfp Tagged Plasmid. Res. Vet. Sci. 2020, 132, 481–484. [Google Scholar] [CrossRef]
- Ingti, B.; Paul, D.; Maurya, A.P.; Bora, D.; Chanda, D.D.; Chakravarty, A.; Bhattacharjee, A. Occurrence of Bla DHA-1 Mediated Cephalosporin Resistance in Escherichia coli and Their Transcriptional Response Against Cephalosporin Stress: A Report from India. Ann. Clin. Microbiol. Antimicrob. 2017, 16, 13. [Google Scholar] [CrossRef] [PubMed]
- Yossapol, M.; Suzuki, K.; Odoi, J.O.; Sugiyama, M.; Usui, M.; Asai, T. Persistence of Extended-Spectrum β-Lactamase Plasmids Among Enterobacteriaceae in Commercial Broiler Farms. Microbiol. Immunol. 2020, 64, 712–718. [Google Scholar] [CrossRef] [PubMed]
- Odoi, J.O.; Takayanagi, S.; Sugiyama, M.; Usui, M.; Tamura, Y.; Asai, T. Prevalence of Colistin-Resistant Bacteria Among Retail Meats in Japan. Food Saf. 2021, 9, 48–56. [Google Scholar] [CrossRef] [PubMed]
- Doloy, A.; Verdet, C.; Gautier, V.; Decré, D.; Ronco, E.; Hammami, A.; Philippon, A.; Arlet, G. Genetic Environment of Acquired Bla(ACC-1) Beta-Lactamase Gene in Enterobacteriaceae Isolates. Antimicrob. Agents Chemother. 2006, 50, 4177–4181. [Google Scholar] [CrossRef]
- Moradigaravand, D.; Boinett, C.J.; Martin, V.; Peacock, S.J.; Parkhill, J. Recent Independent Emergence of Multiple Multidrug-Resistant Serratia marcescens Clones Within the United Kingdom and Ireland. Genome Res. 2016, 26, 1101–1109. [Google Scholar] [CrossRef]
- Nadjar, D.; Rouveau, M.; Verdet, C.; Donay, L.; Herrmann, J.; Lagrange, P.H.; Philippon, A.; Arlet, G. Outbreak of Klebsiella pneumoniae producing transferable AmpC-type beta-lactamase (ACC-1) originating from Hafnia alvei. FEMS Microbiol. Lett. 2000, 187, 35–40. [Google Scholar] [CrossRef]
- Barnaud, G.; Arlet, G.; Verdet, C.; Gaillot, O.; Lagrange, P.H.; Philippon, A. Salmonella enteritidis: AmpC plasmid-mediated inducible beta-lactamase (DHA-1) with an ampR gene from Morganella morganii. Antimicrob. Agents Chemother. 1998, 42, 2352–2358. [Google Scholar] [CrossRef]
- Ruppé, É.; Woerther, P.L.; Barbier, F. Mechanisms of Antimicrobial Resistance in Gram-Negative Bacilli. Ann. Intensive Care 2015, 5, 61. [Google Scholar] [CrossRef] [PubMed]
- Girlich, D.; Bonnin, R.A.; Dortet, L.; Naas, T. Genetics of Acquired Antibiotic Resistance Genes in Proteus spp. Front. Microbiol. 2020, 11, 256. [Google Scholar] [CrossRef] [PubMed]
- Magiorakos, A.P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-Resistant, Extensively Drug-Resistant and Pan Drug-Resistant Bacteria: An International Expert Proposal for Interim Standard Definitions for Acquired Resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef]
- McLellan, J.E.; Pitcher, J.I.; Ballard, S.A.; Grabsch, E.A.; Bell, J.M.; Barton, M.; Grayson, M.L. Superbugs in the Supermarket? Assessing the Rate of Contamination with Third-Generation Cephalosporin-Resistant Gram-Negative Bacteria in Fresh Australian Pork and Chicken. Antimicrob. Resist. Infect. Control 2018, 7, 30. [Google Scholar] [CrossRef]
- Xu, Q.; Fu, Y.; Zhao, F.; Jiang, Y.; Yu, Y. Molecular Characterization of Carbapenem-Resistant Serratia marcescens Clinical Isolates in a Tertiary Hospital in Hangzhou, China. Infect. Drug Resist. 2020, 13, 999–1008. [Google Scholar] [CrossRef]
- Ramos, A.; Dámaso, D. Extraintestinal infection due to Hafnia alvei. Eur. J. Clin. Microbiol. Infect. Dis. 2000, 19, 708–710. [Google Scholar] [CrossRef] [PubMed]
- Schaffer, J.N.; Pearson, M.M. Proteus mirabilis and Urinary Tract Infections. Microbiol. Spectr. 2015, 3, 10. [Google Scholar] [CrossRef]
- Ismaael, T.G.; Zamora, E.M.; Khasawneh, F.A. Cedecea davisae’s Role in a Polymicrobial Lung Infection in a Cystic Fibrosis Patient. Case Rep. Infect. Dis. 2012, 2012, 176864. [Google Scholar] [CrossRef]
- Wie, S.H. Clinical significance of Providencia bacteremia or bacteriuria. Korean J. Intern. Med. 2015, 30, 167–169. [Google Scholar] [CrossRef]
- Liu, H.; Zhu, J.; Hu, Q.; Rao, X. Morganella morganii, a non-negligent opportunistic pathogen. Int. J. Infect. Dis. 2016, 50, 10–17. [Google Scholar] [CrossRef] [PubMed]
- EUCAST (The European Committee on Antimicrobial Susceptibility Testing). Routine and Extended Internal Quality Control for MIC Determination and Disk Diffusion as Recommended by EUCAST. Version 7.0. 2017. Available online: http://www.eucast.org (accessed on 15 June 2021).
- Dallenne, C.; Da Costa, A.; Decré, D.; Favier, C.; Arlet, G. Development of a Set of Multiplex PCR Assays for the Detection of Genes Encoding Important Beta-Lactamases in Enterobacteriaceae. J. Antimicrob. Chemother. 2010, 65, 490–495. [Google Scholar] [CrossRef] [PubMed]
| Bacteria | Total Pos. Samples (%) | Total No. of Isolates | Chicken (n = 103) | Pork (n = 103) | Beef (n = 104) | ||||
|---|---|---|---|---|---|---|---|---|---|
| Genus | Species | Pos. Samples (%) | No. of Isolates | Pos. Samples (%) | No. of Isolates | Pos. Samples (%) | No. of Isolates | ||
| Serratia | liquefaciens | 194 (62.6) | 387 | 63 (61.2) | 118 | 64 (62.1) | 135 | 67 (64.4) | 134 |
| marcescens | 25 (8.1) | 29 | 11 (10.7) | 13 | 6 (5.8) | 7 | 8 (7.7) | 9 | |
| fonticola | 20 (6.5) | 21 | 11 (10.7) | 11 | 4 (3.8) | 5 | 5 (4.8) | 5 | |
| plymuthica | 2 (1) | 3 | 1 (1) | 1 | 1 (1) | 2 | 0 | 0 | |
| subtotal | 202 (65.2) | 440 | 68 (66) | 143 | 67 (65.0) | 149 | 67 (64.4) | 148 | |
| Hafnia | alvei | 89 (28.7) | 136 | 17 (16.5) | 22 | 40 (38.8) | 62 | 32 (30.8) | 52 |
| Proteus | penneri | 14 (4.5) | 18 | 11 (10.7) | 13 | 2 (1.9) | 3 | 1 (1) | 2 |
| hauseri | 7 (2.3) | 9 | 4 (3.8) | 4 | 3 (2.9) | 5 | 0 | 0 | |
| mirabilis | 7 (2.3) | 10 | 6 (5.8) | 9 | 1 (1) | 1 | 0 | 0 | |
| vulgaris | 2 (1) | 2 | 1 (1) | 1 | 1 (1) | 1 | 0 | 0 | |
| subtotal | 29 (9.4) | 39 | 21 (20.4) | 27 | 6 (5.8) | 10 | 1 (1) | 2 | |
| Cedecea | davisae | 14 (4.5) | 22 | 11 (10.7) | 18 | 1 (1) | 2 | 2 (1.9) | 2 |
| Providencia | rustigianii | 12 (3.9) | 12 | 12 (11.7) | 12 | 0 | 0 | 0 | 0 |
| Morganella | morganii | 10 (3.2) | 11 | 7 (6.8) | 7 | 2 (1.9) | 2 | 1 (1) | 2 |
| Bacteria | Resistance Profile | No. of Chicken Samples (No. of Isolates) | No. of Pork Samples (No. of Isolates) | No. of Beef Samples (No. of Isolates) | Total No. of Samples (Total No. of Isolates) |
|---|---|---|---|---|---|
| Serratia liquefaciens | CTX | 2 (3) | 1 (1) | 2 (2) | 5 (6) |
| CTX-CAZ | 1 (1) | 1 (1) | |||
| CTX-CHL | 1 (1) | 1 (1) | |||
| Susceptible | 61 (115) | 61 (132) | 65 (132) | 187 (379) | |
| Subtotal | 63 (118) | 64 (135) | 67 (134) | 199 (387) | |
| Serratia marcescens | CAZ-TET-CIP-LVF | 1 (1) | 1 (1) | ||
| CTX-NAL-CHL | 1 (1) | 1 (1) | |||
| TET | 5 (5) | 4 (4) | 2 (2) | 11 (11) | |
| TET-CHL | 1 (1) | 1 (1) | |||
| Susceptible | 4 (6) | 2 (3) | 5 (6) | 11 (15) | |
| Subtotal | 11 (13) | 6 (7) | 8 (9) | 25 (29) | |
| Serratia fonticola | CTX | 5 (5) | 2 (2) | 3 (3) | 10 (10) |
| Susceptible | 6 (7) | 2 (3) | 2 (2) | 10 (12) | |
| Subtotal | 11 (12) | 4 (5) | 5 (5) | 19 (22) | |
| Serratia plymuthica | Susceptible | 1 (1) | 1 (2) | 2 (3) | |
| Hafnia alvei | CTX | 1 (1) | 4 (4) | 1 (1) | 6 (6) |
| CAZ | 2 (2) | 2 (3) | 4 (5) | ||
| TET | 1 (1) | 1 (1) | |||
| CTX-CAZ | 3 (4) | 1 (1) | 4 (5) | ||
| NAL | 1 (1) | 1 (1) | |||
| CTX-CAZ-TET | 1 (1) | 1 (1) | |||
| Susceptible | 16 (21) | 29 (50) | 27 (46) | 72 (117) | |
| Subtotal | 17 (22) | 40 (62) | 32 (52) | 89 (136) | |
| Proteus penneri | CHL | 1 (1) | 1 (1) | ||
| CTX | 2 (2) | 2 (2) | |||
| Susceptible | 9 (10) | 2 (3) | 1 (2) | 12 (15) | |
| Subtotal | 11 (13) | 2 (3) | 1 (2) | 14 (18) | |
| Proteus mirabilis | CHL | 1 (1) | 1 (1) | ||
| SXT | 1 (2) | 1 (2) | |||
| KAN-CHL-SXT | 1 (1) | 1 (1) | |||
| Susceptible | 4 (6) | 0 | 0 | 4 (6) | |
| Subtotal | 6 (9) | 1 (1) | 0 | 7 (10) | |
| Proteus vulgaris | GEN-KAN-CHL-SXT | 1 (1) | 1 (1) | ||
| Susceptible | 1 (1) | 0 | 1 (1) | ||
| Subtotal | 1 (1) | 1 (1) | 2 (2) | ||
| Proteus hauseri | Susceptible | 4 (4) | 3 (5) | 7 (9) | |
| Cedecea davisae | TET | 1 (2) | 1 (2) | ||
| Susceptible | 10 (16) | 1 (2) | 2 (2) | 13 (20) | |
| Subtotal | 11 (18) | 1 (2) | 2 (2) | 14 (22) | |
| Morganella morganii | CHL | 1 (1) | 1 (1) | ||
| CTX | 1 (1) | 1 (1) | |||
| TET | 2 (2) | 2 (2) | 4 (4) | ||
| Susceptible | 3 (3) | 0 | 1 (2) | 4 (5) | |
| Subtotal | 7 (7) | 2 (2) | 1 (2) | 10 (11) | |
| Providencia rustigianii | Susceptible | 12 (12) | 0 | 0 | 12 (12) |
| Total | 96 (229) | 89 (225) | 67 (206) | 252 (659) |
| Bacteria | Strain No. | Source | Resistance Profile | Beta-Lactamase Gene Detected | |
|---|---|---|---|---|---|
| ESBL | AmpC | ||||
| S. fonticola | CL-320 | Chicken | CTX | blafonA | |
| S. fonticola | CL-398 | Pork | CTX | blafonA | |
| S. fonticola | CL-402 | Pork | CTX | blafonA | |
| S. fonticola | CL-513 | Chicken | CTX | blafonA | |
| S. fonticola | CL-531 | Beef | CTX | blafonA | |
| S. fonticola | CL-537 | Chicken | CTX | blafonA | |
| S. fonticola | CL-559 | Chicken | CTX | blafonA | |
| S. fonticola | CL-586 | Beef | CTX | blafonA | |
| S. fonticola | CL-663 | Chicken | CTX | blafonA | |
| S. fonticola | CL-1126 | Beef | CTX | blafonA | |
| H. alvei | CL-41 | Beef | CTX | blaACC | |
| H. alvei | CL-177 | Beef | CTX-TET | blaACC | |
| H. alvei | CL-208 | Pork | CTX-CAZ | blaACC | |
| H. alvei | CL-209 | Pork | CTX-CAZ | blaACC | |
| H. alvei | CL-215 | Beef | CTX-CAZ | blaACC | |
| H. alvei | CL-284 | Pork | CTX | blaACC | |
| H. alvei | CL-338 | Chicken | CTX | blaACC | |
| H. alvei | CL-607 | Pork | CTX-CAZ | blaACC | |
| H. alvei | CL-698 | Pork | CTX | blaACC | |
| H. alvei | CL-774 | Pork | CTX-CAZ | blaACC | |
| H. alvei | CL-954 | Pork | CTX | blaACC | |
| M. morganii | CL-277 | Chicken | CTX | blaDHA | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Odoi, J.O.; Takayanagi, S.; Yossapol, M.; Sugiyama, M.; Asai, T. Third-Generation Cephalosporin Resistance in Intrinsic Colistin-Resistant Enterobacterales Isolated from Retail Meat. Antibiotics 2021, 10, 1437. https://doi.org/10.3390/antibiotics10121437
Odoi JO, Takayanagi S, Yossapol M, Sugiyama M, Asai T. Third-Generation Cephalosporin Resistance in Intrinsic Colistin-Resistant Enterobacterales Isolated from Retail Meat. Antibiotics. 2021; 10(12):1437. https://doi.org/10.3390/antibiotics10121437
Chicago/Turabian StyleOdoi, Justice Opare, Sayo Takayanagi, Montira Yossapol, Michiyo Sugiyama, and Tetsuo Asai. 2021. "Third-Generation Cephalosporin Resistance in Intrinsic Colistin-Resistant Enterobacterales Isolated from Retail Meat" Antibiotics 10, no. 12: 1437. https://doi.org/10.3390/antibiotics10121437
APA StyleOdoi, J. O., Takayanagi, S., Yossapol, M., Sugiyama, M., & Asai, T. (2021). Third-Generation Cephalosporin Resistance in Intrinsic Colistin-Resistant Enterobacterales Isolated from Retail Meat. Antibiotics, 10(12), 1437. https://doi.org/10.3390/antibiotics10121437






